Synthesis, Crystal Structure, Antibacterial and In Vitro Anticancer Activity of Novel Macroacyclic Schiff Bases and Their Cu (II) Complexes Derived from S-Methyl and S-Benzyl Dithiocarbazate
Abstract
:1. Introduction
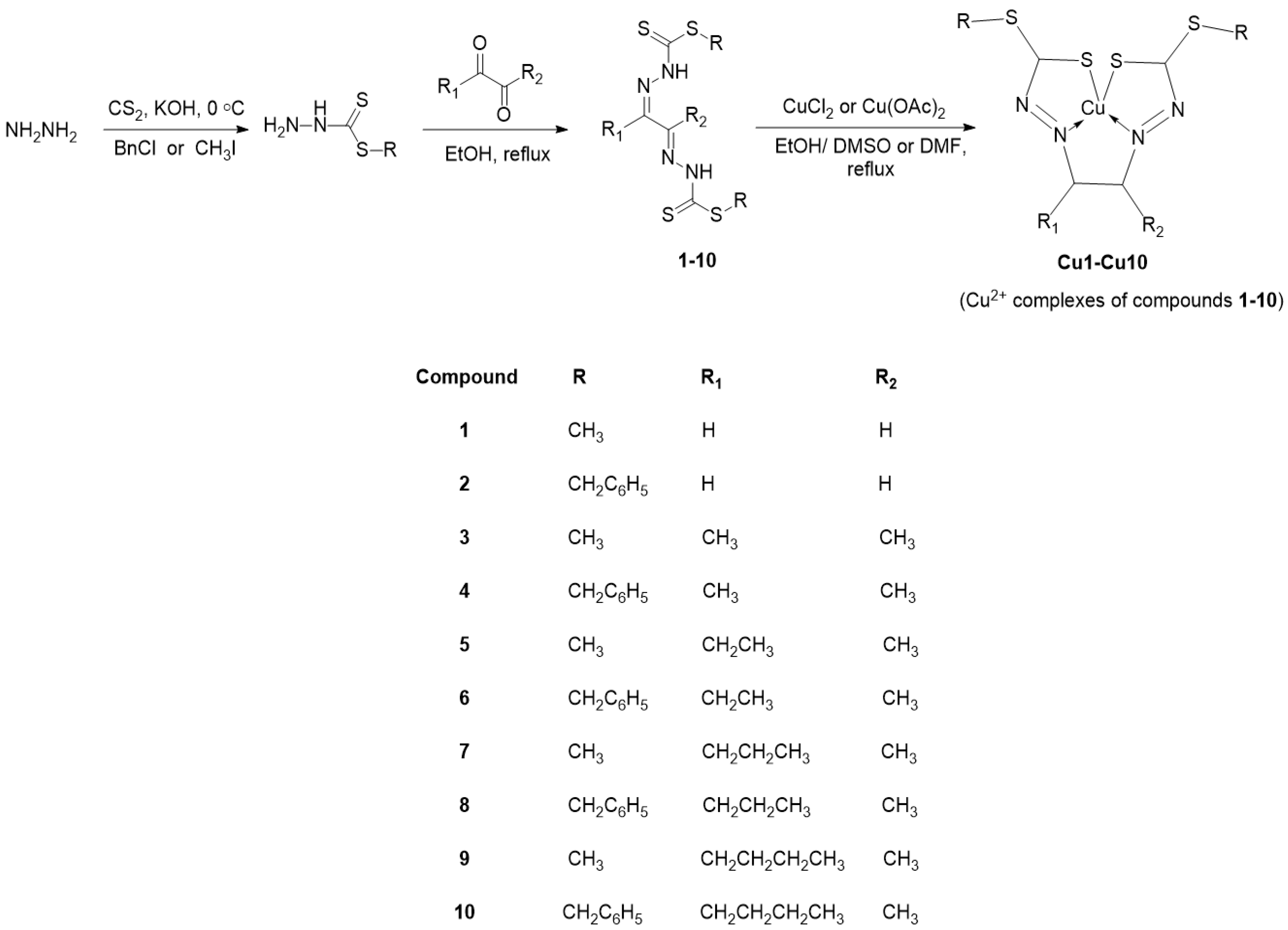
2. Results and Discussion
2.1. Synthesis, Physical and Spectroscopic Characterisation
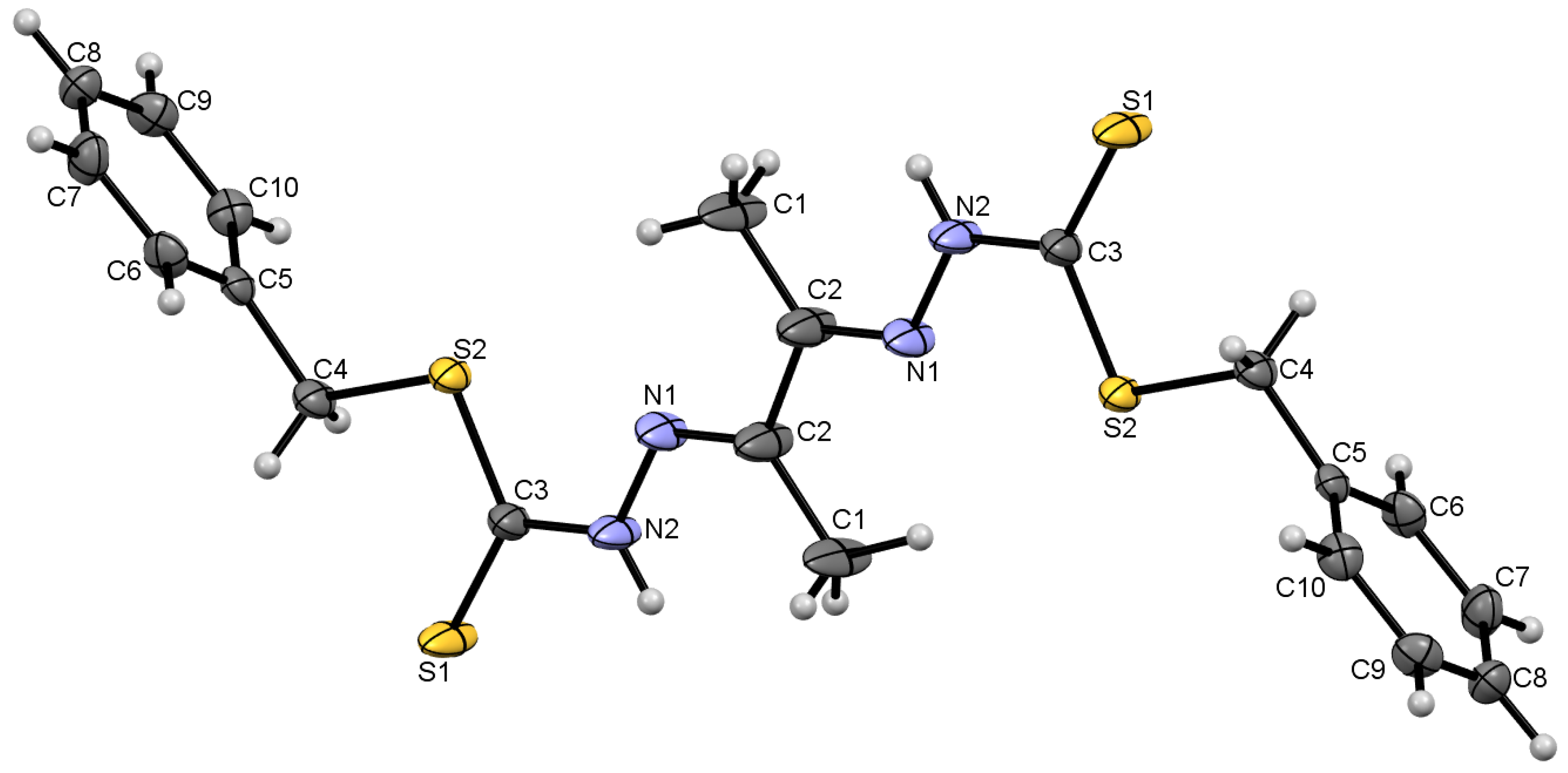
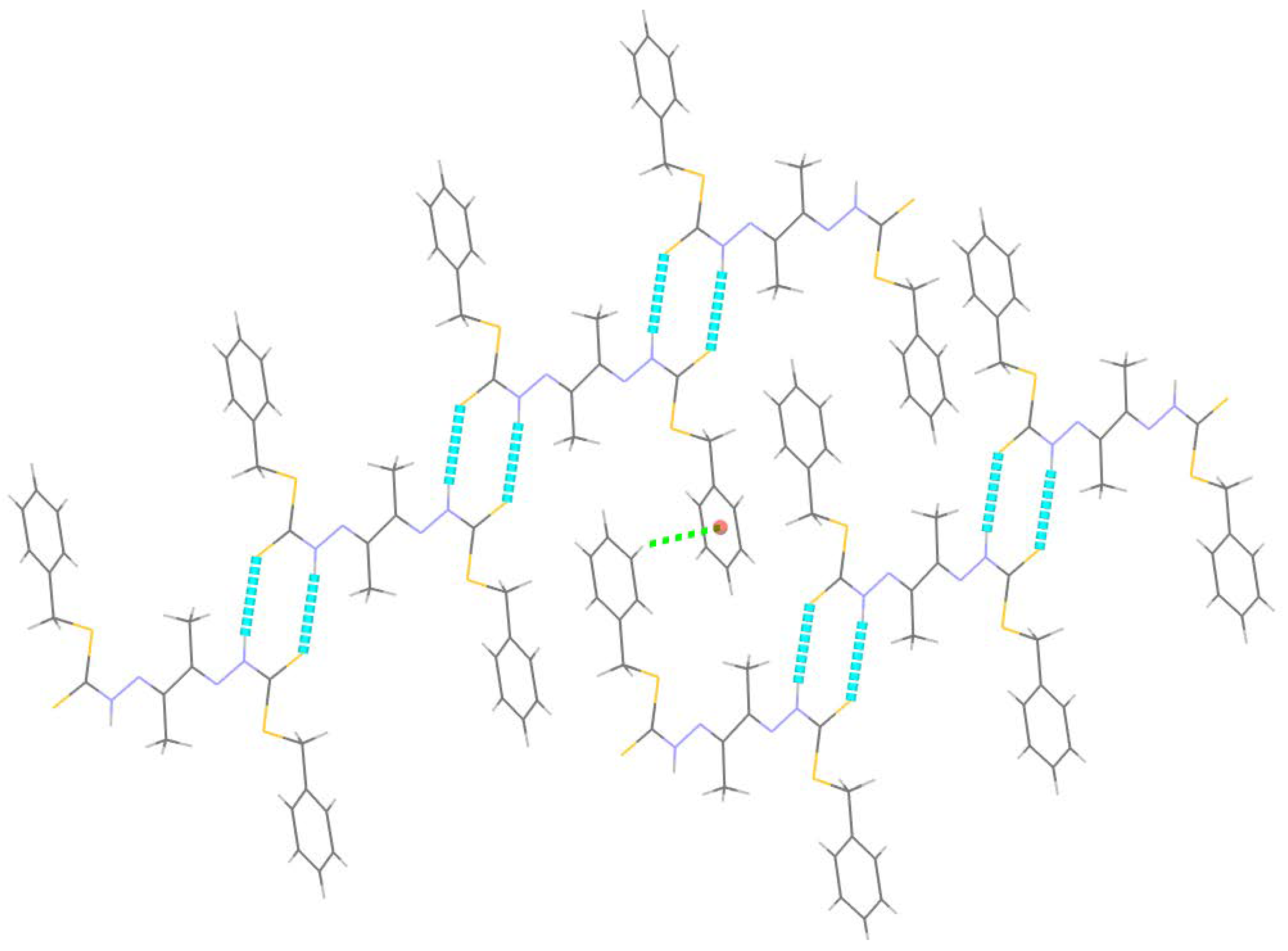
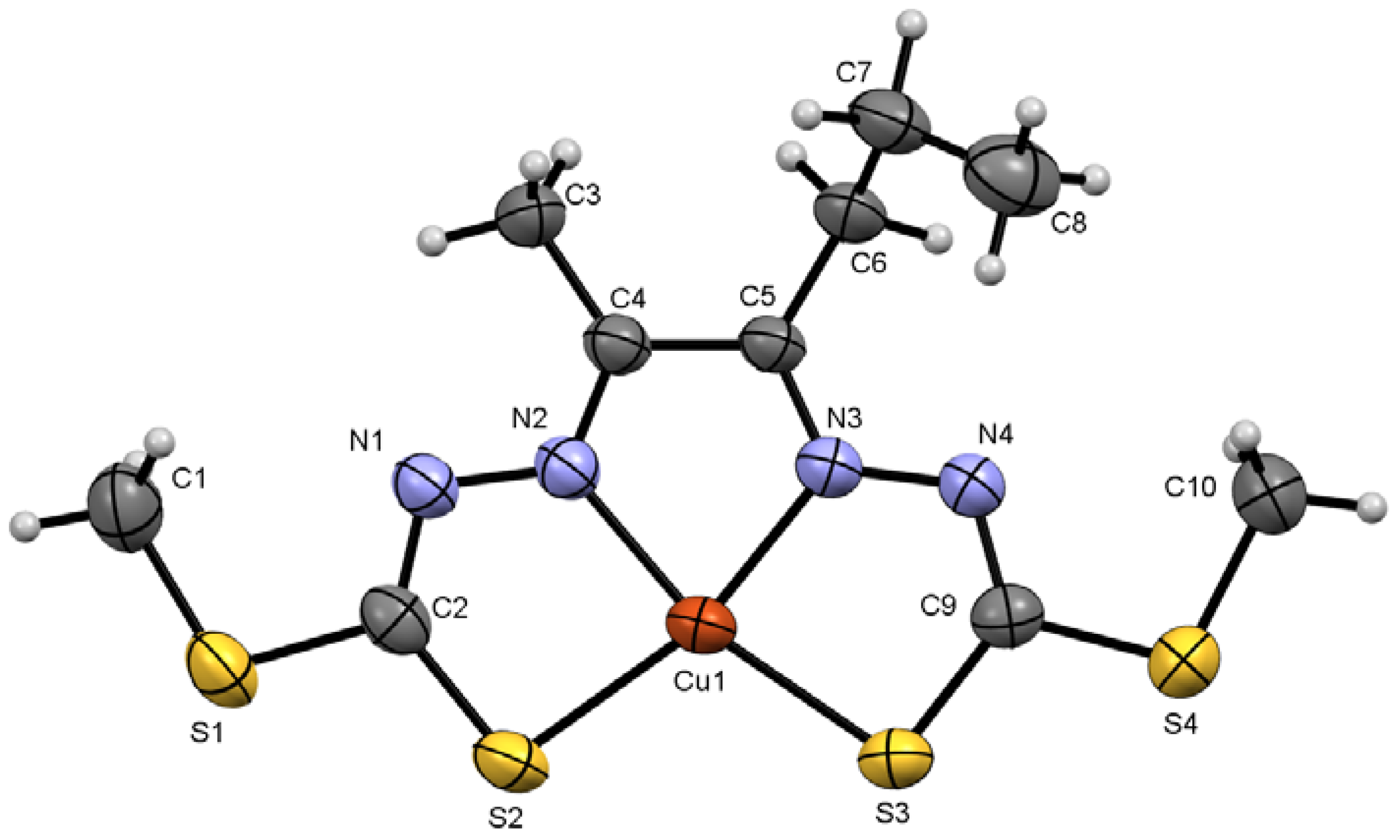
2.2. X-ray Crystal Structure Description of Compound 4 and Metal Complex Cu7
2.2.1. Compound 4 Crystal Structure
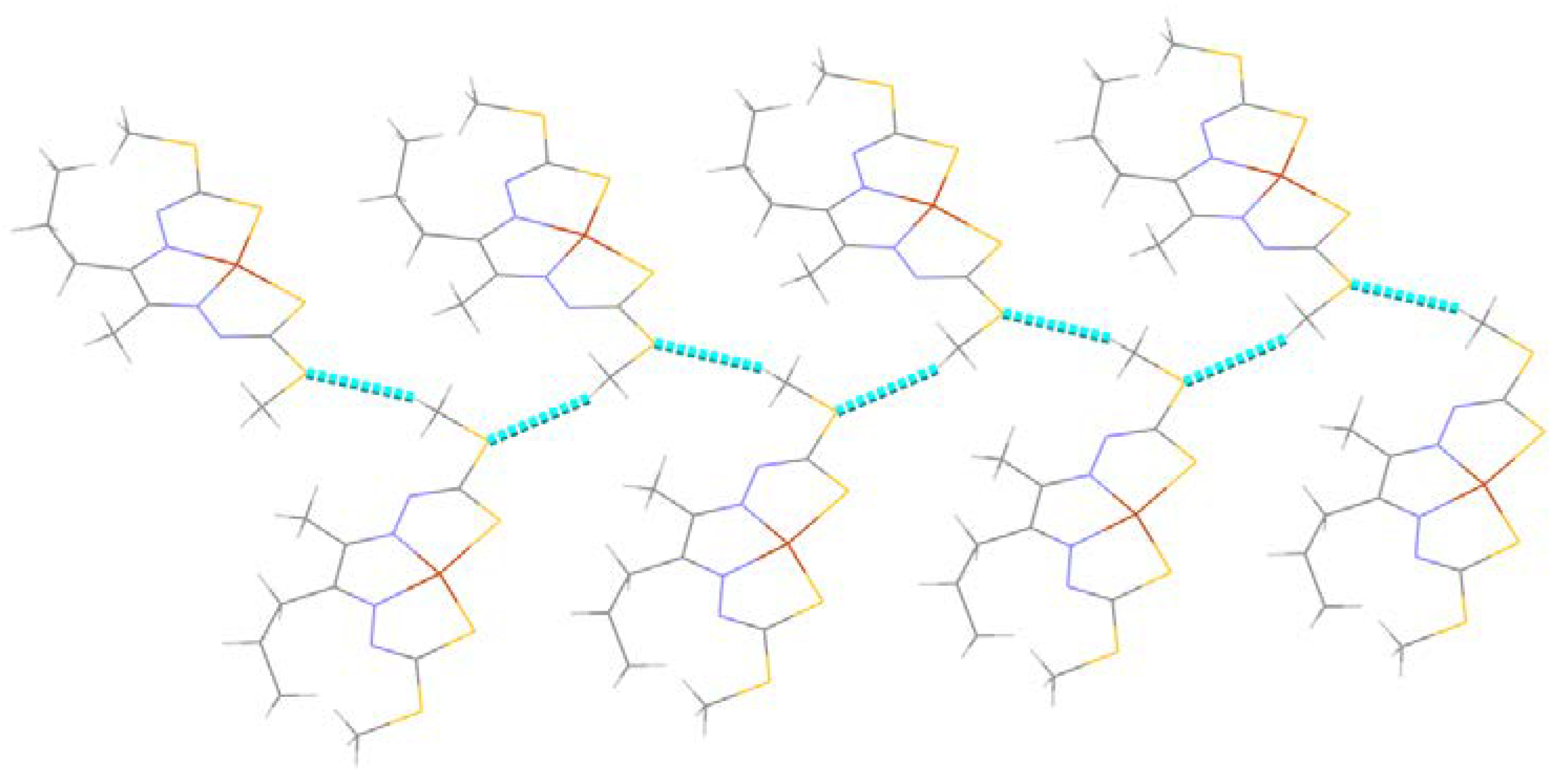
2.2.2. Compound Cu7 Crystal Structure
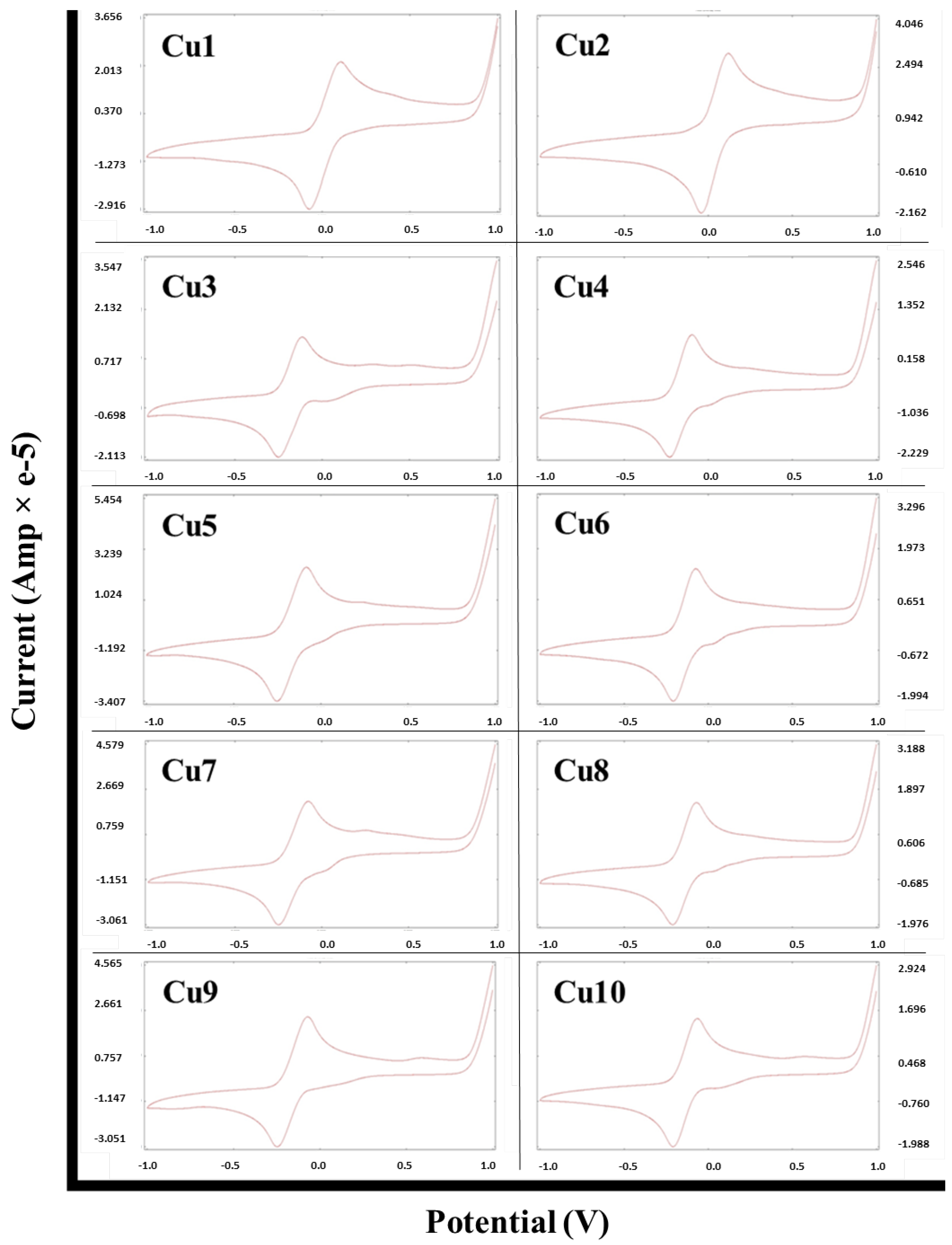
2.3. Cyclic Voltammetry Analysis of the Metal Complexes
2.4. Biological Studies
2.4.1. In Vitro Anticancer Studies
2.4.2. Antibacterial Studies
Qualitative Antibacterial Studies
Quantitative Antibacterial Assay
3. Materials and Methods
3.1. Chemistry
3.1.1. General
3.1.2. Synthesis
General Method for Schiff Base Ligands Synthesis
General Method for Schiff Base Cu (II) Complexes Synthesis
- SMDTC-glyoxal (1)
- Cu–SMDTC–glyoxal (Cu1)
- SBDTC–glyoxal (2)
- Cu–SBDTC–glyoxal (Cu2)
- SMDTC–Butanedione (3)
- Cu–SMDTC–Butanedione (Cu3)
- SBDTC–Butanedione (4)
- Cu–SBDTC–Butanedione (Cu4)
- SMDTC–Pentadione (5)
- Cu–SMDTC–Pentadione (Cu5)
- SBDTC–Pentadione (6)
- Cu–SBDTC–Pentadione (Cu6)
- SMDTC–Hexadione (7)
- Cu–SMDTC–Hexadione (Cu7)
- SBDTC–Hexadione (8)
- Cu–SBDTC–Hexadione (Cu8)
- SMDTC–Heptadione (9)
- Cu–SMDTC–Heptadione (Cu9)
- SBDTC–Heptadione (10)
- Cu–SBDTC–Heptadione (Cu10)
3.1.3. Cyclic Voltammetry
3.1.4. X-ray Crystallography
3.2. Biological Studies
3.2.1. Cell Culture
3.2.2. Cell Viability Assay
3.2.3. Target Microbes Used for Antibacterial Assays
3.2.4. Disc Diffusion Qualitative Antibacterial Assay
3.2.5. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Meenukutty, M.S.; Mohan, A.P.; Vidya, V.G.; Viju Kumar, V.G. Synthesis, Characterization, DFT Analysis and Docking Studies of a Novel Schiff Base Using 5-Bromo Salicylaldehyde and β-Alanine. Heliyon 2022, 8, e09600. [Google Scholar] [CrossRef]
