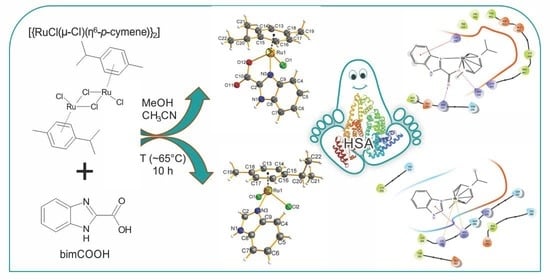
Synthesis, Characterization and Biological Investigations of Half-Sandwich Ruthenium(II) Complexes Containing Benzimidazole Moiety
Abstract
:1. Introduction
2. Results
2.1. Preparation of the Complexes
2.2. Spectroscopic Characterization
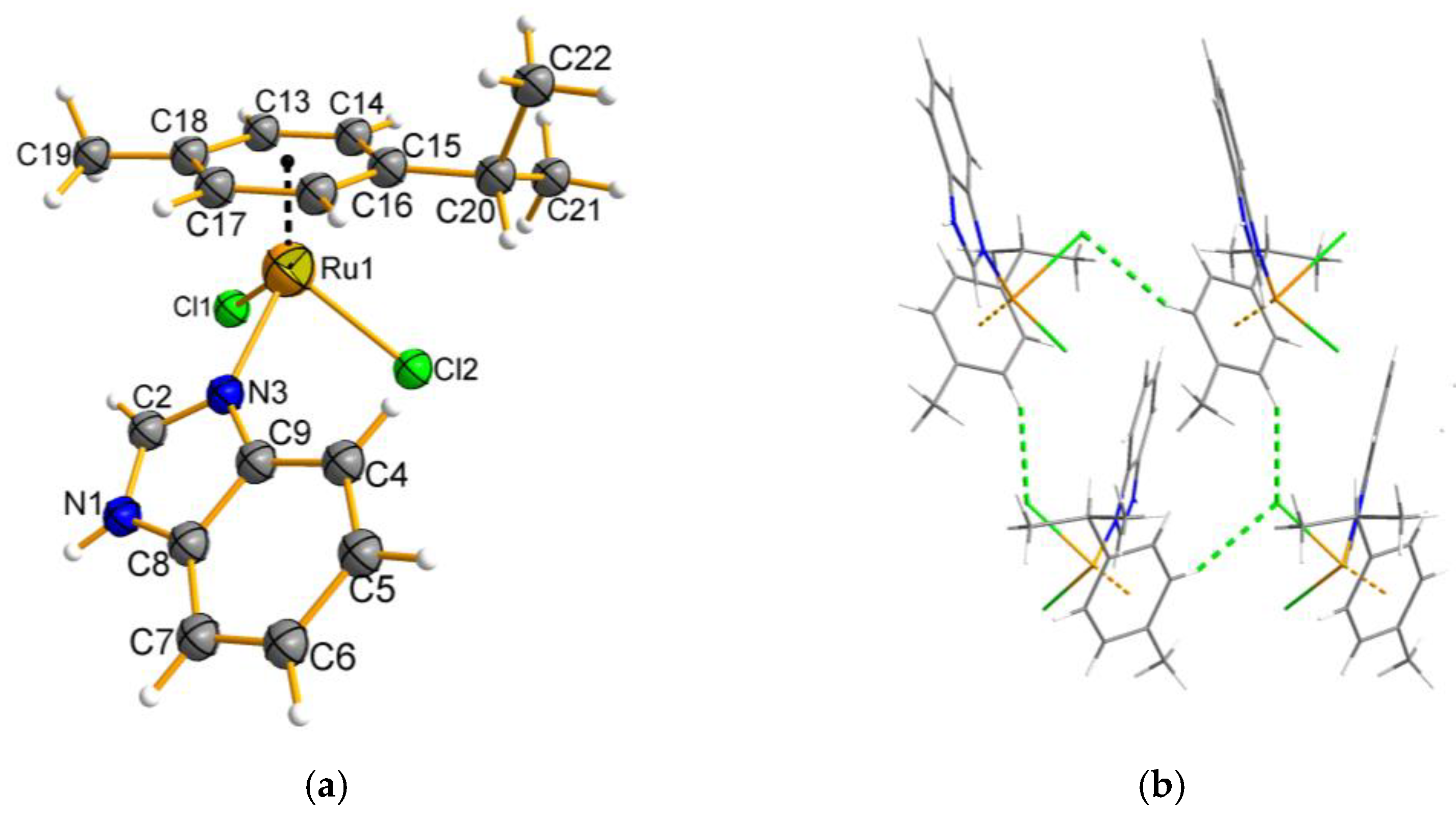
2.3. Description of the Molecular and Crystal Structure of Complexes 1 and 2
2.4. Hirshfeld Surface Analysis
2.5. Electrochemical Behavior of Ruthenium Complexes
2.6. Antimicrobial Assays
2.7. Cytotoxicity Activity
2.8. Solution Stability and Lipophilicity of Complexes
2.9. Binding Ability of the Studied Ru(II) Complexes with HSA
3. Discussion
4. Materials and Methods
4.1. Chemical Experiments
4.1.1. Syntheses of [(η6-p-cymene)RuCl(bimCOO)] (1) and [(η6-p-cymene)RuCl2(bim)] (2)
4.1.2. Physical Measurements
4.1.3. Crystal Structure Setermination and Refinement (SC-XRD)
4.1.4. Hirshfeld Surface Analysis
4.2. Biological Tests
4.2.1. Antibacterial Activity—Minimum Inhibitory Concentration
4.2.2. Inhibition of P. aeruginosa PAO1 Biofilm Formation
4.2.3. Live/Dead Staining of the Bacterial Biofilm
4.2.4. Cytotoxicity Activity (MTS Assay and Apoptosis Test by Flow Cytometry)
4.2.5. Determination of Stability and Lipophilicity
4.2.6. Molecular Docking Study
4.2.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Frezza, M.; Hindo, S.; Chen, D.; Davenport, A.; Schmitt, S.; Tomco, D.; Dou, Q.P. Novel Metals and Metal Complexes as Platforms for Cancer Therapy. Curr. Pharm. Des. 2010, 16, 1813–1825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, K.L.; Franz, K.J. Application of Metal Coordination Chemistry To Explore and Manipulate Cell Biology. Chem. Rev. 2009, 109, 4921–4960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warra, A.A. Transition metal complexes and their application in drugs and cosmetics—A Review. J. Chem. Pharm. Res. 2011, 3, 951–958. [Google Scholar]
- Lucaciu, R.L.; Hangan, A.C.; Sevastre, B.; Oprean, L.S. Metallo-Drugs in Cancer Therapy: Past, Present and Future. Molecules 2022, 27, 6485. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Raheem, N.; Straus, S.K. Mechanisms of action for antimicrobial peptides with antibacterial and antibiofilm functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [Green Version]
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Yarlagadda, V.; Akkapeddi, P.; Manjunath, G.B.; Haldar, J. Membrane active vancomycin analogues: A strategy to combat bacterial resistance. J. Med. Chem. 2014, 57, 4558–4568. [Google Scholar] [CrossRef]
