Evaluation of the Possible Pathways Involved in the Protective Effects of Quercetin, Naringenin, and Rutin at the Gene, Protein and miRNA Levels Using In-Silico Multidimensional Data Analysis
Abstract
:1. Introduction
2. Results
3. Discussion
4. Methods
4.1. Pharmacokinetics and Physicochemical Characteristics
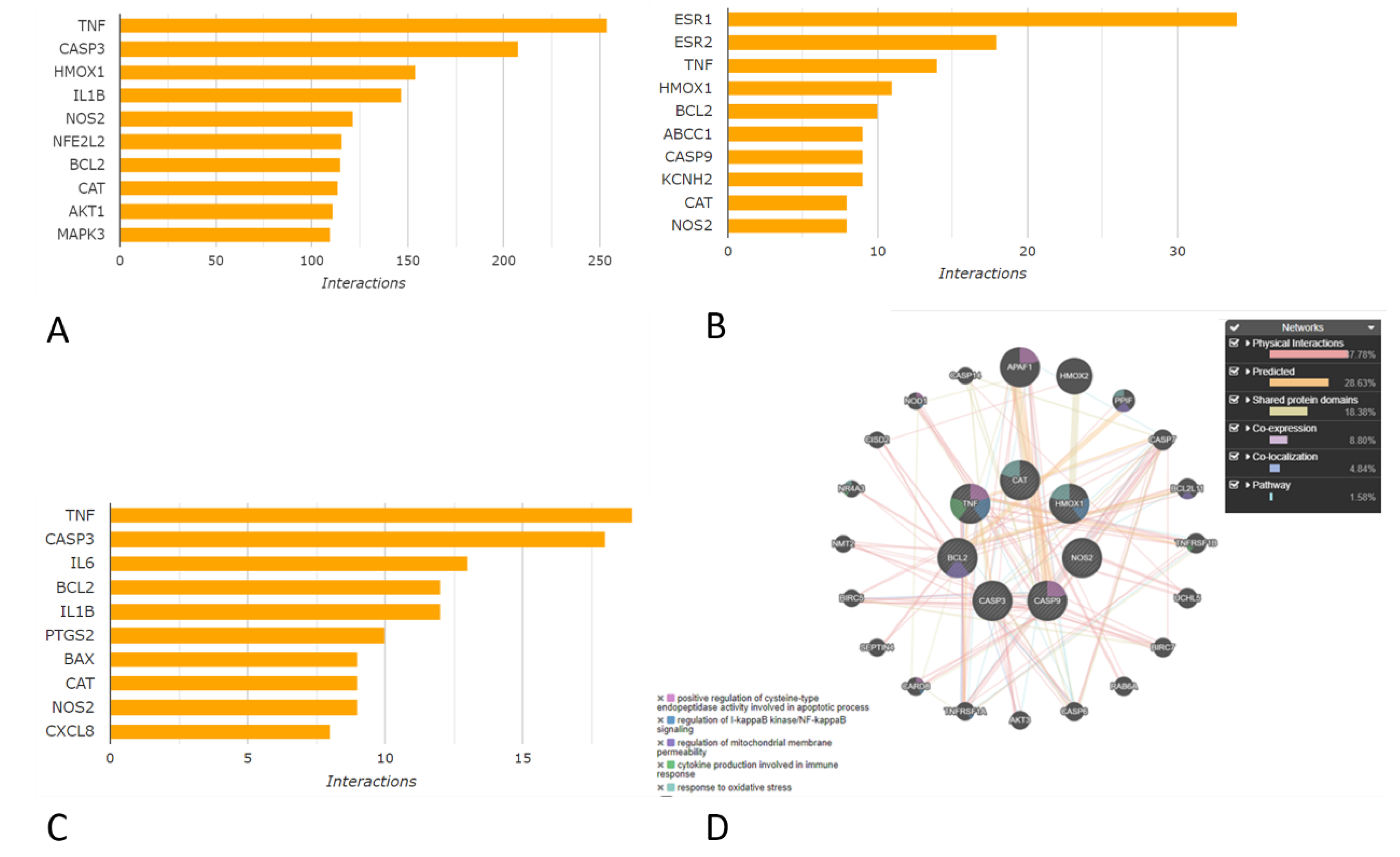
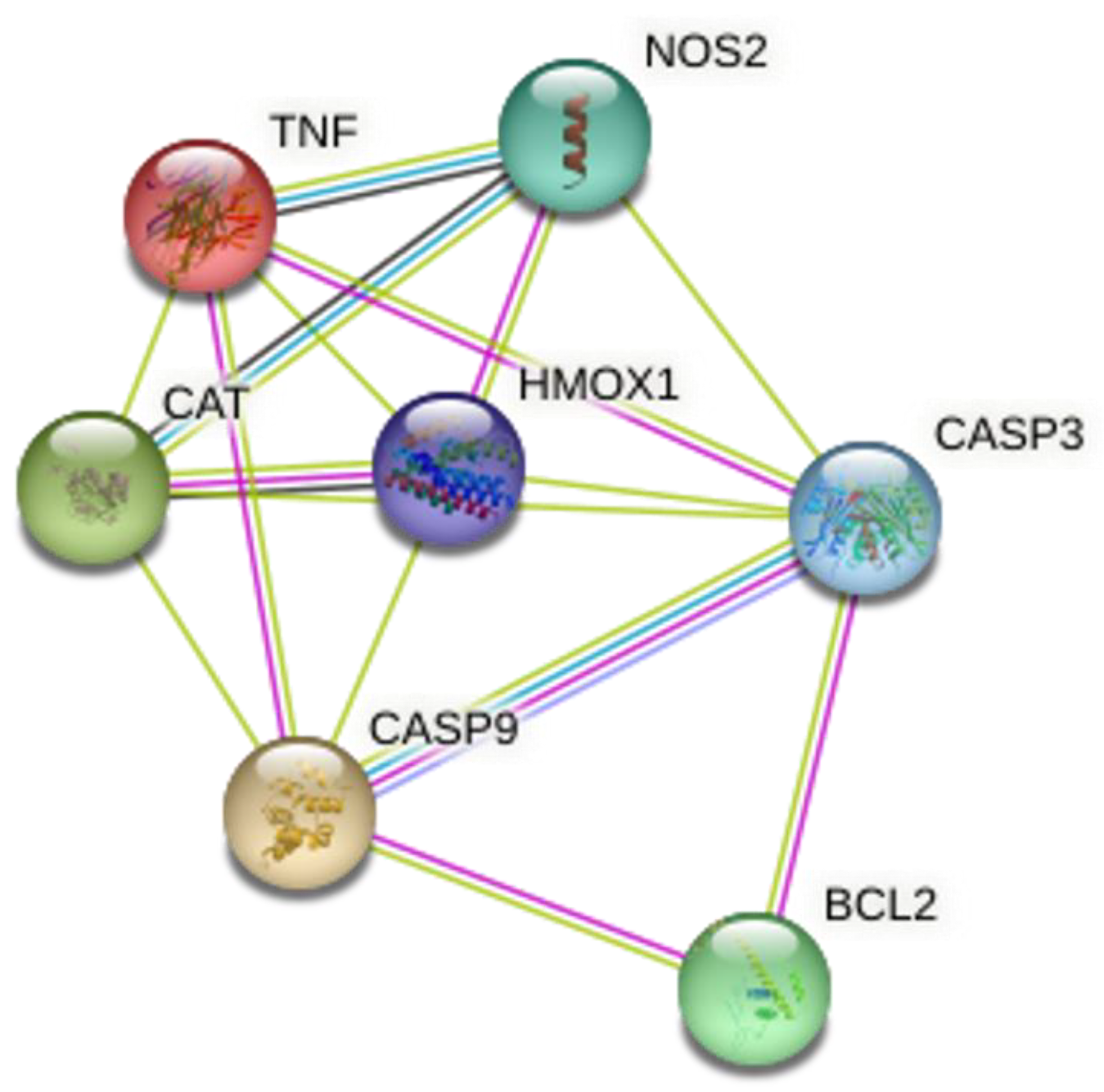
4.2. Gene Association Network Analysis
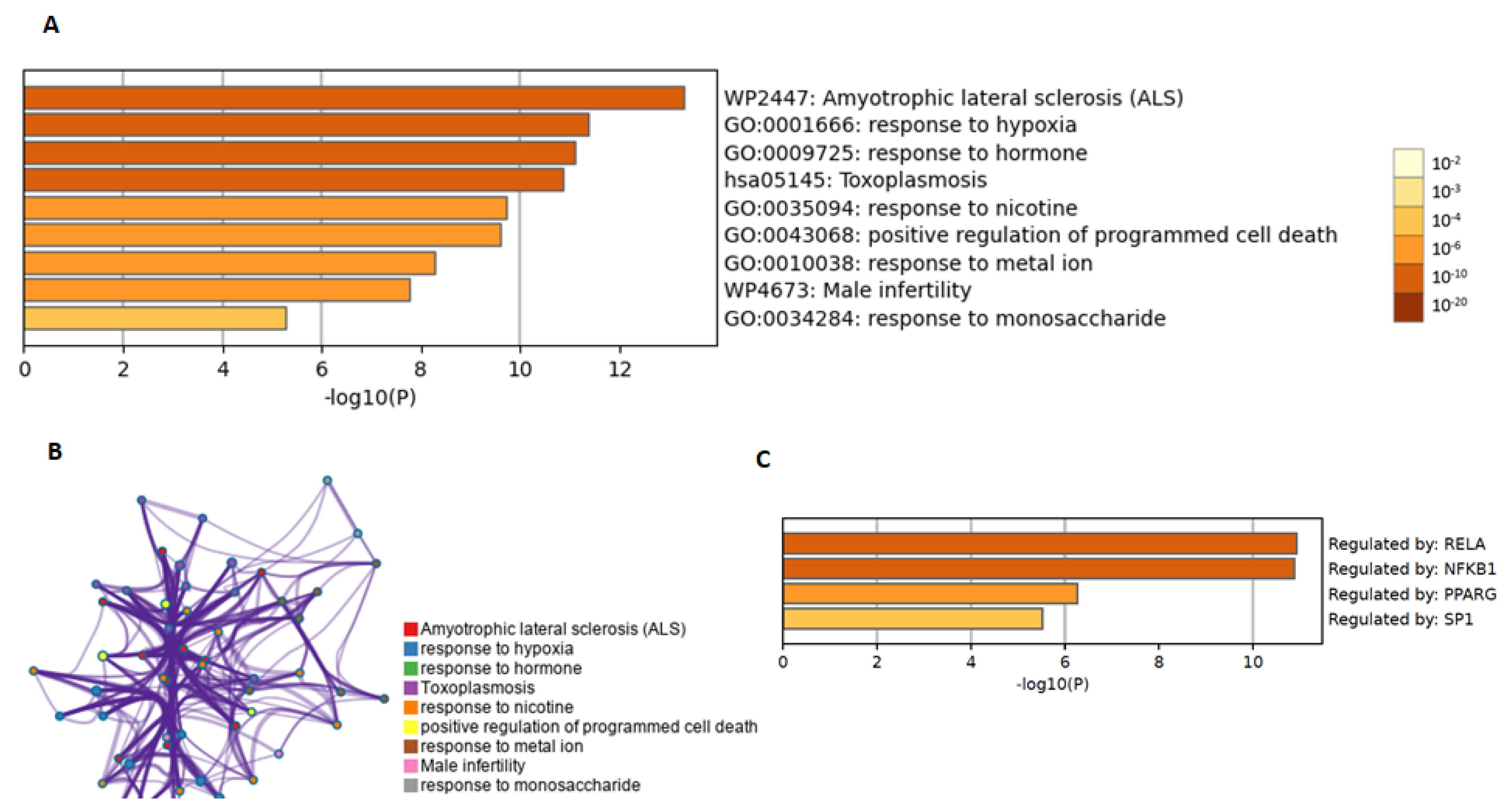
4.3. Enrichment Analysis
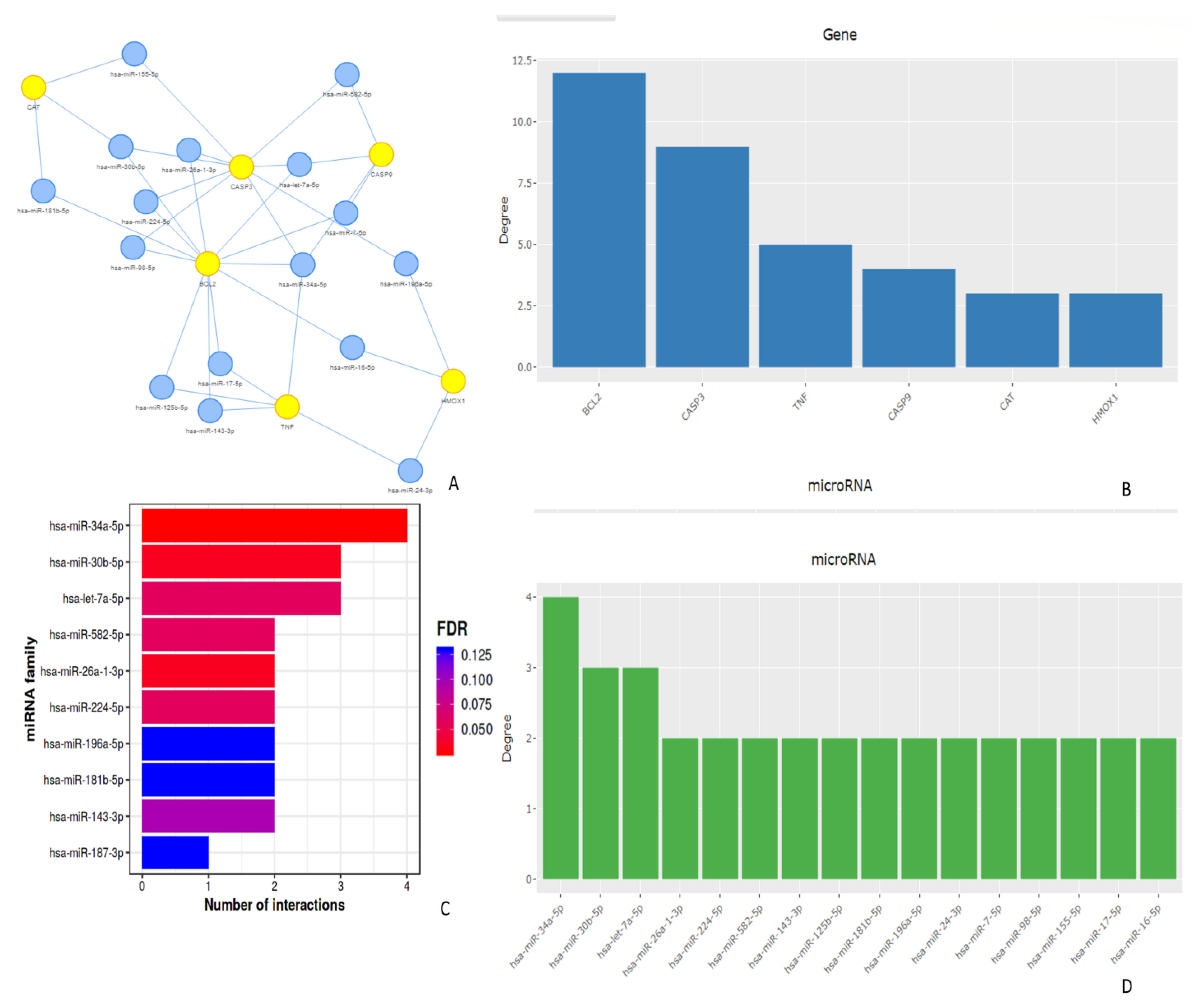
4.4. miRNA Target Interactions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wani, T.A.; Bakheit, A.H.; Zargar, S.; Khayyat, A.I.A.; Al-Majed, A.A. Influence of Rutin, Sinapic Acid, and Naringenin on Binding of Tyrosine Kinase Inhibitor Erlotinib to Bovine Serum Albumin Using Analytical Techniques Along with Computational Approach. Appl. Sci. 2022, 12, 3575. [Google Scholar] [CrossRef]
- Zargar, S.; Alamery, S.; Bakheit, A.H.; Wani, T.A. Poziotinib and bovine serum albumin binding characterization and influence of quercetin, rutin, naringenin and sinapic acid on their binding interaction. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 235, 118335. [Google Scholar] [CrossRef]
- Wani, T.A.; Alanazi, M.M.; Alsaif, N.A.; Bakheit, A.H.; Zargar, S.; Alsalami, O.M.; Khan, A.A. Interaction Characterization of a Tyrosine Kinase Inhibitor Erlotinib with a Model Transport Protein in the Presence of Quercetin: A Drug–Protein and Drug–Drug Interaction Investigation Using Multi-Spectroscopic and Computational Approaches. Molecules 2022, 27, 1265. [Google Scholar] [CrossRef] [PubMed]
- Zargar, S.; Al-Majed, A.R.; Wani, T.A. Potentiating and synergistic effect of grapefruit juice on the antioxidant and anti-inflammatory activity of aripiprazole against hydrogen peroxide induced oxidative stress in mice. BMC Complement. Altern. Med. 2018, 18, 106. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, U.; Gorlach, S.; Owczarek, K.; Hrabec, E.; Szewczyk, K. Synergistic interactions between anticancer chemothera-peutics and phenolic compounds and anticancer synergy between polyphenols. Adv. Hyg. Exp. Med. 2014, 68, 528–540. [Google Scholar]