- Yusof, E.N.M.; Azam, M.; Sirat, S.S.; Ravoof, T.B.S.A.; Page, A.J.; Veerakumarasivam, A.; Karunakaran, T.; Razali, M.R. Dithiocarbazate Ligand-Based Cu(II), Ni(II), and Zn(II) Complexes: Synthesis, Structural Investigations, Cytotoxicity, DNA Binding, and Molecular Docking Studies. Bioinorg. Chem. Appl. 2022, 2022, 2004052. [Google Scholar] [CrossRef] [PubMed]
- Samy, F.; Omar, F.M. Synthesis, Characterization, Antitumor Activity, Molecular Modeling and Docking of New Ligand, (2,5-Pyrrole)-Bis(5,6-Diphenyl-[1,2,4]-Triazin-3-Yl)Hydrazone and Its Complexes. J. Mol. Struct. 2020, 1222, 128910. [Google Scholar] [CrossRef]
- Lima, F.C.; Só, Y.A.O.; Gargano, R.; de Oliveira, D.M.; Gatto, C.C. Structural, Theoretical and Biological Activity of Mono and Binuclear Nickel(II) Complexes with Symmetrical and Asymmetrical 4,6-Diacetylresorcinol-Dithiocarbazate Ligands. J. Inorg. Biochem. 2021, 224, 111559. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, C.d.Q.O.; da Mota, T.H.A.; de Oliveira, D.M.; Nascimento, É.C.M.; Martins, J.B.L.; Pittella-Silva, F.; Gatto, C.C. Dithiocarbazate Ligands and Their Ni(II) Complexes with Potential Biological Activity: Structural, Antitumor and Molecular Docking Study. Front. Mol. Biosci. 2023, 10, 1146820. [Google Scholar] [CrossRef]
- bin Break, M.K.; Tahir, M.I.M.; Crouse, K.A.; Khoo, T.-J. Synthesis, Characterization, and Bioactivity of Schiff Bases and Their Cd2+, Zn2+, Cu2+, and Ni2+ Complexes Derived from Chloroacetophenone Isomers with S-Benzyldithiocarbazate and the X-ray Crystal Structure of S-Benzyl-β-N-(4-Chlorophenyl) Methylene. Bioinorg. Chem. Appl. 2013, 2013, 362513. [Google Scholar]
- Priya Gogoi, H.; Singh, A.; Barman, P.; Choudhury, D. A New Potential ONO Schiff-Base Ligand and Its Cu(II), Zn(II) and Cd(II) Complexes: Synthesis, Structural Elucidation, Theoretical and Bioactivity Studies. Inorg. Chem. Commun. 2022, 146, 110153. [Google Scholar] [CrossRef]
