Copper(II) Complexes of 5–Fluoro–Salicylaldehyde: Synthesis, Characterization, Antioxidant Properties, Interaction with DNA and Serum Albumins
Abstract
:1. Introduction
2. Results and Discussion
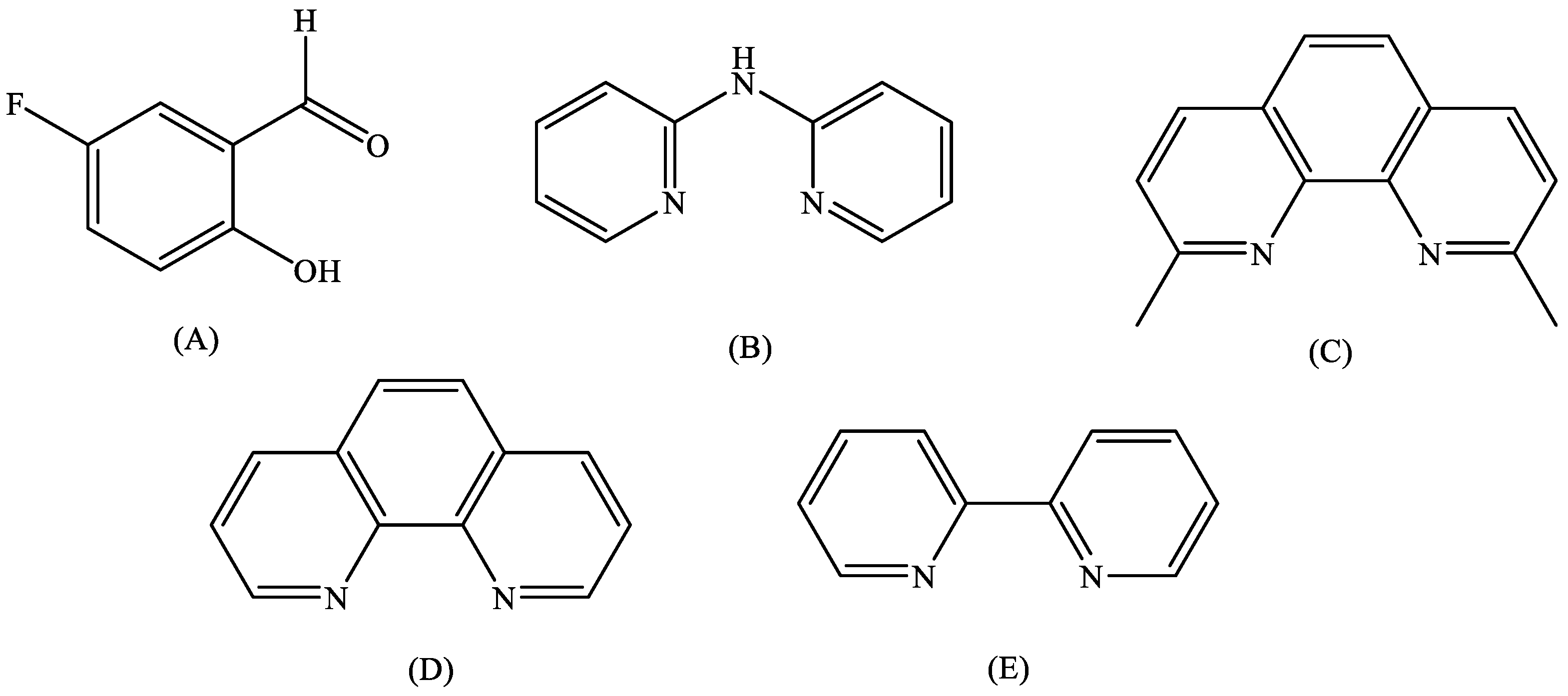
2.1. Synthesis and Characterization
2.2. Structures of the Complexes
2.2.1. Description of the Structure of Complex 1
2.2.2. Description of the Structures of Complexes 2–4
2.2.3. Proposed Structure for Complex 5
2.3. Study of the Antioxidant Activity
2.4. Interaction of the Complexes with CT DNA
2.5. Study of the Interaction with Serum Albumins
3. Materials and Methods
3.1. Materials—Instrumentation—Physical Measurements
3.2. Synthesis of the Complexes
3.2.1. Synthesis of Complex [Cu(5–F–salo)2], 1
3.2.2. Synthesis of Complexes 2–5
3.3. X-ray Crystal Structure Determination
3.4. Study of the Biological Profile of the Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
References
- Romano, S.; Guerreiro, J.P.; Rodrigues, A.T. Drug shortages in community pharmacies: Impact on patients and on the health system. J. Am. Pharm. Assoc. 2022, 62, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Balsano, C.; Porcu, C.; Sideri, S. Is copper a new target to counteract the progression of chronic diseases? Metallomics 2018, 10, 1712–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uauy, R.; Olivares, M.; Gonzalez, M. Essentiality of copper in humans. Am. J. Clin. Nutr. 1998, 67, 952S–959S. [Google Scholar] [CrossRef] [Green Version]
- Milanino, R.; Buchner, V. Copper: Role of the ‘Endogenous’ and ‘Exogenous’ Metal on the Development and Control of Inflammatory Processes. Rev. Environ. Health 2006, 21, 153–215. [Google Scholar] [CrossRef] [PubMed]
- Psomas, G. Copper(II) and zinc(II) coordination compounds of non-steroidal anti-inflammatory drugs: Structural features and antioxidant activity. Coord. Chem. Rev. 2020, 412, 213259. [Google Scholar] [CrossRef]
- Khalid, H.; Hanif, M.; Hashmi, M.; Mahmood, T.; Ayub, K.; Monim-ul-Mehboob, M. Copper Complexes of Bioactive Ligands with Superoxide Dismutase Activity. Mini-Rev. Med. Chem. 2013, 13, 1944–1956. [Google Scholar] [CrossRef] [PubMed]
- Zalevskaya, O.A.; Gur’eva, Y.A. Recent Studies on the Antimicrobial Activity of Copper Complexes. Russ. J. Coord. Chem. 2021, 47, 861–880. [Google Scholar] [CrossRef]
- McGuire, K.L.; Smit, P.; Ess, D.H.; Hill, J.T.; Harrison, R.G.; Busath, D.D. Mechanism and Kinetics of Copper Complexes Binding to the Influenza A M2 S31N and S31N/G34E Channels. Biophys. J. 2021, 120, 168–177. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, D.; Huang, M.; Huang, J.; Wang, D.; Liu, X.; Nguyen, M.; Vendier, L.; Mazères, S.; Robert, A.; et al. Preparation of Tetradentate Copper Chelators as Potential Anti-Alzheimer Agents. ChemMedChem 2018, 13, 684–704. [Google Scholar] [CrossRef]
- Leuthauser, S.W.C.; Oberley, L.W.; Oberley, T.D.; Sorenson, J.R.J.; Ramakrishna, K. Antitumor Effect of a Copper Coordination Compound With Superoxide Dismutase-Like Activity. J. Nat. Cancer Inst. 1981, 66, 1077–1081. [Google Scholar] [CrossRef]