- Gao, X.; Cassidy, A.; Schwarzschild, M.A.; Rimm, E.B.; Ascherio, A. Habitual intake of dietary flavonoids and risk of Parkinson disease. Neurology 2012, 78, 1138–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansuri, M.L.; Parihar, P.; Solanki, I.; Parihar, M.S. Flavonoids in modulation of cell survival signalling pathways. Genes Nutr. 2014, 9, 400. [Google Scholar] [CrossRef]
- Duan, N.; Hu, X.; Zhou, R.; Li, Y.; Wu, W.; Liu, N. A review on dietary flavonoids as modulators of the tumor microenvi-ronment. Mol. Nutr. Food Res. 2023, 67, 2200435. [Google Scholar] [CrossRef]
- Sousa, C.; Duarte, D.; Silva-Lima, B.; Videira, M. Repurposing natural dietary flavonoids in the modulation of cancer tumor-igenesis: Decrypting the molecular targets of naringenin, hesperetin and myricetin. Nutr. Cancer 2022, 74, 1188–1202. [Google Scholar] [CrossRef]
- Basu, A. The interplay between apoptosis and cellular senescence: Bcl-2 family proteins as targets for cancer therapy. Pharmacol. Ther. 2021, 230, 107943. [Google Scholar] [CrossRef]
- Wong, S.C.; Kamarudin, M.N.A.; Naidu, R. Anticancer Mechanism of Flavonoids on High-Grade Adult-Type Diffuse Gliomas. Nutrients 2023, 15, 797. [Google Scholar] [CrossRef]
- D’Arcy, M.S. A review of biologically active flavonoids as inducers of autophagy and apoptosis in neoplastic cells and as cytoprotective agents in non-neoplastic cells. Cell Biol. Int. 2022, 46, 1179–1195. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ren, X.; Zhang, X.; Griffin, N.; Liu, H.; Wang, L. Rutin ameliorates perfluorooctanoic acid-induced testicular injury in mice by reducing oxidative stress and improving lipid metabolism. Drug Chem. Toxicol. 2022. ahead of print. [Google Scholar] [CrossRef]
- Liu, Q.; Pan, R.; Ding, L.; Zhang, F.; Hu, L.; Ding, B.; Zhu, L.; Xia, Y.; Dou, X. Rutin exhibits hepatoprotective effects in a mouse model of non-alcoholic fatty liver disease by reducing hepatic lipid levels and mitigating lipid-induced oxidative injuries. Int. Immunopharmacol. 2017, 49, 132–141. [Google Scholar] [CrossRef]