- Low, M.L.; Paulus, G.; Dorlet, P.; Guillot, R.; Rosli, R.; Delsuc, N.; Crouse, K.A.; Policar, C. Synthesis, Characterization and Biological Activity of Cu(II), Zn(II) and Re(I) Complexes Derived from S-Benzyldithiocarbazate and 3-Acetylcoumarin. BioMetals 2015, 28, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Low, M.L.; Maigre, L.; Tahir, M.I.M.; Tiekink, E.R.T.; Dorlet, P.; Guillot, R.; Ravoof, T.B.; Rosli, R.; Pagès, J.-M.; Policar, C.; et al. New Insight into the Structural, Electrochemical and Biological Aspects of Macroacyclic Cu(II) Complexes Derived from S-Substituted Dithiocarbazate Schiff Bases. Eur. J. Med. Chem. 2016, 120, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chah, C.K.; Ravoof, T.; Veerakumarasivam, A. Synthesis, Characterization and Biological Activities of Ru (Iii), Mo (v), Cd (Ii), Zn (Ii) and Cu (Ii) Complexes Containing a Novel Nitrogen-Sulphur Macrocyclic Schiff Base Derived from Glyoxal. Pertanika J. Sci. Technol. 2018, 26, 653–670. [Google Scholar]
- Kudrat-E-Zahan, M.; Islam, M.S.; Abul Bashar, M. Synthesis, Characteristics, and Antimicrobial Activity of Some Complexes of Mn(II), Fe(III) Co(II), Ni(II), Cu(II), and Sb(III) Containing Bidentate Schiff Base of SMDTC. Russ. J. Gen. Chem. 2015, 85, 667–672. [Google Scholar] [CrossRef]
- Abdul Mumit, M.; Pal, T.K.; Alam, M.A.; Islam, M.A.-A.-A.-A.; Paul, S.; Sheikh, M.C. DFT Studies on Vibrational and Electronic Spectra, HOMO–LUMO, MEP, HOMA, NBO and Molecular Docking Analysis of Benzyl-3-N-(2,4,5-Trimethoxyphenylmethylene)Hydrazinecarbodithioate. J. Mol. Struct. 2020, 1220, 128715. [Google Scholar] [CrossRef] [PubMed]
- Ngarivhume, T.; Díaz, A.; Cao, R.; Ortiz, M.; Sánchez, I. Association Capacity of Ribose Bis(Thiosemicarbazonato)Copper(II) with Nitric Oxide. Synth. React. Inorg. Met. Nano-Met. Chem. 2005, 35, 795–800. [Google Scholar] [CrossRef]
