Investigation of the Influence of Charge State and Collision Energy on Oligonucleotide Fragmentation by Tandem Mass Spectrometry
Abstract
:1. Introduction
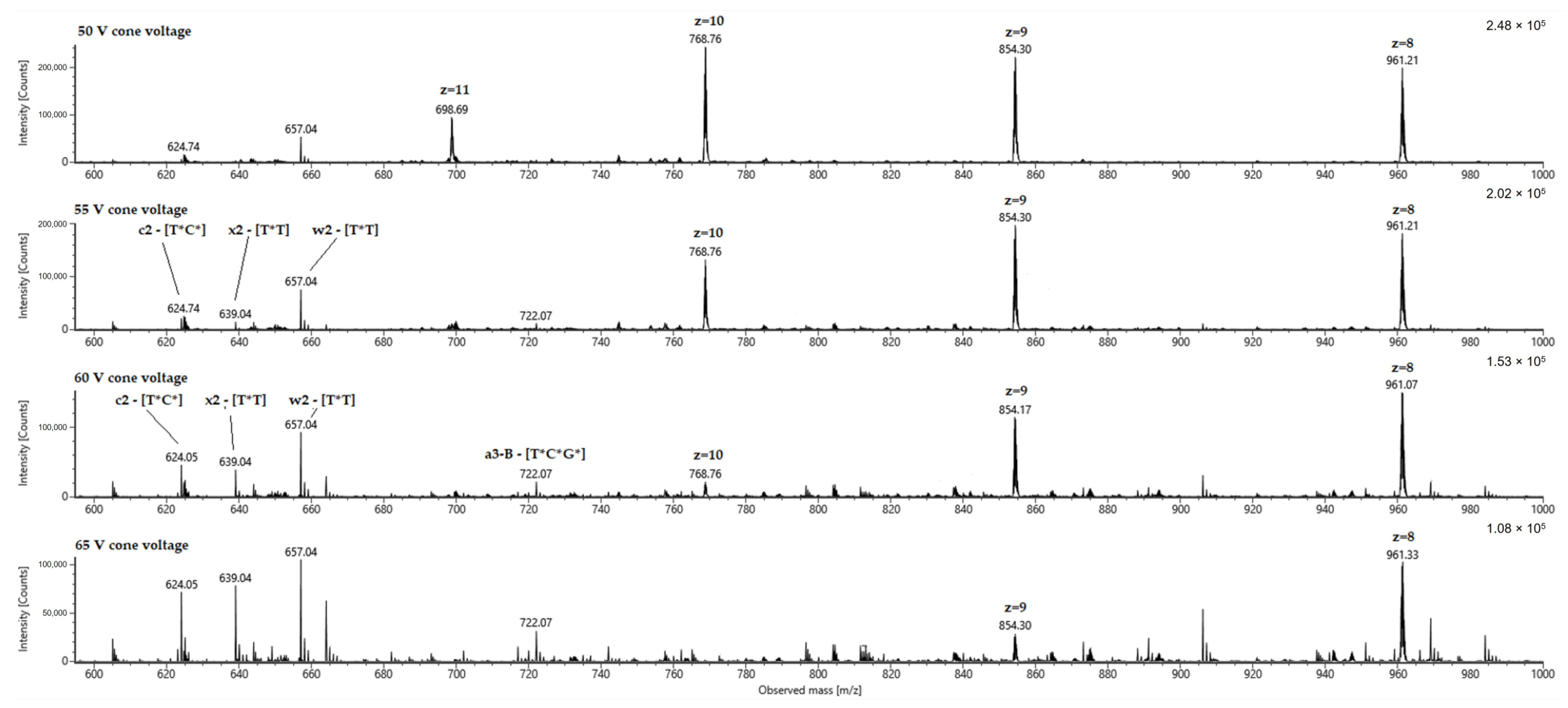
2. Results
3. Materials and Methods
3.1. LC-MS/MS Instrumentation
3.2. Chemicals and Reagents
3.3. Sample Preparation
3.4. LC-MS/MS Measurement