- Weng, C.-J.; Chen, M.-J.; Yeh, C.-T.; Yen, G.-C. Hepatoprotection of quercetin against oxidative stress by induction of metal-lothionein expression through activating MAPK and PI3K pathways and enhancing Nrf2 DNA-binding activity. New Biotechnol. 2011, 28, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Thabet, N.M.; Moustafa, E.M. Protective effect of rutin against brain injury induced by acrylamide or gamma radiation: Role of PI3K/AKT/GSK-3β/NRF-2 signalling pathway. Arch. Physiol. Biochem. 2017, 124, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Tamatani, M.; Che, Y.H.; Matsuzaki, H.; Ogawa, S.; Okado, H.; Miyake, S.-I.; Mizuno, T.; Tohyama, M. Tumor Necrosis Factor Induces Bcl-2 and Bcl-x Expression through NFκB Activation in Primary Hippocampal Neurons. J. Biol. Chem. 1999, 274, 8531–8538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erlund, I. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr. Res. 2004, 24, 851–874. [Google Scholar] [CrossRef]
- Arinç, E.; Yilmaz, D.; Bozcaarmutlu, A. Mechanism of Inhibition of CYP1A1 and Glutathione S-Transferase Activities in Fish Liver by Quercetin, Resveratrol, Naringenin, Hesperidin, and Rutin. Nutr. Cancer 2014, 67, 137–144. [Google Scholar] [CrossRef]
- Zargar, S.; Wani, T.A.; Ahamad, S.R. An Insight into Wheat Germ Oil Nutrition, Identification of Its Bioactive Constituents and Computer-Aided Multidimensional Data Analysis of Its Potential Anti-Inflammatory Effect via Molecular Connections. Life 2023, 13, 526. [Google Scholar] [CrossRef]
- Miron, A.; Aprotosoaie, A.C.; Trifan, A.; Xiao, J. Flavonoids as modulators of metabolic enzymes and drug transporters. Ann. N. Y. Acad. Sci. 2017, 1398, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Lee, M.-S.; Chang, E.; Shin, Y.; Oh, S.; Kim, I.-H.; Kim, Y. Rutin Increases Muscle Mitochondrial Biogenesis with AMPK Activation in High-Fat Diet-Induced Obese Rats. Nutrients 2015, 7, 8152–8169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stabrauskiene, J.; Kopustinskiene, D.M.; Lazauskas, R.; Bernatoniene, J. Naringin and Naringenin: Their Mechanisms of Action and the Potential Anticancer Activities. Biomedicines 2022, 10, 1686. [Google Scholar] [CrossRef]