- Paterson, B.M.; Donnelly, P.S. Copper Complexes of Bis(Thiosemicarbazones): From Chemotherapeutics to Diagnostic and Therapeutic Radiopharmaceuticals. Chem. Soc. Rev. 2011, 40, 3005–3018. [Google Scholar] [CrossRef] [PubMed]
- Gaber, A.; Refat, M.S.; Belal, A.A.M.; El-Deen, I.M.; Hassan, N.; Zakaria, R.; Alhomrani, M.; Alamri, A.S.; Alsanie, W.F.; Saied, E.M. New Mononuclear and Binuclear Cu(II), Co(II), Ni(II), and Zn(II) Thiosemicarbazone Complexes with Potential Biological Activity: Antimicrobial and Molecular Docking Study. Molecules 2021, 26, 2288. [Google Scholar] [CrossRef]
- Hossan, M.S.; Break, M.K.; Bradshaw, T.D.; Collins, H.M.; Wiart, C.; Khoo, T.-J.; Alafnan, A. Novel Semi-Synthetic Cu (II)–Cardamonin Complex Exerts Potent Anticancer Activity against Triple-Negative Breast and Pancreatic Cancer Cells via Inhibition of the Akt Signaling Pathway. Molecules 2021, 26, 2166. [Google Scholar] [CrossRef]
- Khoo, T.-J.; bin Break, M.K.; Crouse, K.A.; Tahir, M.I.M.; Ali, A.M.; Cowley, A.R.; Watkin, D.J.; Tarafder, M.T.H. Synthesis, Characterization and Biological Activity of Two Schiff Base Ligands and Their Nickel(II), Copper(II), Zinc(II) and Cadmium(II) Complexes Derived from S-4-Picolyldithiocarbazate and X-Ray Crystal Structure of Cadmium(II) Complex Derived from Pyr. Inorg. Chim. Acta 2014, 413, 68–76. [Google Scholar] [CrossRef]
- Tiwari, A.D.; Mishra, A.K.; Mishra, S.B.; Mamba, B.B.; Maji, B.; Bhattacharya, S. Synthesis and DNA Binding Studies of Ni (II), Co (II), Cu (II) and Zn (II) Metal Complexes of N1, N5-Bis [Pyridine-2-Methylene]-Thiocarbohydrazone Schiff-Base Ligand. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 79, 1050–1056. [Google Scholar] [CrossRef]
- Saha, S.; Jana, S.; Gupta, S.; Ghosh, A.; Nayek, H.P. Syntheses, Structures and Biological Activities of Square Planar Ni (II), Cu (II) Complexes. Polyhedron 2016, 107, 183–189. [Google Scholar] [CrossRef]