- Denoyer, D.; Masaldan, S.; La Fontaine, S.; Cater, M.A. Targeting copper in cancer therapy: ‘Copper That Cancer’. Metallomics 2015, 7, 1459. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Muller, A.; Vizcaya-Ruiz, A.; Plant, N.; Ruiz, L.; Dobrota, M. Mixed chelate copper complex, Casiopeina IIgly®, binds and degrades nucleic acids: A mechanism of cytotoxicity. Chem.Biol. Interact. 2007, 165, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Pauls, G.; Becker, T.; Rahfeld, P.; Gretscher, R.R.; Paetz, C.; Pasteels, J.; von Reuss, S.H.; Burse, A.; Boland, W. Two Defensive Lines in Juvenile Leaf Beetles; Esters of 3-nitropropionic Acid in the Hemolymph and Aposematic Warning. J. Chem. Ecol. 2016, 42, 240–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelttari, E.; Karhumaki, E.; Langshaw, J.; Perakyla, H.; Elo, H. Antimicrobial Properties of Substituted Salicylaldehydes and Related Compounds. Z. Nat. C 2007, 62, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Pelttari, E.; Lehtinen, M.; Elo, H. Substituted Salicylaldehydes as Potential Antimicrobial Drugs: Minimal Inhibitory and Microbicidal Concentrations. Z. Nat. C 2011, 66, 571–580. [Google Scholar]
- Kordestani, N.; Rudbari, H.A.; Fernandes, A.R.; Raposo, L.R.; Baptista, P.V.; Ferreira, D.; Bruno, G.; Bella, G.; Scopelliti, R.; Braun, J.D.; et al. Antiproliferative Activities of Diimine–Based Mixed Ligand Copper(II) Complexes. ACS Comb. Sci. 2020, 22, 89–99. [Google Scholar] [CrossRef]
- Zianna, A.; Psomas, G.; Hatzidimitriou, A.; Lalia-Kantouri, M. Copper(II) complexes of salicylaldehydes and benzophenones: Synthesis, Structure, Thermal decomposition study and Interaction with calf-thymus DNA and albumins. RSC Adv. 2015, 5, 37495–37511. [Google Scholar] [CrossRef]
- Zianna, A.; Psomas, G.; Hatzidimitriou, A.; Lalia-Kantouri, M. Ni(II) complexes with 2,2′-dipyridylamine and salicylaldehydes: Synthesis, crystal structure and interaction with calf-thymus DNA and albumins. J. Inorg. Biochem. 2016, 163, 131–142. [Google Scholar] [CrossRef]
- Ntanatsidis, S.; Perontsis, S.; Konstantopoulou, S.; Kalogiannis, S.; Hatzidimitriou, A.G.; Papadopoulos, A.N.; Psomas, G. Manganese(II) complexes of substituted salicylaldehydes and α-diimines: Synthesis, characterization and biological activity. J. Inorg. Biochem. 2022, 227, 111693. [Google Scholar] [CrossRef]
- Zianna, A.; Geromichalou, E.; Geromichalos, G.; Fiotaki, A.-M.; Hatzidimitriou, A.G.; Kalogiannis, S.; Psomas, G. Zinc(II) complexes of 3,5-dibromo-salicylaldehyde and α-diimines: Synthesis, characterization and in vitro and in silico biological profile. J. Inorg. Biochem. 2022, 226, 111659. [Google Scholar] [CrossRef]
- Zianna, A.; Geromichalos, G.; Fiotaki, A.-M.; Hatzidimitriou, A.G.; Kalogiannis, S.; Psomas, G. Palladium(II) complexes of substituted salicylaldehydes: Synthesis, characterization and investigation of their biological profile. Pharmaceuticals 2022, 15, 886. [Google Scholar] [CrossRef] [PubMed]
- Zianna, A.; Vradi, E.; Hatzidimitriou, A.G.; Kalogiannis, S.; Psomas, G. Zinc(II) complexes of 3-bromo-5-chloro-salicylaldehyde: Characterization and biological activity. Dalton Trans. 2022, 51, 17629–17641. [Google Scholar] [CrossRef] [PubMed]