- Baranowska, M.; Koziara, Z.; Suliborska, K.; Chrzanowski, W.; Wormstone, M.; Namieśnik, J.; Bartoszek, A. Interactions between polyphenolic antioxidants quercetin and naringenin dictate the distinctive redox-related chemical and biological behaviour of their mixtures. Sci. Rep. 2021, 11, 12282. [Google Scholar] [CrossRef] [PubMed]
- Celik, H.; Arinç, E. Evaluation of the protective effects of quercetin, rutin, naringenin, resveratrol and trolox against idarubicin-induced DNA damage. J. Pharm. Pharm. Sci. 2010, 13, 231–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cione, E.; La Torre, C.; Cannataro, R.; Caroleo, M.C.; Plastina, P.; Gallelli, L. Quercetin, Epigallocatechin Gallate, Curcumin, and Resveratrol: From Dietary Sources to Human MicroRNA Modulation. Molecules 2019, 25, 63. [Google Scholar] [CrossRef] [Green Version]
- Spagnuolo, C.; Moccia, S.; Russo, G.L. Anti-inflammatory effects of flavonoids in neurodegenerative disorders. Eur. J. Med. Chem. 2017, 153, 105–115. [Google Scholar] [CrossRef]
- Sun, B.; Ross, S.M.; Trask, O.J.; Carmichael, P.L.; Dent, M.; White, A.; Andersen, M.E.; Clewell, R.A. Assessing dose-dependent differences in DNA-damage, p53 response and genotoxicity for quercetin and curcumin. Toxicol. In Vitro 2013, 27, 1877–1887. [Google Scholar] [CrossRef]
- Licursi, V.; Conte, F.; Fiscon, G.; Paci, P. MIENTURNET: An interactive web tool for microRNA-target enrichment and net-work-based analysis. BMC Bioinform. 2019, 20, 545. [Google Scholar] [CrossRef] [Green Version]
- Spencer, J.P.; Vafeiadou, K.; Rendeiro, C. Flavonoids and cognition: The molecular mechanisms underlying their behavioural effects. Arch. Biochem. Biophys. 2008, 476, 133–143. [Google Scholar] [CrossRef]