- Parrino, B.; Schillaci, D.; Carnevale, I.; Giovannetti, E.; Diana, P.; Cirrincione, G.; Cascioferro, S. Synthetic small molecules as anti-biofilm agents in the struggle against antibiotic resistance. Eur. J. Med. Chem. 2019, 161, 154–178. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Agents that inhibit bacterial biofilm formation. Future Med. Chem. 2015, 7, 647–671. [Google Scholar] [CrossRef]
- Festa, R.A.; Helsel, M.E.; Franz, K.J.; Thiele, D.J. Exploiting innate immune cell activation of a copper-dependent antimicrobial agent during infection. Chem. Biol. 2014, 21, 977–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djoko, K.Y.; Goytia, M.M.; Donnelly, P.S.; Schembri, M.A.; Shafer, W.M.; McEwan, A.G. Copper(II)-bis(thiosemicarbazonato) complexes as antibacterial agents: Insights into their mode of action and potential as therapeutics. Antimicrob. Agents Chemother. 2015, 59, 6444–6453. [Google Scholar] [CrossRef] [PubMed]
- Frei, A.; Zuegg, J.; Elliott, A.G.; Baker, M.; Braese, S.; Brown, C.; Chen, F.; Dowson, C.G.; Dujardin, G.; Jung, N.; et al. Metal complexes as a promising source for new antibiotics. Chem. Sci. 2020, 11, 2627–2639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claudel, M.; Schwarte, J.V.; Fromm, K.M. New antimicrobial strategies based on metal complexes. Chemistry 2020, 2, 849–899. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Nurchi, V.M.; Lachowicz, J.I.; Crisponi, G.; Zoroddu, M.A. Noble metals in medicine: Latest advances. Coord. Chem. Rev. 2015, 284, 329–350. [Google Scholar] [CrossRef]
- Ndagi, U.; Mhlongo, N.; Soliman, M.E. Metal complexes in cancer therapy—An update from drug design perspective. Drug Des. Devel. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Gust, R. Update on metal N-heterocyclic carbene complexes as potential anti-tumor metallodrugs. Coord. Chem. Rev. 2016, 329, 191–213. [Google Scholar] [CrossRef]
- Li, F.; Collins, J.G.; Keene, F.R. Ruthenium complexes as antimicrobial agents. Chem. Soc. Rev. 2015, 44, 2529–2542. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Kim, C.Y.K.; Nam, T.-G. Ruthenium Complexes as Anticancer Agents: A Brief History and Perspectives. Drug Des. Devel. Ther. 2020, 14, 5375–5392. [Google Scholar] [CrossRef]
- Fromantin, I.; Watson, S.; Baffie, A.; Rivat, A.; Falcou, M.-C.; Kriegel, I.; De Rycke Ingenior, Y. A prospective, descriptive cohort study of malignant wound characteristics and wound care strategies in patients with breast cancer. Ostomy Wound Manag. 2014, 60, 38–48. [Google Scholar]
- Payne, W.G.; Naidu, D.K.; Wheeler, C.K.; Barkoe, D.; Mentis, M.; Salas, R.E.; Smith, D.J.; Robson, M.C. Wound healing in patients with cancer. Eplasty 2008, 8, e9. [Google Scholar] [PubMed]