4. Conclusions
- (I)
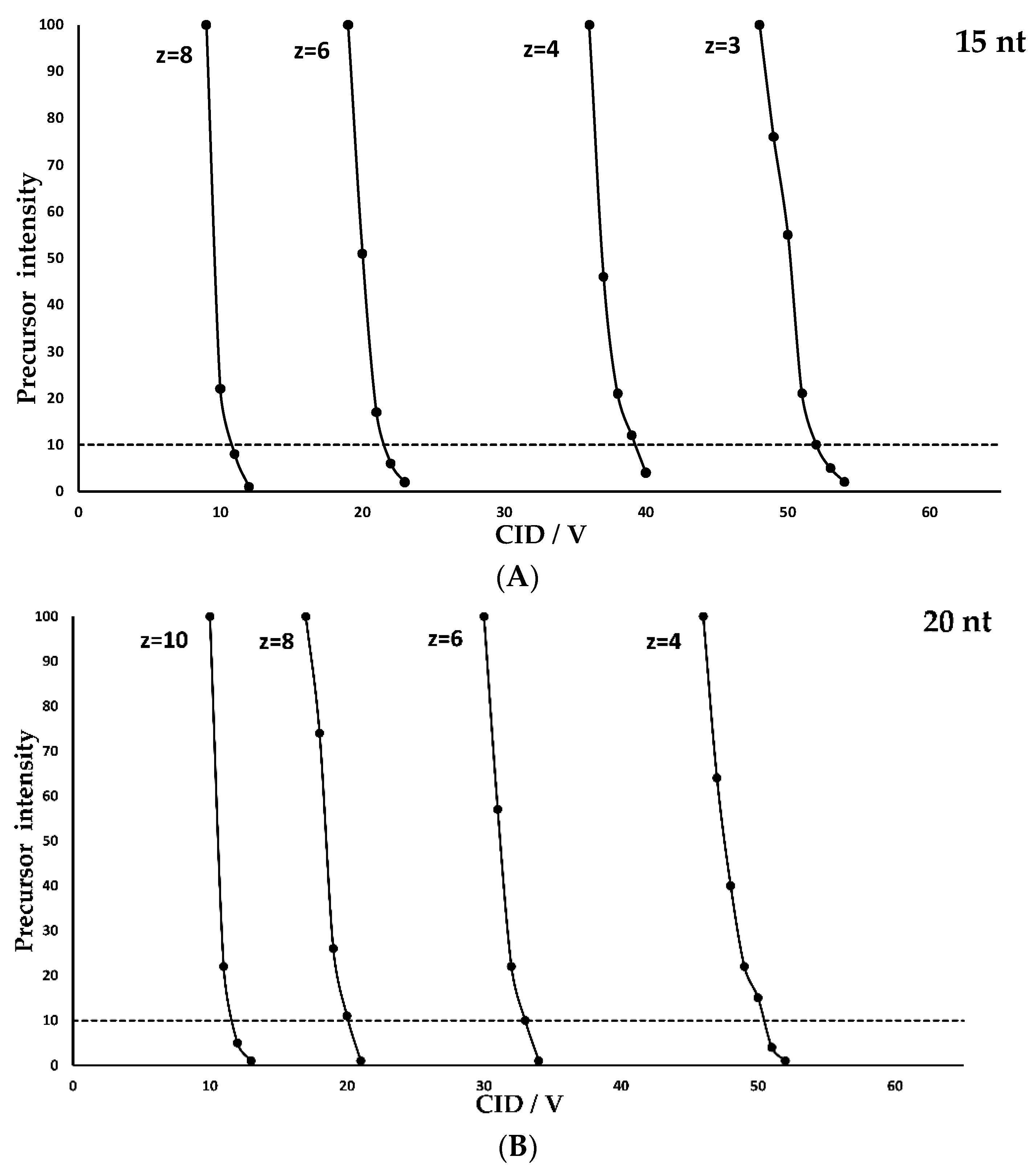
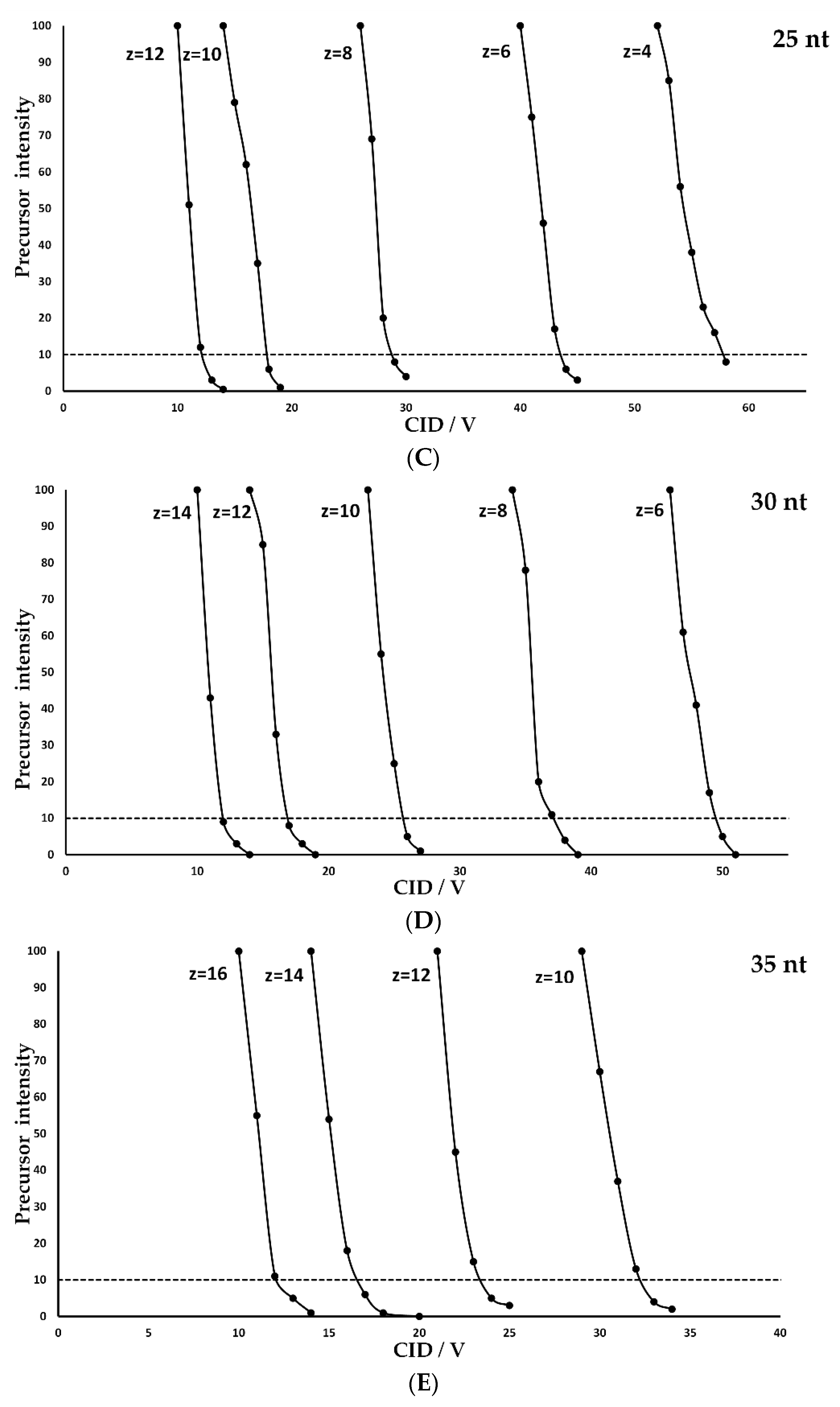
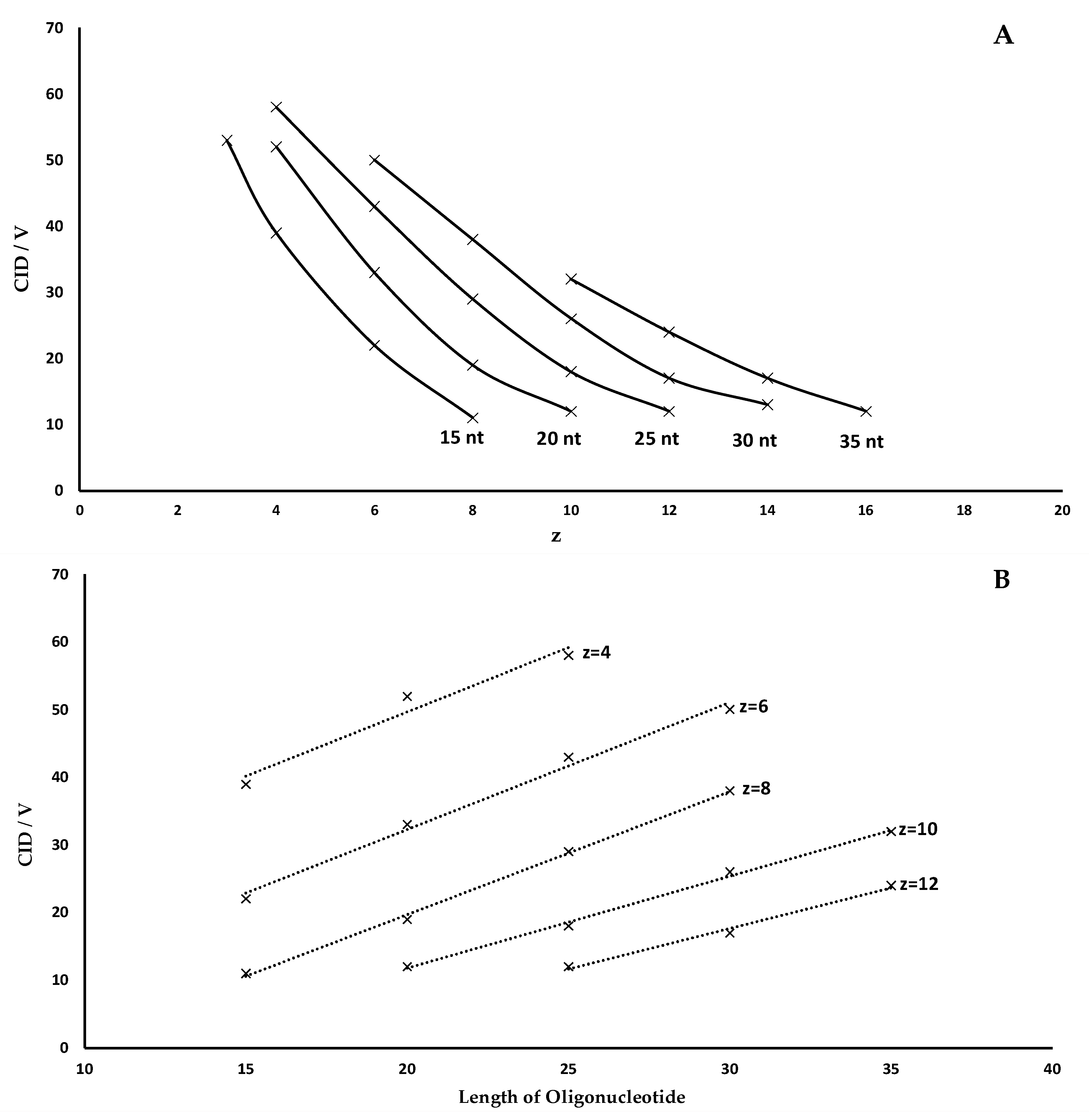
- The optimal CID voltage required for the fragmentation of an oligonucleotide carrying a specific charge state increases linearly with oligonucleotide length. This relationship is shown graphically in Figure 2 as described for peptides in various publications and for oligonucleotides by Ickert et al. [21] A more robust fragmentation is observed at lower charge states with higher individual fragmentation energy, while higher charge states need less energy for successful fragmentation but bear the risk of internal fragmentation, an undesired effect in sequence confirmation or multiple reaction monitoring (MRM) measurements. Additionally, high charge states have a smaller fragmentation energy window and are therefore more susceptible to variations in the fragmentation voltage, which is important for a successful method validation.