- Chang, S.C.; Cassidy, A.; Willett, W.C.; Rimm, E.R.; O’Reilly, E.J.; Okereke, O.I. Dietary flavonoid intake and risk of incident depression in midlife and older women. Am. J. Clin. Nutr. 2020, 112, 343–353. [Google Scholar] [CrossRef] [Green Version]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Al-Khayri, J.M.; Sahana, G.R.; Nagella, P.; Joseph, B.V.; Alessa, F.M.; Al-Mssallem, M.Q. Flavonoids as Potential Anti-Inflammatory Molecules: A Review. Molecules 2022, 27, 2901. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.J.; Crespo, J.F.; Cabanillas, B. Anti-inflammatory effects of flavonoids. Food Chem. 2019, 299, 125124. [Google Scholar] [CrossRef]
- Wang, C.; Chen, H.; Jiang, H.-H.; Mao, B.-B.; Yu, H. Total Flavonoids of Chuju Decrease Oxidative Stress and Cell Apoptosis in Ischemic Stroke Rats: Network and Experimental Analyses. Front. Neurosci. 2021, 15, 772401. [Google Scholar] [CrossRef]
- Zhang, H.-W.; Hu, J.-J.; Fu, R.-Q.; Liu, X.; Zhang, Y.-H.; Li, J.; Liu, L.; Li, Y.-N.; Deng, Q.; Luo, Q.-S.; et al. Flavonoids inhibit cell proliferation and induce apoptosis and autophagy through downregulation of PI3Kγ mediated PI3K/AKT/mTOR/p70S6K/ULK signaling pathway in human breast cancer cells. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Wang, C.; Klabnik, J.; O’Donnell, J.M. Novel therapeutic targets in depression and anxiety: Antioxidants as a candidate treatment. Curr. Neuropharmacol. 2014, 12, 108–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, T.V.A.; McMahon, J.; Howley, B.A.; Stanley, A.; Ritter, T.; Mohr, A.; Zwacka, R.; Fearnhead, H. A non-apoptotic role for caspase-9 in muscle differentiation. J. Cell Sci. 2008, 121, 3786–3793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinberg, J.B. Nitric Oxide Production and Nitric Oxide Synthase Type 2 Expression by Human Mononuclear Phagocytes: A Review. Mol. Med. 1998, 4, 557–591. [Google Scholar] [CrossRef] [Green Version]
- Su, S.; Panmanee, W.; Wilson, J.J.; Mahtani, H.K.; Li, Q.; VanderWielen, B.D.; Makris, T.; Rogers, M.; McDaniel, C.; Lipscomb, J.; et al. Catalase (KatA) Plays a Role in Protection against Anaerobic Nitric Oxide in Pseudomonas aeruginosa. PLoS ONE 2014, 9, e91813. [Google Scholar] [CrossRef]
- de Oliveira, L.M.G.; Carreira, R.B.; de Oliveira, J.V.R.; Nascimento, R.P.D.; Souza, C.d.S.; Trias, E.; da Silva, V.D.A.; Costa, S.L. Impact of Plant-Derived Compounds on Amyotrophic Lateral Sclerosis. Neurotox. Res. 2023, 41, 288–309. [Google Scholar] [CrossRef] [PubMed]