- Kuate, M.; Conde, M.A.; Ngandung Mainsah, E.; Paboudam, A.G.; Tchieno, F.M.M.; Ketchemen, K.I.Y.; Tonle Kenfack, I.; Ndifon, P.T. Synthesis, Characterization, Cyclic Voltammetry, and Biological Studies of Co(II), Ni(II), and Cu(II) Complexes of a Tridentate Schiff Base, 1-((E)-(2-Mercaptophenylimino) Methyl) Naphthalen-2-Ol (H2L1). J. Chem. 2020, 2020, 5238501. [Google Scholar] [CrossRef]
- Deghadi, R.G.; Mohamed, G.G. Can New Series of Half-Sandwich Lanthanum(III), Erbium(III), and Ytterbium(III) Complexes of Organometallic Ferrocenyl Schiff Base Ligands Display Biological Activities as Antibacterial and Anticancer Drugs? Comments Inorg. Chem. 2022, 42, 368–401. [Google Scholar] [CrossRef]
- Plech, T.; Wujec, M.; Siwek, A.; Kosikowska, U.; Malm, A. Synthesis and Antimicrobial Activity of Thiosemicarbazides, s-Triazoles and Their Mannich Bases Bearing 3-Chlorophenyl Moiety. Eur. J. Med. Chem. 2011, 46, 241–248. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; Streek, J.V.D.; Wood, P.A. Mercury CSD 2.0–New Features for the Visualization and Investigation of Crystal Structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Khan, M.S.; Ranjani, S.; Hemalatha, S. Synthesis and Characterization of Kappaphycus Alvarezii Derived Silver Nanoparticles and Determination of Antibacterial Activity. Mater. Chem. Phys. 2022, 282, 125985. [Google Scholar] [CrossRef]
- Thapa, A.; Kaushik, R.; Arora, S.; Jaglan, S.; Jaswal, V.; Yadav, V.K.; Singh, M.; Bains, A.; Chawla, P.; Khan, A.; et al. Biological Activity of Picrorhiza Kurroa: A Source of Potential Antimicrobial Compounds against Yersinia Enterocolitica. Int. J. Mol. Sci. 2022, 23, 14090. [Google Scholar] [CrossRef]
- Khedr, A.M.; Gaber, M.; Abd El-Zaher, E.H. Synthesis, Structural Characterization, and Antimicrobial Activities of Mn (II), Co (II), Ni (II), Cu (II) and Zn (II) Complexes of Triazole-based Azodyes. Chin. J. Chem. 2011, 29, 1124–1132. [Google Scholar] [CrossRef]
| Compounds | 4 | Cu7 |
|---|---|---|
| Chemical formula | C20H22N4S4 | CuC10H16 N4S4 |
| Mr | 446.66 | 384.05 |
| Crystal colour | Dark brown | Brown |