- Rolston, K.V.I.; Nesher, L.; Tarrand, J.T. Current Microbiology of Surgical Site Infections in Patients with Cancer: A Retrospective Review. Infect. Dis. Ther. 2014, 3, 245–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabłońska-Wawrzycka, A.; Rogala, P.; Michałkiewicz, S.; Hodorowicz, M.; Barszcz, B. Ruthenium complexes in different oxidation states: Synthesis, crystal structure, spectra and redox properties. Dalton Trans. 2013, 42, 6092–6101. [Google Scholar] [CrossRef] [PubMed]
- Rogala, P.; Jabłońska-Wawrzycka, A.; Kazimierczuk, K.; Borek, A.; Błażejczyk, A.; Wietrzyk, J.; Barszcz, B. Synthesis, crystal structure and cytotoxic activity of ruthenium(II) piano-stool complex with N,N-chelating ligand. J. Mol. Struct. 2016, 1126, 74–82. [Google Scholar] [CrossRef]
- Rogala, P.; Czerwonka, G.; Michałkiewicz, S.; Hodorowicz, M.; Barszcz, B.; Jabłońska-Wawrzycka, A. Synthesis, structural characterization and antimicrobial evaluation of ruthenium complexes with heteroaromatic carboxylic acids. Chem. Biodivers. 2019, 16, e1900403. [Google Scholar] [CrossRef]
- Jabłońska-Wawrzycka, A.; Rogala, P.; Czerwonka, G.; Michałkiewicz, S.; Hodorowicz, M.; Kowalczyk, P. Ruthenium(IV) Complexes as Potential Inhibitors of Bacterial Biofilm Formation. Molecules 2020, 25, 4938. [Google Scholar] [CrossRef]
- Jabłońska-Wawrzycka, A.; Rogala, P.; Czerwonka, G.; Gałczyńska, K.; Drabik, M.; Dańczuk, M. Ruthenium Complexes with 2-Pyridin-2-yl-1H-benzimidazole as Potential Antimicrobial Agents: Correlation between Chemical Properties and Anti-Biofilm Effects. Int. J. Mol. Sci. 2021, 22, 10113. [Google Scholar] [CrossRef]
- Trynda-Lemiesz, L.; Kozłowski, H.; Keppler, B.K. Effect of cis-, trans-diamminedichloroplatinum(II) and DBP on human serum albumin. J. Inorg. Biochem. 1999, 77, 141–146. [Google Scholar] [CrossRef]
- Khalaila, I.; Bergamo, A.; Bussy, F.; Sava, G.; Dyson, P.J. The role of cisplatin and NAMI-A plasma-protein interactions in relation to combination therapy. Int. J. Oncol. 2006, 29, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Webb, M.I.; Wu, B.; Jang, T.; Chard, R.A.; Wong, E.W.Y.; Wong, M.Q.; Yapp, D.T.T.; Walsby, C.J. Increasing the Bioavailability of RuIII Anticancer Complexes through Hydrophobic Albumin Interactions. Chem. Eur. J. 2013, 19, 17031–17042. [Google Scholar] [CrossRef]
- Bijelic, A.; Theiner, S.; Keppler, B.K.; Rompel, A. X-ray Structure Analysis of Indazolium trans-[Tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019) Bound to Human Serum Albumin Reveals Two Ruthenium Binding Sites and Provides Insights into the Drug Binding Mechanism. J. Med. Chem. 2016, 59, 5894–5903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Liu, Z.; Wang, J. Uncovering the molecular and physiological processes of anticancer leads binding human serum albumin: A physical insight into drug efficacy. PLoS ONE 2017, 12, e0178660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoogenboezem, E.N.; Duvall, C.L. Harnessing albumin as a carrier for cancer therapies. Adv. Drug Deliv. Rev. 2018, 130, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Kalidhar, U.; Kaur, A. An overview on some benzimidazole and sulfonamide derivatives with antimicrobial activity. Res. J. Pharm. Biol. Chem. Sci. 2011, 2, 1116–1135. [Google Scholar]