- Fernando, W.; Vasantha Rupasinghe, H.P.; Hoskin, D.W. Regulation of hypoxia-inducible factor-1α and vascular endothelial growth factor signaling by plant flavonoids. Mini Rev. Med. Chem. 2015, 15, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Tao, R.; He, C.; Liu, S.; Wang, Y.; Zhang, X. The Risk Factors of the Alcohol Use Disorders—Through Review of Its Comorbidities. Front. Neurosci. 2018, 12, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Manson, J.E.; Buring, J.E.; Sesso, H.D.; Liu, S. Associations of Dietary Flavonoids with Risk of Type 2 Diabetes, and Markers of Insulin Resistance and Systemic Inflammation in Women: A Prospective Study and Cross-Sectional Analysis. J. Am. Coll. Nutr. 2005, 24, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.; Ramos, S. Dietary Flavonoids and Insulin Signaling in Diabetes and Obesity. Cells 2021, 10, 1474. [Google Scholar] [CrossRef]
- Sinyani, A.; Idowu, K.; Shunmugam, L.; Kumalo, H.M.; Khan, R. A molecular dynamics perspective into estrogen receptor inhibition by selective flavonoids as alternative therapeutic options. J. Biomol. Struct. Dyn. 2022, 41, 4093–4105. [Google Scholar] [CrossRef]
- Xiao, Z.-P.; Peng, Z.-Y.; Peng, M.-J.; Yan, W.-B.; Ouyang, Y.-Z.; Zhu, H.-L. Flavonoids Health Benefits and Their Molecular Mechanism. Mini-Rev. Med. Chem. 2011, 11, 169–177. [Google Scholar] [CrossRef]
- Tsao, R. Chemistry and Biochemistry of Dietary Polyphenols. Nutrients 2010, 2, 1231–1246. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Chen, F.; Hua, M.; Guo, J.; Nong, Y.; Tang, Q.; Zhong, F.; Qin, L. Knockdown of lncRNA MIR31HG inhibits cell proliferation in human HaCaT keratinocytes. Biol. Res. 2018, 51, 30. [Google Scholar] [CrossRef] [Green Version]
- Shinjyo, N.; Nakayama, H.; Li, L.; Ishimaru, K.; Hikosaka, K.; Suzuki, N.; Yoshida, H.; Norose, K. Hypericum perforatum extract and hyperforin inhibit the growth of neurotropic parasite Toxoplasma gondii and infection-induced inflammatory responses of glial cells in vitro. J. Ethnopharmacol. 2020, 267, 113525. [Google Scholar] [CrossRef]