- Stamou, P.; Hatzidimitriou, A.G.; Psomas, G. Manganese(II) complexes with 5-nitro-2-hydroxy-benzaldehyde or substituted 2-hydroxy-phenones: Structure and interaction with bovine serum albumin and calf-thymus DNA. J. Inorg. Biochem. 2022, 235, 111923. [Google Scholar] [CrossRef] [PubMed]
- Zianna, A.; Geromichalos, G.; Psoma, E.; Kalogiannis, S.; Hatzidimitriou, A.G.; Psomas, G. Structure and in vitro and in silico biological activity of zinc(II) complexes with 3,5-dichloro-salicylaldehyde. J. Inorg. Biochem. 2022, 229, 111727. [Google Scholar] [CrossRef]
- Christidou, A.; Zavalani, K.; Hatzidimitriou, A.G.; Psomas, G. Copper(II) complexes with 3,5-dihalogeno-salicylaldehydes: Synthesis, structure and interaction with DNA and albumins. J. Inorg. Biochem. 2023, 238, 112049. [Google Scholar] [CrossRef]
- Vitomirov, T.; Dimiza, F.; Matić, I.Z.; Stanojković, T.; Pirković, A.; Živković, L.; Spremo-Potparević, B.; Novaković, I.; Anđelković, K.; Milčić, M.; et al. Copper(II) complexes with 4-(diethylamino)salicylaldehyde and α-diimines: Anticancer, antioxidant, antigenotoxic effects and interaction with DNA and albumins. J. Inorg. Biochem. 2022, 235, 111942. [Google Scholar] [CrossRef]
- Geary, W.J. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds, Part B: Applications in Coordination, Organometallic, and Bioinorganic Chemistry, 6th ed.; Wiley: Hoboken, NJ, USA, 2009. [Google Scholar]
- Nakamoto, Κ. Infrared and Raman Spectra of Inorganic and Coordination Compounds, 4th ed.; Wiley–Interscience: New York, NY, USA, 1986; pp. 254–257. [Google Scholar]
- Hathaway, B.J. Comprehensive Coordination Chemistry; Wilkinson, G., Ed.; Pergamon Press: Oxford, UK, 1987; Volume 5, pp. 533–773. [Google Scholar]
- Jageja, R.N.; Vyas, K.M.; Gupta, V.K.; Joshi, R.G.; Prabha, C.R. Syntheses, characterization and molecular structures of calcium(II) and copper(II) complexes bearing O2-chelate ligands: DNA binding, DNA cleavage and anti-microbial study. Polyhedron 2012, 31, 767–778. [Google Scholar]
- Battaglia, L.P.; Bonamartini-Corradi, A.; Marcotrigiano, G.; Menabue, L.; Pellacani, G.C. Halocuprates(II) of the N-phenylpiperazinium mono- and dications: Crystal and molecular structure of N-phenylpiperazinium tetrachloro cuprate(II). Correlation of the electronic spectrum vs. distortion of the CuCl42– anions from tetrahedral symmetry. Inorg. Chem. 1979, 18, 148–152. [Google Scholar] [CrossRef]
- Yang, L.; Powell, D.R.; Houser, R.P. Structural variation in copper(I) complexes with pyridylmethylamide ligands: Structural analysis with a new four-coordinate geometry index, τ4. Dalton Trans. 2007, 955–964. [Google Scholar] [CrossRef]
- Okuniewski, A.; Rosiak, D.; Chojnacki, J.; Becker, B. Coordination polymers and molecular structures among complexes of mercury(II) halides with selected 1-benzoylthioureas. Polyhedron 2015, 90, 47–57. [Google Scholar] [CrossRef]
- Li, G.Z.; Zhang, S.H.; Liu, Z. Bis (2, 4-dibromo-6-formylphenolato-κ2O, O′) copper (II). Acta Cryst. 2008, E64, m52. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q. Synthesis and crystal structure of bis(5-methyl-2-aldehyde-phenolato-κ2O1, O2)copper(II), C16H14CuO4. Z. Kristallogr. NCS 2020, 235, 1359–1360. [Google Scholar]