- Deacon, G.B.; Phillips, R.J. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord. Chem. Rev. 1980, 33, 227–250. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Montgomery, J.A.; Vreven, T., Jr.; Kudin, K.N.; Burant, J.C.; et al. Gaussian 03, Revision E.01; Gaussian, Inc.: Wallingford CT, UK, 2004. [Google Scholar]
- Tähtinen, P.; Bagno, A.; Klika, K.D.; Pihlaja, K. Modeling NMR Parameters by DFT Methods as an Aid to the Conformational Analysis of cis-Fused 7a(8a)-Methyl Octa(hexa)hydrocyclopenta[d][1,3]oxazines and [3,1]benzoxazines. J. Am. Chem. Soc. 2003, 125, 4609–4618. [Google Scholar] [CrossRef]
- Lever, A.B.P. Inorganic Electronic Spectroscopy, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 1984. [Google Scholar]
- Kumar, K.N.; Venkatachalam, G.; Ramesh, R.; Liu, Y. Half-sandwich para-cymene ruthenium(II) naphthylazophenolato complexes: Synthesis, molecular structure, light emission, redox behavior and catalytic oxidation properties. Polyhedron 2008, 27, 157–166. [Google Scholar] [CrossRef]
- Govindaswamy, P.; Therrien, B.; Süss-Fink, G.; Štěpnička, P.; Ludvík, J. Mono and dinuclear iridium, rhodium and ruthenium complexes containing chelating carboxylato pyrazine ligands: Synthesis, molecular structure and electrochemistry. J. Organomet. Chem. 2007, 692, 1661–1671. [Google Scholar] [CrossRef] [Green Version]
- Matveevskaya, V.V.; Pavlov, D.I.; Samsonenko, D.G.; Ermakova, E.A.; Klyushova, L.S.; Baykov, S.V.; Boyarskiy, V.P.; Potapov, A.S. Synthesis and Structural Characterization of Half-Sandwich Arene–Ruthenium(II) Complexes with Bis(imidazol-1-yl)methane, Imidazole and Benzimidazole. Inorganics 2021, 9, 34. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Purkait, K.; Mukherjee, A. Ruthenium(II) p-cymene complexes of a benzimidazole-based ligand capable of VEGFR2 inhibition: Hydrolysis, reactivity and cytotoxicity studies. Dalton Trans. 2017, 46, 8539–8554. [Google Scholar] [CrossRef] [Green Version]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Marken, F.; Neudeck, A.; Bond, A.M. Cyclic Voltammetry. In Electroanalytical Methods. Guide to Experiments and Applications; Scholz, F., Ed.; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2010; pp. 57–102. ISBN 978-3-642-02914-1. [Google Scholar]
- Gosser, D.K. Cyclic Voltammetry: Simulation and Analysis of Reaction Mechanisms; Wiley-VCH: New York, NY, USA, 1993; ISBN 1560810262. [Google Scholar]