- Alsohaimi, I.H.; Khan, M.R.; Ali, H.M.; Azam, M. Emergence of mutagenic/carcinogenic heterocyclic amines in traditional Saudi chicken dishes prepared from local restaurants. Food Chem. Toxicol. 2019, 132, 110677. [Google Scholar] [CrossRef] [PubMed]
- Mejía-Pedroza, R.; Espinal-Enríquez, J.; Hernández-Lemus, E. Pathway-Based Drug Repositioning for Breast Cancer Molecular Subtypes. Front. Pharmacol. 2018, 9, 905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Li, N.; Dong, Y.; Li, S.; Xu, L.; Li, X.; Li, Y.; Li, Z.; Ng, S.S.; Sung, J.J.; et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene 2014, 34, 4142–4152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, N.; Wu, H.; Tao, T.; Peng, E. NEAT1 regulates cell proliferation and apoptosis of ovarian cancer by miR-34a-5p/BCL2. OncoTargets Ther. 2017, 10, 4905–4915. [Google Scholar] [CrossRef] [Green Version]
- Erdogan, S.; Doganlar, O.; Doganlar, Z.B.; Turkekul, K. Naringin sensitizes human prostate cancer cells to paclitaxel therapy. Prostate Int. 2017, 6, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.-Y.; Li, J.-Y.; Hao, G.-F.; Yang, G.-F. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov. Today 2019, 25, 248–258. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. Application of the Swiss Drug Design Online Resources in Virtual Screening. Int. J. Mol. Sci. 2019, 20, 4612. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [Green Version]
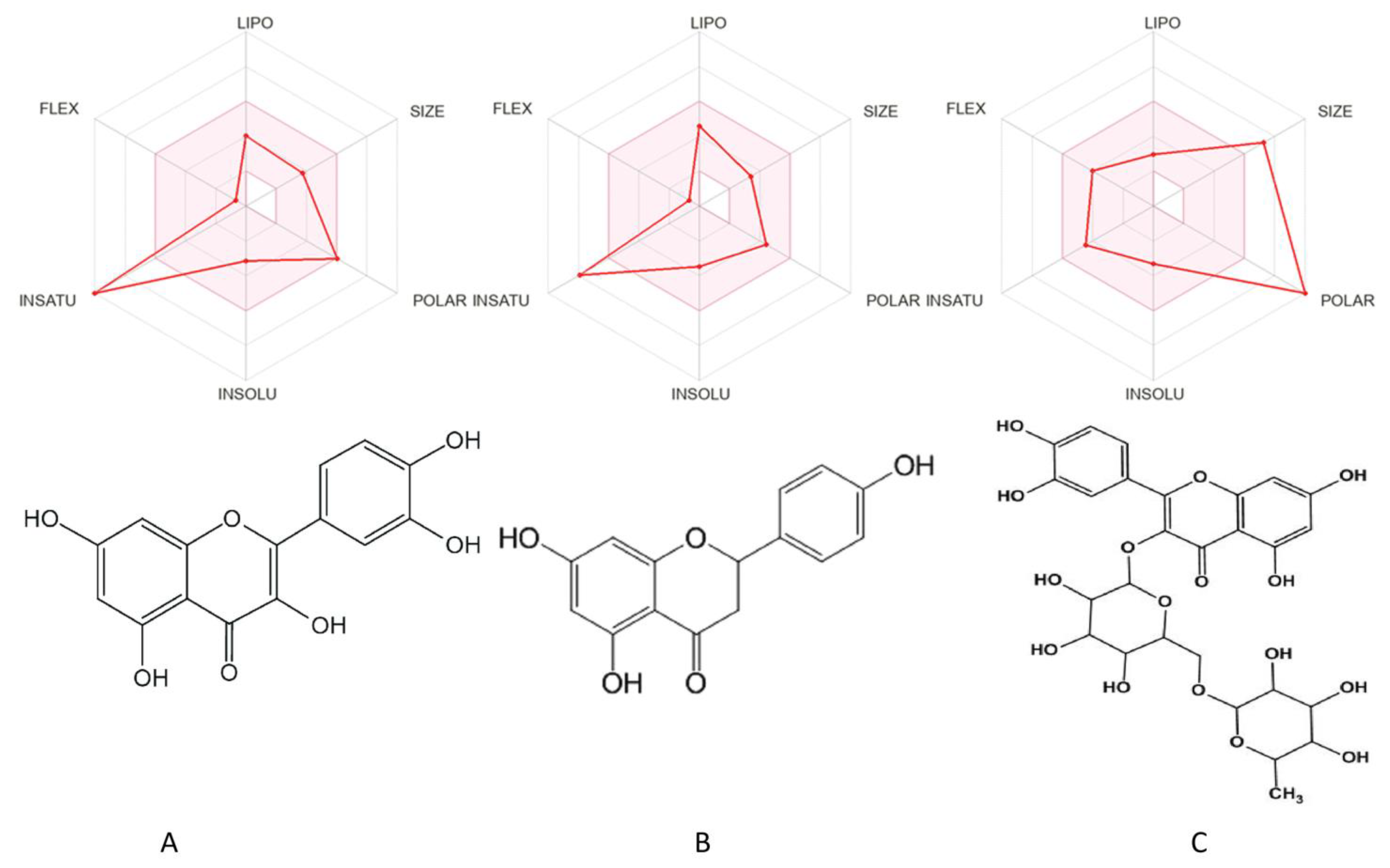


| NAME | Quercetin | Naringenin | Rutin |
|---|---|---|---|
| Molecular weight | 302.24 | 272.25 | 610.52 |
| Num. heavy atoms | 22 | 20 | 43 |
| Num. aromatic heavy atoms | 16 | 12 | 16 |
| Fraction Csp3 | 0 | 0.13 | 0.44 |
| Num. rotatable bonds | 1 | 1 | 6 |
| Num. H-bond acceptors | 7 | 5 | 16 |
| Num. H-bond donors | 5 | 3 | 10 |
| Molar refractivity | 78.03 | 71.57 | 141.38 |
| TPSA | 131.36 | 86.99 | 269.43 |
| Consensus Log Po/w | 1.23 | 1.84 | −1.29 |
| ESOL Log S | −3.16 | −3.49 | −3.3 |
| ESOL solubility (mg/mL) | 0.211 | 0.0874 | 0.308 |
| ESOL solubility (mol/l) | 0.000698 | 0.000321 | 0.000505 |