| Crystal habit | Rod | Needle |
| Crystal size (mm3) | 0.50 × 0.25 × 0.16 | 0.21 × 0.19 × 0.14 |
| Temperature (K) | 293 | 100 |
| λ (Å) | 0.71073 | 1.54178 |
| Crystal system | monoclinic | orthorhombic |
| Space group | P 21/n | Pbca |
| a (Å) | 8.2086 (5) | 9.2009 (4) |
| b (Å) | 5.4551 (3) | 9.5250 (4) |
| c (Å) | 24.6907 (12) | 35.8100 (13) |
| α (°) | 90.000 | 90.000 |
| β (°) | 95.035 (5) | 90.000 |
| γ (°) | 90.000 | 90.000 |
| V (Å3) | 1101.34 (10) | 3138.3 (2) |
| Z | 1 | 8 |
| Temperature (K) | 293 (2) | 100 (2) |
| F(000) | 468 | 1576 |
| μ (mm−1) | 0.445 | 6.870 |
| θmin (°) | 3.1042 | 4.940 |
| θmax (°) | 29.2305 | 71.371 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 11384, 2049, 1759 | 6741, 2965, 2460 |
| Rint | 0.127 | 0.031 |
| Goodness of fit (GOF) on F2 | 1.09 | 1.061 |
| R[F2 > 2σ(F2)], wR(F2), S | 0.084, 0.227, 1.09 | 0.036, 0.093, 1.06 |
| No. of reflections | 2049 | 2965 |
| No. of parameters | 128 | 173 |
| Δρmax (e Å−3) | 2.20 | 0.34 |
| Δρmin (e Å−3) | −1.50 | −0.27 |
| Compound 4 | |||
| C3–S1 | 1.659 (5) | C3–N2 | 1.352 (7) |
| C3–S2 | 1.735 (5) | N1–N2 | 1.364 (6) |
| C2–N1 | 1.294 (9) | C1–C2 | 1.517 (9) |
| C3–S2–C4 | 100.3 (2) | S2–C4–C5 | 110.3 (3) |
| C3–N2–N1 | 118.2 (4) | N2–C3–S1 | 121.4 (4) |
| Compound Cu7 | |||
| Cu1–S3 | 2.2432 (9) | Cu1–N3 | 1.963 (3) |
| N1–N2 | 1.382 (4) | C5–N3 | 1.293 (4) |
| C2–N1 | 1.309 (4) | C9–S3 | 1.752 (3) |
| N3–Cu1–S2 | 164.42 (9) | C9–N4–N3 | 111.1 (3) |
| N2–Cu1–S3 | 164.99 (9) | S4–C9–S3 | 113.4 (2) |
| Cg is the Centroid of the Ring (C5–C6–C7–C8–C9–C10) | ||||
|---|---|---|---|---|
| D–H···A | D–H | H···A | D···A | D–H···A |
| N2–H2···S1 i | 0.86 | 2.63 | 3.480 (5) | 171 |
| C9–H9···Cg ii | 0.93 | 2.74 | 3.466 (5) | 136 |
| Cg is the Centroid of the Ring (C5–C6–C7–C8–C9–C10) | ||||
|---|---|---|---|---|
| D–H···A | D–H | H···A | D···A | D–H···A |
| C1–H1C···S1 i | 0.98 | 2.86 | 3.652 (4) | 138 |
| Compound | Epc (V) | Epa (V) | ΔEp (mV) |
|---|---|---|---|
| Cu1 | −0.072 | 0.107 | 179 |