- Vock, C.A.; Scolaro, C.; Phillips, A.D.; Scopelliti, R.; Sava, G.; Dyson, P.J. Synthesis, Characterization, and in Vitro Evaluation of Novel Ruthenium(II) η6-Arene Imidazole Complexes. J. Med. Chem. 2006, 49, 5552–5561. [Google Scholar] [CrossRef] [PubMed]
- Tsopelas, F.; Giaginis, C.; Tsantili-Kakoulidou, A. Lipophilicity and biomimetic properties to support drug discovery. Expert Opin. Drug Discov. 2017, 12, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Horobin, R.; Stockert, J.; Rashid-Doubell, F. Uptake and localization mechanisms of fluorescent and colored lipid probes. Part 2. QSAR models that predict localization of fluorescent probes used to identify (”specifically stain”) various biomembranes and membranous organelles. Biotech. Histochem. 2015, 90, 241–254. [Google Scholar] [CrossRef]
- Parsekar, S.U.; Velankanni, P.; Sridhar, S.; Haldar, P.; Mate, N.A.; Banerjee, A.; Sudhadevi Antharjanam, P.K.; Koley, A.P.; Kumar, M. Protein binding studies with human serum albumin, molecular docking and in vitro cytotoxicity studies using HeLa cervical carcinoma cells of Cu(II)/Zn(II) complexes containing a carbohydrazone ligand. Dalton Trans. 2020, 49, 2947–2965. [Google Scholar] [CrossRef]
- Zsila, F.; Bikadi, Z.; Malik, D.; Hari, P.; Pechan, I.; Berces, A.; Hazai, E. Evaluation of drug–human serum albumin binding interactions with support vector machine aided online automated docking. Bioinformatics 2011, 27, 1806–1813. [Google Scholar] [CrossRef] [Green Version]
- Babur Şaş, E.; Çifçi, S.; Kurt, M. Spectroscopic Characterization and DFT Calculations on 1H-benzimidazole-2-carboxylic acid monohydrate Molecule. Sak. Univ. J. Sci. 2022, 26, 1879–1891. [Google Scholar] [CrossRef]
- Clavel, C.M.; Paăunescu, E.; Nowak-Sliwinska, P.; Griffioen, A.W.; Scopelliti, R.; Dyson, P.J. Modulating the Anticancer Activity of Ruthenium(II)−Arene Complexes. J. Med. Chem. 2015, 58, 3356–3365. [Google Scholar] [CrossRef]
- Clavel, C.M.; Păunescu, E.; Nowak-Sliwinska, P.; Griffioen, A.W.; Scopelliti, R.; Dyson, P.J. Discovery of a Highly Tumor-Selective Organometallic Ruthenium(II)−Arene Complex. J. Med. Chem. 2014, 57, 3546–3558. [Google Scholar] [CrossRef]
- Czerwonka, G.; Gmiter, D.; Guzy, A.; Rogala, P.; Jabłońska-Wawrzycka, A.; Borkowski, A.; Cłapa, T.; Narożna, D.; Kowalczyk, P.; Syczewski, M.; et al. A benzimidazole-based ruthenium(IV) complex inhibits Pseudomonas aeruginosa biofilm formation by interacting with siderophores and the cell envelope, and inducing oxidative stress. Biofouling 2019, 35, 59–74. [Google Scholar] [CrossRef]
- Klika, K.D. The Application of Simple and Easy to Implement Decoupling Pulse Scheme Combinations to Effect Decoupling of Large J Values with Reduced Artifacts. Int. J. Spectrosc. 2014, 2014, 289638. [Google Scholar] [CrossRef] [Green Version]