- Hathaway, B.J. The evidence for “out-of-the-plane” bonding in axial complexes of the copper(II) ion. Struct. Bond. 1973, 14, 49–67. [Google Scholar]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verchoor, G.C. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen-sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 1349–1356. [Google Scholar] [CrossRef]
- Wu, H.; Feng, Y.; Wang, S.; Huang, W. (4-Bromo-2-formylphenolato)perchlorato(1,10-phenanthroline)copper(II). Acta Crystallogr. Sect. C Cryst. Struct. Commun. 2006, 62, m358–m359. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zhao, G.; Wang, X.; Peng, Q.; Pan, Z. (2-Acetylphenolato)(2,2′-bipyridine)nitratocopper(II). Acta Crystallogr. Sect. E Struct. Rep. Online 2009, 65, m1427. [Google Scholar] [CrossRef] [Green Version]
- Marchi, R.C.; Campos, I.A.S.; Santana, V.T.; Carlos, R.M. Chemical implications and considerations on techniques used to assess the in vitro antioxidant activity of coordination compounds. Coord. Chem. Rev. 2022, 451, 2142752. [Google Scholar] [CrossRef]
- Dairi, S.; Carbonneau, M.; Galeano-Diaz, T.; Remini, H.; Dahmoune, F.; Aoun, O.; Belbahi, A.; Lauret, C.; Cristol, J.; Madani, K. Antioxidant effects of extra virgin olive oil enriched by myrtle phenolic extracts on iron-mediated lipid peroxidation under intestinal conditions model. Food Chem. 2017, 237, 297–304. [Google Scholar] [CrossRef]
- Slavova–Kazakova, A.; Karamac, M.; Kancheva, V.; Amarowicz, R. Antioxidant Activity of Flaxseed Extracts in Lipid Systems. Molecules 2016, 21, 17. [Google Scholar] [CrossRef] [Green Version]
- Kontogiorgis, C.; Ntella, M.; Mpompou, L.; Karallaki, F.; Papadopoulos, A.; Hadjipavlou-Litina, D.; Lazari, D. Study of the antioxidant activity of Thymus sibthorpii Bentham (Lamiaceae). J. Enz. Inhib. Med. Chem. 2016, 31, 154–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blois, M.S. Antioxidant Determinations by the Use of a Stable Free Radical. Nature 1958, 181, 1199–1200. [Google Scholar] [CrossRef]
- Kontogiorgis, C.; Hadjipavlou-Litina, D. Biological Evaluation of Several Coumarin Derivatives Designed as Possible Anti–inflammatory/Antioxidant Agents. J. Enzym. Inhib. Med. Chem. 2003, 180, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Gulcin, I.; Beydemır, S.; Alici, H.A.; Elmasta, M.; Buyukokuroglu, M.E. In vitro antioxidant properties of morphine. Pharmacol. Res. 2004, 49, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Jayaprakasha, G.K.; Rao, L.; Sakariah, K. Antioxidant activities of flavidin in different in vitro model systems. Bioorg. Med. Chem. 2004, 12, 5141–5146. [Google Scholar] [CrossRef]
- Zeglis, B.M.; Pierre, V.C.; Barton, J.K. Metallo-intercalators and metallo-insertors. Chem. Commun. 2007, 44, 4565–4576. [Google Scholar] [CrossRef] [Green Version]
- Pratviel, G.; Bernadou, J.; Meunier, B. DNA And RNA Cleavage by Metal Complexes. Adv. Inorg. Chem. 1998, 45, 251–262. [Google Scholar]