| Cu2 | −0.043 | 0.120 | 163 |
| Cu3 | −0.250 | −0.115 | 135 |
| Cu4 | −0.231 | −0.096 | 135 |
| Cu5 | −0.261 | −0.065 | 196 |
| Cu6 | −0.205 | −0.091 | 114 |
| Cu7 | −0.250 | −0.078 | 172 |
| Cu8 | −0.212 | −0.078 | 134 |
| Cu9 | −0.249 | −0.070 | 179 |
| Cu10 | −0.210 | −0.064 | 146 |
| Compound | IC50 (µM) a | |
|---|---|---|
| MCF-7 | MDA-MB-231 | |
| 1 | 6.9 ± 0.3 | >50 |
| Cu1 | 1.7 ± 0.1 | 1.4 ± 0.1 |
| 2 | 2.6 ± 0.8 | 4.5 ± 0.7 |
| Cu2 | >50 | >50 |
| 3 | 20 ± 0.3 | 4.2 ± 0.7 |
| Cu3 | 46 ± 1.0 | 19 ± 4.1 |
| 4 | 49 ± 5.4 | >50 |
| Cu4 | 11 ± 1.9 | 38 ± 7.5 |
| 5 | 17 ± 1.5 | 22 ± 2.0 |
| Cu5 | 14 ± 2.1 | 16 ± 4.6 |
| 6 | 22 ± 1.2 | >50 |
| Cu6 | >50 | 9 ± 1.2 |
| 7 | 12 ± 0.4 | 4 ± 0.2 |
| Cu7 | 45 ± 2.3 | >50 |
| 8 | 7.3 ± 0.8 | 12 ± 1.1 |
| Cu8 | 7.3 ± 2.8 | 12 ± 0.5 |
| 9 | 9.8 ± 0.1 | 6 ± 0.2 |
| Cu9 | 20 ± 1.5 | >50 |
| 10 | 5.7 ± 0.1 | 22 ± 0.8 |
| Cu10 | >50 | >50 |
| Cisplatin | 25 ± 0.3 | 48 ± 3.5 |
| Compound | Zone of Inhibition Diameter (mm) a | |||||
|---|---|---|---|---|---|---|
| S. aureus | B. cereus | K. rhizophila | E. coli | P. aeruginosa | C. freundii | |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cu1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 7.5 | 8.5 | 8 | 0 | 0 |
| Cu2 | 0 | 8 | 8 | 0 | 0 | 0 |
| 3 | 0 | 0 | 25 | 0 | 0 | 0 |
| Cu3 | 12 | 9 | 9 | 0 | 0 | 0 |
| 4 | 13 | 11 | 15 | 0 | 0 | 0 |
| Cu4 | 0 | 10 | 10 | 0 | 0 | 0 |
| 5 | 17 | 11 | 22 | 0 | 0 | 0 |
| Cu5 | 9 | 0 | 8 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cu6 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cu7 | 0 | 0 | 15 | 0 | 7 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cu8 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cu9 | 0 | 0 | 15 | 0 | 8 | 0 |
| 10 | 10.5 | 0 | 0 | 0 | 0 | 0 |
| Cu10 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chloramphenicol | 25 | 18.7 | 19.3 | 24.3 | 27.3 | 21 |
| Compound | MIC (µg/mL) | MBC (µg/mL) | ||
|---|---|---|---|---|
| S. aureus | K. rhizophila | S. aureus | K. rhizophila | |