- Virta, P.; Koch, A.; Roslund, M.U.; Mattjus, P.; Kleinpeter, E.; Kronberg, L.; Sjöholm, R.; Klika, K.D. Synthesis, characterisation and theoretical calculations of 2,6-diaminopurine etheno derivatives. Org. Biomol. Chem. 2005, 3, 2924–2929. [Google Scholar] [CrossRef] [PubMed]
- Klika, K.D.; Bernát, J.; Imrich, J.; Chomča, I.; Sillanpää, R.; Pihlaja, K. Unexpected Formation of a Spiro Acridine and Fused Ring System from the Reaction between an N-Acridinylmethyl-Substituted Thiourea and Bromoacetonitrile under Basic Conditions. J. Org. Chem. 2001, 66, 4416–4418. [Google Scholar] [CrossRef] [PubMed]
- Balentová, E.; Imrich, J.; Bernát, J.; Suchá, L.; Vilková, M.; Prónayová, N.; Kristian, P.; Pihlaja, K.; Klika, K.D. Stereochemistry, Tautomerism, and Reactions of Acridinyl Thiosemicarbazides in the Synthesis of 1,3-Thiazolidines. J. Heterocycl. Chem. 2006, 43, 645–656. [Google Scholar] [CrossRef]
- Mäki, J.; Tähtinen, P.; Kronberg, L.; Klika, K.D. Restricted rotation/tautomeric equilibrium and determination of the site and extent of protonation in bi-imidazole nucleosides by multinuclear NMR and GIAO-DFT calculations. J. Phys. Org. Chem. 2005, 18, 240–249. [Google Scholar] [CrossRef]
- Program Stoe, & Cie GmbH. X-Area 1.75, Software Package for Collecting Single-Crystal Data on STOE Area-Detector Diffractometers, for Image Processing, ScalingReflection Intensities and for Outlier Rejection, Darmstad. 2015. Available online: https://www.stoe.com/product/software-x-area/ (accessed on 30 November 2022).
- Sheldrick, G.M. SHELXL-17, Program for Crystal Structure Refinement; University of Göttingen: Göttingen, Germany, 2017. [Google Scholar]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Brandenburg, K.; Putz, H. Diamond—Crystal and Molecular Structure Visualization Crystal Impact, version 3.1 f; CRYSTAL IMPACT: Bonn, Germany, 1997–2000.
- Wolff, S.K.; Grimwood, D.J.; McKinnon, J.J.; Turner, M.J.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer, version 3.1.; University of Western Australia: Perth, WA, Australia, 2013.
- De Soyza, A.; Hall, A.J.; Mahenthiralingam, E.; Drevinek, P.; Kaca, W.; Drulis-Kawa, Z.; Stoitsova, S.R.; Toth, V.; Coenye, T.; Zlosnik, J.E.A.; et al. Developing an international Pseudomonas aeruginosa reference panel. MicrobiologyOpen 2013, 2, 1010–1023. [Google Scholar] [CrossRef]
- OECD. Test No. 107: Partition Coefficient (n-octanol/water): Shake Flask Method; OECD Guidelines for the Testing of Chemicals, Section 1: Paris, France, 1995. Available online: https://www.oecd.org/chemicalsafety/testing/oecdguidelinesforthetestingofchemicals.htm (accessed on 30 November 2022).