- Pyle, A.M.; Rehmann, J.P.; Meshoyrer, R.; Kumar, C.V.; Turro, N.J.; Barton, J.K. Mixed-ligand complexes of ruthenium(II): Factors governing binding to DNA. J. Am. Chem. Soc. 1989, 111, 3053–3063. [Google Scholar] [CrossRef]
- Wolfe, A.; Shimer, G.; Meehan, T. Polycyclic Aromatic Hydrocarbons Physically Intercalate into Duplex Regions of Denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef]
- Dimitrakopoulou, A.; Dendrinou-Samara, C.; Pantazaki, A.A.; Alexiou, M.; Nordlander, E.; Kessissoglou, D.P. Synthesis, structure and interactions with DNA of novel tetranuclear, [Mn4(II/II/II/IV)] mixed valence complexes. J. Inorg. Biochem. 2008, 102, 618–628. [Google Scholar] [CrossRef]
- Garcia-Gimenez, J.L.; Gonzalez-Alvarez, M.; Liu-Gonzalez, M.; Macias, B.; Borras, J.; Alzuet, G. Toward the development of metal-based synthetic nucleases: DNA binding and oxidative DNA cleavage of a mixed copper(II) complex with N-(9H-purin-6-yl)benzenesulfonamide and 1,10-phenantroline. Antitumor activity in human Caco-2 cells and Jurkat T lymphocytes. Evaluation of p53 and Bcl-2 proteins in the apoptotic mechanism. J. Inorg. Biochem. 2009, 103, 923–934. [Google Scholar] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Plenum Press: New York, NY, USA, 2006. [Google Scholar]
- Zhao, G.; Lin, H.; Zhu, S.; Sun, H.; Chen, Y. Dinuclear palladium (II) complexes containing two monofunctional [Pd(en)(pyridine) Cl]+ units bridged by Se or S. Synthesis, characterization, cytotoxicity and kinetic studies of DNA-binding. J. Inorg. Biochem. 1998, 70, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Heller, D.P.; Greenstock, C.L. Fluorescence lifetime analysis of DNA intercalated ethidium bromide and quenching by free dye. Biophys. Chem. 1994, 50, 305–312. [Google Scholar] [CrossRef] [PubMed]
- He, X.M.; Carter, D.C. Atomic structure and chemistry of human serum albumin. Nature 1992, 358, 209–215. [Google Scholar] [CrossRef] [Green Version]
- Olson, R.E.; Christ, D.D. Chapter 33. Plasma Protein Binding of Drugs. Ann. Rep. Med. Chem. 1996, 31, 327–336. [Google Scholar]
- Tan, C.; Liu, J.; Li, H.; Zheng, W.; Shi, S.; Chen, L.; Ji, L. Differences in structure, physiological stability, electrochemistry, cytotoxicity, DNA and protein binding properties between two Ru(III) complexes. J. Inorg. Biochem. 2008, 102, 347–358. [Google Scholar] [CrossRef]
- Stella, L.; Capodilupo, A.L.; Bietti, M. A reassessment of the association between azulene and [60]fullerene. Possible pitfalls in the determination of binding constants through fluorescence spectroscopy. Chem. Commun. 2008, 39, 4744–4746. [Google Scholar] [CrossRef]
- Rajendiran, V.; Karthik, R.; Palaniandavar, M.; Stoeckli-Evans, H.; Periasamy, V.S.; Akbarsha, M.A.; Srinag, B.S.; Krishnamurthy, H. Mixed-Ligand Copper(II)-phenolate Complexes: Effect of Coligand on Enhanced DNA and Protein Binding, DNA Cleavage, and Anticancer Activity. Inorg. Chem. 2007, 46, 8208–8221. [Google Scholar] [CrossRef]
- Laitinen, O.H.; Hytönen, V.P.; Nordlund, H.R.; Kulomaa, M.S. Genetically engineered avidins and streptavidins. Cell. Mol. Life Sci. 2006, 63, 2992–3017. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961, 3, 208–211. [Google Scholar] [CrossRef]