| 3 | <24.4 | <24.4 | <24.4 | 48.8 |
| 4 | - | 1562.5 | - | 3125 |
| 5 | - | 781.25 | - | 1562.5 |
| Cu7 | - | <24.4 | - | 48.8 |
| Cu9 | - | <24.4 | - | 48.8 |
| Chloramphenicol | <24.4 | <24.4 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Break, M.K.B.; Fung, T.Y.; Koh, M.Z.; Ho, W.Y.; Tahir, M.I.M.; Elfar, O.A.; Syed, R.U.; Khojali, W.M.A.; Alluhaibi, T.M.; Huwaimel, B.; et al. Synthesis, Crystal Structure, Antibacterial and In Vitro Anticancer Activity of Novel Macroacyclic Schiff Bases and Their Cu (II) Complexes Derived from S-Methyl and S-Benzyl Dithiocarbazate. Molecules 2023, 28, 5009. https://doi.org/10.3390/molecules28135009
Break MKB, Fung TY, Koh MZ, Ho WY, Tahir MIM, Elfar OA, Syed RU, Khojali WMA, Alluhaibi TM, Huwaimel B, et al. Synthesis, Crystal Structure, Antibacterial and In Vitro Anticancer Activity of Novel Macroacyclic Schiff Bases and Their Cu (II) Complexes Derived from S-Methyl and S-Benzyl Dithiocarbazate. Molecules. 2023; 28(13):5009. https://doi.org/10.3390/molecules28135009
Chicago/Turabian StyleBreak, Mohammed Khaled Bin, Tan Yew Fung, May Zie Koh, Wan Yong Ho, Mohamed Ibrahim Mohamed Tahir, Omar Ashraf Elfar, Rahamat Unissa Syed, Weam M. A. Khojali, Turki Mubarak Alluhaibi, Bader Huwaimel, and et al. 2023. "Synthesis, Crystal Structure, Antibacterial and In Vitro Anticancer Activity of Novel Macroacyclic Schiff Bases and Their Cu (II) Complexes Derived from S-Methyl and S-Benzyl Dithiocarbazate" Molecules 28, no. 13: 5009. https://doi.org/10.3390/molecules28135009
APA StyleBreak, M. K. B., Fung, T. Y., Koh, M. Z., Ho, W. Y., Tahir, M. I. M., Elfar, O. A., Syed, R. U., Khojali, W. M. A., Alluhaibi, T. M., Huwaimel, B., Wiart, C., & Khoo, T. -J. (2023). Synthesis, Crystal Structure, Antibacterial and In Vitro Anticancer Activity of Novel Macroacyclic Schiff Bases and Their Cu (II) Complexes Derived from S-Methyl and S-Benzyl Dithiocarbazate. Molecules, 28(13), 5009. https://doi.org/10.3390/molecules28135009