- Petitpas, I.; Bhattacharya, A.A.; Twine, S.; East, M.; Curry, S. Crystal Structure Analysis of Warfarin Binding to Human Serum Albumin. J. Biol. Chem. 2001, 276, 22804–22809. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger Release 2022—3:Maestro. 2021. Available online: https://www.schrodinger.com/products/maestro (accessed on 30 November 2022).
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved Protein–Ligand Docking Using GOLD. Proteins Struct. Funct. Genet. 2003, 52, 609–623. [Google Scholar] [CrossRef]
- Sapundzhi, F.; Prodanova, K.; Lazarova, M. Survey of the scoring functions for protein-ligand docking. AIP Conf. Proc. 2019, 2172, 100008. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. Available online: https://www.schrodinger.com/products/pymol (accessed on 19 January 2018).









| Compound | Transition λ, nm (ε, dm3/mol∙cm) | |
|---|---|---|
| π→π*/n→π* | MLCT dπ(Ru)→π*(L)/d–d | |
| bimCOOH | 234 (6936), 259 (7159), 291 (6665) | |
| Complex 1 | 227 (8064), 293 (7153) | 332 (1275), ~410 (405) |
| Complex 2 | 213 (6554), 267 (3159), 273 (3119), 278 (2328) | 326 (495), ~405 (260) |
| Bond lengths (Å) | |||
|---|---|---|---|
| Ru1-O12 | 2.1163(1) | Ru1-Cl1 | 2.4015(5) |
| Ru1-N3 | 2.0876(2) | Ru1-Ct1 | 1.6583(6) |
| Valence angles (°) | |||
| O12-Ru1-N3 | 76.68(2) | Ct1-Ru1-Cl1 | 128.43(1) |
| O12-Ru1-Cl1 | 84.49(4) | Ct1-Ru1-N3 | 134.97(4) |
| N3-Ru1-Cl1 | 83.49(5) | Ct1-Ru1-O12 | 129.74(4) |
| Bond lengths (Å) | |||
|---|---|---|---|
| Ru1-N3 | 2.1120(2) | Ru1-Cl2 | 2.4297(6) |
| Ru1-Cl1 | 2.4185(6) | Ru1-Ct1 | 1.6574(3) |
| Valence angles (°) | |||
| N3-Ru1-Cl1 | 86.83(6) | Ct1-Ru1-N3 | 128.25(6) |
| N3-Ru1-Cl2 | 83.76(6) | Ct1-Ru1-Cl1 | 125.50(2) |
| Cl1-Ru1-Cl2 | 86.95(2) | Ct1-Ru1-Cl2 | 130.72(2) |
| Complex | D-H⋅⋅⋅A | D-H (Å) | H⋅⋅⋅A (Å) | D···A (Å) | ∠D-H⋅⋅⋅A (°) |
|---|---|---|---|---|---|
| 1 | N1-H1⋅⋅⋅O11 (2−x,1−y,−z) | 0.8796(2) | 1.8622(2) | 2.6795(2) | 153.75(1) |
| C4-H4⋅⋅⋅Cl1 (−1+x,y,z) | 0.9498(2) | 2.8195(5) | 3.6588(2) | 147.88(1) | |
| C21-H21C⋅⋅⋅Cl1(1.5−x,0.5+y,0.5−z) | 0.9795(3) | 2.805(5) | 3.7293(3) | 157.60(2) | |
| C5-H5⋅⋅⋅Cg2 | 0.9505(2) | 2.6858(1) | 3.5023(2) | 144.36(2) | |
| C13-H13⋅⋅⋅Cg3 | 0.9502(2) | 2.5874(1) | 3.505(2) | 162.43(2) | |
| 2 | N1-H1⋅⋅⋅Cl1 (x+0.5,−y+0.5,z) | 0.88 | 1.91 | 3.387(2) | 115.90 |
| N1-H1⋅⋅⋅Cl2 (x+0.5,−y+0,5,z) | 0.88 | 2.37 | 3.224(2) | 163.80 | |