- Reichmann, M.F.; Rice, S.A.; Thomas, C.A.; Doty, P. A Further Examination of the Molecular Weight and Size of Desoxypentose Nucleic Acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Zianna, A.; Psomas, G.; Hatzidimitriou, A.; Coutouli-Argyropoulou, E.; Lalia-Kantouri, M. Zinc complexes of salicylaldehydes: Synthesis, characterization and DNA-binding properties. J. Inorg. Biochem. 2013, 127, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Bruker Analytical X-ray Systems, Inc. Apex2, Version 2 User Manual, M86-E01078; Madison, WI, USA, 2006. [Google Scholar]
- Siemens Industrial Automation, Inc. SADABS: Area-Detector Absorption Correction; Madison, WI, USA, 1996. [Google Scholar]
- Palatinus, L.; Chapuis, G. SUPERFLIP—A computer program for the solution of crystal structures by charge flipping in arbitrary dimensions. J. Appl. Cryst. 2007, 40, 786–790. [Google Scholar] [CrossRef] [Green Version]
- Betteridge, P.W.; Carruthers, J.R.; Cooper, R.I.; Prout, K.; Watkin, D.J. CRYSTALS version 12: Software for guided crystal structure analysis. J. Appl. Cryst. 2003, 36, 1487. [Google Scholar] [CrossRef]
- Watkin, D.J.; Prout, C.K.; Pearce, L.J. CAMERON Program; Chemical Crystallographic Laboratory, Oxford University: Oxford, UK, 1996. [Google Scholar]
- Ruch, R.J.; Cheng, C.; Klaunig, J.E. Prevention of cytotoxicity and inhibition of intercellular communication by antioxidant catechins isolated from Chinese green tea. Carcinogenesis 1989, 10, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Zhang, G.; Tao, W.; Tang, S. Interaction of the flavonoid hesperidin with bovine serum albumin: A fluorescence quenching study. J. Lumin. 2007, 126, 211–218. [Google Scholar] [CrossRef]









| 1 | 2 | 3 | 4 | |
|---|---|---|---|---|
| Crystal Data | ||||
| Chemical formula | C14H8CuF2O4 | C17H13ClCuFN3O2 | C22H20ClCuFN2O3 | C19H12CuFN3O5 |
| Moiety formula | C21H16ClCuFN2O2·CH4O | |||
| Mr | 341.76 | 409.31 | 478.41 | 444.87 |
| Crystal system | Orthorhombic | Monoclinic | Monoclinic | Triclinic |
| Space group | Pca21 | P21/n | P21/n | P–1 |
| Temperature (K) | 295 | 295 | 295 | 295 |
| a (Å) | 12.5045 (17) | 10.3584 (4) | 9.0506 (11) | 7.6719 (9) |
| b (Å) | 3.8457 (6) | 15.4548 (8) | 17.2678 (16) | 9.4646 (11) |
| c (Å) | 25.277 (3) | 10.3842 (5) | 13.4078 (15) | 13.1039 (15) |
| α (°) | 90 | 90 | 90 | 92.412 (3) |
| β (°) | 90 | 105.4055 (16) | 95.523 (3) | 106.764 (4) |
| γ (°) | 90 | 90 | 90 | 106.721 (4) |
| V (Å3) | 1215.5 (3) | 1602.65 (13) | 2085.7 (4) | 864.38 (18) |
| Z | 4 | 4 | 4 | 2 |
| µ (mm−1) | 1.84 | 1.56 | 1.21 | 1.31 |
| Crystal size (mm) | 0.21 × 0.02 × 0.02 | 0.18 × 0.10 × 0.09 | 0.19 × 0.15 × 0.12 | 0.17 × 0.15 × 0.14 |
| Data Collection | ||||
| Diffractometer | Bruker Kappa Apex2 | |||
| Radiation type | Mo Kα (λ = 0.71073 Å, source operating at 50 kV and 30 mA) | |||
| Absorption correction | Numerical, Analytical Absorption (De Meulenaer and Tompa, 1965) | |||
| Tmin, Tmax | 0.95, 0.96 | 0.86, 0.87 | 0.85, 0.86 | 0.81, 0.83 |
| Measured reflections | 9456 | 12675 | 18728 | 10790 |
| Independent reflections | 3671 | 3048 | 3968 | 3284 |
| Observed reflections with [I > 2.0σ(I)] | 2454 | 2496 | 3029 | 2964 |