| ESOL class | Soluble | Soluble | Soluble |
| GI absorption | High | High | Low |
| BBB permeant | No | No | No |
| Pgp substrate | No | Yes | Yes |
| CYP1A2 inhibitor | Yes | Yes | No |
| CYP2C19 inhibitor | No | No | No |
| CYP2C9 inhibitor | No | No | No |
| CYP2D6 inhibitor | Yes | No | No |
| CYP3A4 inhibitor | Yes | Yes | No |
| Log Kp (cm/s) | −7.05 | −6.17 | −10.26 |
| Lipinski violations | 0 | 0 | 3 |
| Ghose violations | 0 | 0 | 4 |
| Veber violations | 0 | 0 | 1 |
| Egan violations | 0 | 0 | 1 |
| Muegge violations | 0 | 0 | 4 |
| Bioavailability score | 0.55 | 0.55 | 0.17 |
| PAINS alerts | 1 | 0 | 1 |
| Brenk alerts | 1 | 0 | 1 |
| Lead-likeness violations | 0 | 0 | 1 |
| Synthetic accessibility | 3.23 | 3.01 | 6.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zargar, S.; Altwaijry, N.; Wani, T.A.; Alkahtani, H.M. Evaluation of the Possible Pathways Involved in the Protective Effects of Quercetin, Naringenin, and Rutin at the Gene, Protein and miRNA Levels Using In-Silico Multidimensional Data Analysis. Molecules 2023, 28, 4904. https://doi.org/10.3390/molecules28134904
Zargar S, Altwaijry N, Wani TA, Alkahtani HM. Evaluation of the Possible Pathways Involved in the Protective Effects of Quercetin, Naringenin, and Rutin at the Gene, Protein and miRNA Levels Using In-Silico Multidimensional Data Analysis. Molecules. 2023; 28(13):4904. https://doi.org/10.3390/molecules28134904
Chicago/Turabian StyleZargar, Seema, Nojood Altwaijry, Tanveer A. Wani, and Hamad M. Alkahtani. 2023. "Evaluation of the Possible Pathways Involved in the Protective Effects of Quercetin, Naringenin, and Rutin at the Gene, Protein and miRNA Levels Using In-Silico Multidimensional Data Analysis" Molecules 28, no. 13: 4904. https://doi.org/10.3390/molecules28134904
APA StyleZargar, S., Altwaijry, N., Wani, T. A., & Alkahtani, H. M. (2023). Evaluation of the Possible Pathways Involved in the Protective Effects of Quercetin, Naringenin, and Rutin at the Gene, Protein and miRNA Levels Using In-Silico Multidimensional Data Analysis. Molecules, 28(13), 4904. https://doi.org/10.3390/molecules28134904