| C2-H2⋅⋅⋅Cl1 | 0.9498(3) | 2.7816(6) | 3.2087(3) | 108.27(4) | |
| C21-H21A⋅⋅⋅Cl(2) | 0.9799(3) | 2.2726(6) | 3.4858(3) | 134.73(2) | |
| C17-H17⋅⋅⋅Cl(2) (x+1,y,z) | 0.9502(3) | 2.8013(6) | 3.5254(3) | 133.71(2) | |
| C13-H13⋅⋅⋅Cl(2) (−x+1,−y+1, z) | 0.9501(3) | 2.7195(7) | 3.5293(3) | 143.57(2) |
| Complex | Scan Rate V·s−1 | Ru(II)↔Ru(III) | |||
|---|---|---|---|---|---|
| Epa/V | Epc/V | ΔEp/V | E1/2/V | ||
| 1 | 0.05 | ~0.635 | ~0.565 | ~0.07 | ~0.60 |
| 0.10 | ~0.640 | ~0.560 | ~0.08 | ~0.60 | |
| 0.20 | ~0.645 | ~0.555 | ~0.09 | ~0.60 | |
| 2 | 0.05 | ~0.780 | ~0.720 | ~0.06 | ~0.75 |
| 0.10 | ~0.780 | ~0.720 | ~0.06 | ~0.75 | |
| 0.20 | ~0.790 | ~0.730 | ~0.06 | ~0.76 | |
| Compound | BACTERIA | |||||||
|---|---|---|---|---|---|---|---|---|
| S. aureus ATCC 6538P | E. coli ATCC 8739 | P. aeruginosa PAO1 | P. aeruginosa LES B58 | |||||
| mM | μg/mL | mM | μg/mL | mM | μg/mL | mM | μg/mL | |
| Ru(II) precursor | >1 | >612 | >1 | >612 | >1 | >612 | >1 | >612 |
| bimCOOH | >1 | >162 | >1 | >162 | 1 | 162 | >1 | >162 |
| 1 | >1 | >432 | >1 | >432 | 1 | 432 | 1 | 432 |
| 2 | >1 | >424 | 1 | 424 | 1 | 424 | >1 | >424 |
| Streptomycin | 0.0625 | 36 | 0.125 | 73 | 0.0625 | 36 | 0.5 | 291 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rogala, P.; Jabłońska-Wawrzycka, A.; Czerwonka, G.; Kazimierczuk, K.; Gałczyńska, K.; Michałkiewicz, S.; Kalinowska-Tłuścik, J.; Karpiel, M.; Klika, K.D. Synthesis, Characterization and Biological Investigations of Half-Sandwich Ruthenium(II) Complexes Containing Benzimidazole Moiety. Molecules 2023, 28, 40. https://doi.org/10.3390/molecules28010040
Rogala P, Jabłońska-Wawrzycka A, Czerwonka G, Kazimierczuk K, Gałczyńska K, Michałkiewicz S, Kalinowska-Tłuścik J, Karpiel M, Klika KD. Synthesis, Characterization and Biological Investigations of Half-Sandwich Ruthenium(II) Complexes Containing Benzimidazole Moiety. Molecules. 2023; 28(1):40. https://doi.org/10.3390/molecules28010040
Chicago/Turabian StyleRogala, Patrycja, Agnieszka Jabłońska-Wawrzycka, Grzegorz Czerwonka, Katarzyna Kazimierczuk, Katarzyna Gałczyńska, Sławomir Michałkiewicz, Justyna Kalinowska-Tłuścik, Marta Karpiel, and Karel D. Klika. 2023. "Synthesis, Characterization and Biological Investigations of Half-Sandwich Ruthenium(II) Complexes Containing Benzimidazole Moiety" Molecules 28, no. 1: 40. https://doi.org/10.3390/molecules28010040