| Rint | 0.030 | 0.026 | 0.018 | 0.024 |
| (sin θ/λ)max (Å−1) | 0.725 | 0.612 | 0.611 | 0.616 |
| Refinement | ||||
| R[F2 > 2σ(F2)] | 0.033 | 0.026 | 0.030 | 0.034 |
| wR(F2) | 0.066 | 0.052 | 0.056 | 0.054 |
| S | 1.00 | 1.00 | 1.00 | 1.00 |
| No. of reflections | 2454 | 2496 | 3029 | 2964 |
| No. of parameters | 191 | 226 | 269 | 280 |
| No. of restraints | 1 | - | 2 | 35 |
| Δρmax, Δρmin (e Å−3) | 0.47, −0.51 | 0.21, −0.29 | 0.51, −0.37 | 0.43, −0.43 |
| Absolute structure | Flack (1983), 1725 Friedel–pairs | |||
| Absolute structure parameter | 0.19 (2) | |||
| Bond | Length (Å) | Bond | Length (Å) |
|---|---|---|---|
| Cu1—O1 | 1.934 (2) | Cu1—O3 | 1.925 (2) |
| Cu1—O2 | 1.893 (4) | Cu1—O4 | 1.902 (4) |
| Bonds | Angle (°) | Bond | Angle (°) |
| O1—Cu1—O2 | 92.50 (13) | O1—Cu1—O4 | 86.56 (14) |
| O1—Cu1—O3 | 178.72 (14) | O2—Cu1—O4 | 177.6 (2) |
| O2—Cu1—O3 | 87.60 (13) | O3—Cu1—O4 | 93.39 (13) |
| Complex 2 | Complex 3 | Complex 4 | |
|---|---|---|---|
| Bond | Length (Å) | Length (Å) | Length (Å) |
| Cu1—O1 | 1.9901 (17) | 2.0471 (18) | 1.9708 (16) |
| Cu1—O2 | 1.9225 (17) | 1.910 (2) | 1.8980 (15) |
| Cu1—N1 | 2.0151 (19) | 2.247 (2) | 2.007 (2) |
| Cu1—N2 | 2.0244 (19) | 2.011 (2) | 1.9915 (17) |
| Cu1—X 1 | 2.5531 (7) | 2.3055 (7) | 2.3672 (19) |
| Bonds | Angles (°) | Angles (°) | Angles (°) |
| Ν1—Cu1—N2 | 90.04 (8) | 79.59 (9) | 82.82 (8) |
| N1—Cu1—O1 | 88.56 (7) | 95.80 (8) | 175.29 (7) |
| N1—Cu1—O2 | 161.04 (8) | 106.24 (9) | 90.87 (7) |
| N1—Cu1—X 1 | 100.25 (6) | 106.51 (6) | 100.23 (7) |
| N2—Cu1—O1 | 176.00 (8) | 85.45 (8) | 93.12 (7) |
| N2—Cu1—O2 | 90.15 (7) | 172.93 (9) | 168.61 (8) |
| N2—Cu1—X 1 | 91.35 (6) | 92.15 (6) | 96.66 (7) |
| O1—Cu1—O2 | 89.95 (7) | 89.92 (8) | 92.65(12) |
| O1—Cu1—X 1 | 92.59 (6) | 156.77 (6) | 82.57 (7) |
| O2—Cu1—X 1 | 98.70 (6) | 90.00 (6) | 93.78 (7) |
| Trigonality index τ5 | 0.25 | 0.27 | 0.11 |
| Tetragonality, T5 | 0.79 | 0.92 | 0.83 |
| Complex | DPPH% (30 min) | DPPH% (60 min) | ABTS% | H2O2% |
|---|---|---|---|---|
| 5–F–saloH [21] | 3.96 ± 1.16 | 5.56 ± 1.06 | 19.57 ± 0.58 | 71.84 ± 0.95 |
| [Cu(5–F–salo)2], 1 | 9.05 ± 0.54 | 10.79 ± 0.20 | 78.89 ± 0.18 | 71.61 ± 0.35 |
| [Cu(5–F–salo)(bipyam)Cl], 2 | 7.54 ± 0.20 | 7.42 ± 0.50 | 48.89 ± 0.38 | 99.69 ± 0.29 |
| [Cu(5–F–salo)(neoc)Cl], 3 | 7.42 ± 0.54 | 14.15 ± 0.13 | 46.03 ± 0.60 | 26.10 ± 0.66 |
| [Cu(5–F–salo)(phen)(NO3)], 4 | 6.15 ± 0.33 | 4.64 ± 0.10 | 48.14 ± 0.35 | 25.90 ± 0.76 |
| [Cu(5–F–salo)(bipy)(NO3)], 5 | 7.19 ± 0.26 | 4.06 ± 0.20 | 7.36 ± 0.08 | 69.21 ± 1.10 |
| NDGA | 87.08 ± 0.12 | 87.47 ± 0.12 | Not tested | Not tested |
| BHT | 61.30 ± 1.16 | 79.78 ± 1.12 | Not tested | Not tested |
| Trolox | Not tested | Not tested | 98.10 ± 0.48 | Not tested |
| L–ascorbic acid | Not tested | Not tested | Not tested | 60.80 ± 0.20 |
| Compound | λ (nm) (ΔA/Aο (%) a, Δλ (nm) b) | Kb (Μ−1) |
|---|---|---|
| 5–F–saloH [21] | 334 (−30, +1); 421 (>+50,c 0) | 8.37 (±0.47) × 104 |
| [Cu(5–F–salo)2], 1 | 320 (−34, +16); 398 (−20, +20) | 2.37 (±0.07) × 106 |
| [Cu(5–F–salo)(bipyam)Cl], 2 | 319 (−12, −3); 401 (>+50, +20) | 6.35 (±0.30) × 105 |
| [Cu(5–F–salo)(neoc)Cl], 3 | 334 (−41, +5); 421 (+34, +4) | 2.69 (±0.45) × 105 |
| [Cu(5–F–salo)(phen)(NO3)], 4 | 295 (−38, +4); 330 (−29, −4), 405 (+44, +19) | 1.09 (±0.14) × 106 |
| [Cu(5–F–salo)(bipy)(NO3)], 5 | 312 (−72, +3); 345 (−28, 0) | 9.32 (±0.29) × 105 |
| Compound | ΔΙ/Ιο (%) | Ksv (M−1) | kq (M−1 s−1) |
|---|---|---|---|
| 5–F–saloH [21] | 51.3 | 3.79 (±0.11) × 104 | 1.73 (±0.05) × 1012 |
| [Cu(5–F–salo)2], 1 | 44.8 | 1.56 (±0.02) × 105 | 6.78 (±0.09) × 1012 |
| [Cu(5–F–salo)(bipyam)Cl], 2 | 44.5 | 6.23 (±0.11) × 104 | 2.71 (±0.05) × 1012 |
| [Cu(5–F–salo)(neoc)Cl], 3 | 49.1 | 3.35 (±0.05) × 104 | 1.46 (±0.02) × 1012 |
| [Cu(5–F–salo)(phen)(NO3)], 4 | 48.4 | 6.77 (±0.07) × 104 | 2.94 (±0.03) × 1012 |
| [Cu(5–F–salo)(bipy)(NO3)], 5 | 42.2 | 3.14 (±0.06) × 104 | 1.36 (±0.03) × 1012 |
| Compound | ΔΙ/Ιο (%) | kq (M−1 s−1) | K (M−1) |
|---|---|---|---|
| BSA | |||
| 5–F–saloH [21] | 25.7 | 1.98 (±0.08) × 1012 | 4.31 (±0.31) × 104 |
| [Cu(5–F–salo)2], 1 | 62.5 | 9.38 (±0.28) × 1012 | 4.05 (±0.02) × 104 |
| [Cu(5–F–salo)(bipyam)Cl], 2 | 54.0 | 6.49 (±0.14) × 1012 | 3.15 (±0.18) × 104 |
| [Cu(5–F–salo)(neoc)Cl], 3 | 84.3 | 2.75 (±0.10) × 1013 | 2.51 (±0.10) × 105 |
| [Cu(5–F–salo)(phen)(NO3)], 4 | 75.3 | 1.62 (±0.06) × 1013 | 2.35 (±0.10) × 105 |
| [Cu(5–F–salo)(bipy)(NO3)], 5 | 68.3 | 1.87 (±0.09) × 1013 | 3.23 (±0.12) × 105 |
| HSA | |||
| 5–F–saloH [21] | 25.7 | 1.96 (±0.07) × 1012 | 4.65 (±0.35) × 104 |
| [Cu(5–F–salo)2], 1 | 61.3 | 4.37 (±0.18) × 1012 | 1.58 (±0.09) × 105 |
| [Cu(5–F–salo)(bipyam)Cl], 2 | 62.7 | 6.27 (±0.21) × 1012 | 5.09 (±0.29) × 105 |
| [Cu(5–F–salo)(neoc)Cl], 3 | 68.5 | 1.25 (±0.05) × 1013 | 7.16 (±0.40) × 105 |
| [Cu(5–F–salo)(phen)(NO3)], 4 | 74.6 | 8.63 (±0.33) × 1012 | 1.70 (±0.09) × 106 |
| [Cu(5–F–salo)(bipy)(NO3)], 5 | 63.0 | 3.01 (±0.10) × 1012 | 1.31 (±0.05) × 106 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadopoulos, Z.; Doulopoulou, E.; Zianna, A.; Hatzidimitriou, A.G.; Psomas, G. Copper(II) Complexes of 5–Fluoro–Salicylaldehyde: Synthesis, Characterization, Antioxidant Properties, Interaction with DNA and Serum Albumins. Molecules 2022, 27, 8929. https://doi.org/10.3390/molecules27248929
Papadopoulos Z, Doulopoulou E, Zianna A, Hatzidimitriou AG, Psomas G. Copper(II) Complexes of 5–Fluoro–Salicylaldehyde: Synthesis, Characterization, Antioxidant Properties, Interaction with DNA and Serum Albumins. Molecules. 2022; 27(24):8929. https://doi.org/10.3390/molecules27248929
Chicago/Turabian StylePapadopoulos, Zisis, Efstratia Doulopoulou, Ariadni Zianna, Antonios G. Hatzidimitriou, and George Psomas. 2022. "Copper(II) Complexes of 5–Fluoro–Salicylaldehyde: Synthesis, Characterization, Antioxidant Properties, Interaction with DNA and Serum Albumins" Molecules 27, no. 24: 8929. https://doi.org/10.3390/molecules27248929
APA StylePapadopoulos, Z., Doulopoulou, E., Zianna, A., Hatzidimitriou, A. G., & Psomas, G. (2022). Copper(II) Complexes of 5–Fluoro–Salicylaldehyde: Synthesis, Characterization, Antioxidant Properties, Interaction with DNA and Serum Albumins. Molecules, 27(24), 8929. https://doi.org/10.3390/molecules27248929