- (II)
- For a given oligonucleotide length, high charge states require lower collision energies for an optimal fragmentation while low charge states require higher collision energies.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Al Musaimi, O.; Al Shaer, D.; Albericio, F.; De la Torre, B.G. 2020 FDA TIDES (Peptides and Oligonucleotides) Harvest. Pharmaceuticals 2021, 14, 145. [Google Scholar] [CrossRef] [PubMed]
- Al Shaer, D.; Al Musaimi, O.; Albericio, F.; De la Torre, B.G. 2021 FDA TIDES (Peptides and Oligonucleotides) Harvest. Pharmaceuticals 2022, 15, 222. [Google Scholar] [CrossRef] [PubMed]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef] [PubMed]
- Rossi, J.J.; Rossi, D.J. SiRNA Drugs: Here to Stay. Mol. Ther. 2021, 29, 431–432. [Google Scholar] [CrossRef]
- Ni, S.; Zhuo, Z.; Pan, Y.; Yu, Y.; Li, F.; Liu, J.; Wang, L.; Wu, X.; Li, D.; Wan, Y.; et al. Recent Progress in Aptamer Discoveries and Modifications for Therapeutic Applications. ACS Appl. Mater. Interfaces 2021, 13, 9500–9519. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. MRNA Vaccines—A New Era in Vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Park, J.W.; Lagniton, P.N.P.; Liu, Y.; Xu, R.-H. MRNA Vaccines for COVID-19: What, Why and How. Int. J. Biol. Sci. 2021, 17, 1446–1460. [Google Scholar] [CrossRef]
- Gilar, M. Analysis and Purification of Synthetic Oligonucleotides by Reversed-Phase High-Performance Liquid Chromatography with Photodiode Array and Mass Spectrometry Detection. Anal. Biochem. 2001, 298, 196–206. [Google Scholar] [CrossRef] [Green Version]
- Cook, K.; Thayer, J. Advantages of Ion-Exchange Chromatography for Oligonucleotide Analysis. Bioanalysis 2011, 3, 1109–1120. [Google Scholar] [CrossRef]
- Shimoyama, A.; Fujisaka, A.; Obika, S. Evaluation of Size-Exclusion Chromatography for the Analysis of Phosphorothioate Oligonucleotides. J. Pharm. Biomed. Anal. 2017, 136, 55–65. [Google Scholar] [CrossRef]
- Sutton, J.M.; Guimaraes, G.J.; Annavarapu, V.; Van Dongen, W.D.; Bartlett, M.G. Current State of Oligonucleotide Characterization Using Liquid Chromatography–Mass Spectrometry: Insight into Critical Issues. J. Am. Soc. Mass Spectrom. 2020, 31, 1775–1782. [Google Scholar] [CrossRef] [PubMed]
- Pourshahian, S. Therapeutic Oligonucleotides, Impurities, Degradants, and Their Characterization by Mass Spectrometry. Mass Spectrom. Rev. 2021, 40, 75–109. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.J.; Li, W.; Dai, G. Application of LC–MS for Quantitative Analysis and Metabolite Identification of Therapeutic Oligonucleotides. J. Pharm. Biomed. Anal. 2007, 44, 330–341. [Google Scholar] [CrossRef]
- Mcluckey, S.A.; Berkel, G.J.V.; Glish, G.L. Tandem Mass Spectrometry of Small, Multiply Charged Oligonucleotides. J. Am. Soc. Mass Spectrom. 1992, 3, 60–70. [Google Scholar] [CrossRef] [Green Version]
- McLuckey, S.A.; Habibi-Goudarzi, S. Ion Trap Tandem Mass Spectrometry Applied to Small Multiply Charged Oligonucleotides with a Modified Base. J. Am. Soc. Mass Spectrom. 1994, 5, 740–747. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, A.M.; Sommers, C.; Hawes, J.; Rodriguez, J.D.; Yang, K. Tandem Mass Spectrometric Sequence Characterization of Synthetic Thymidine-Rich Oligonucleotides. J. Mass Spectrom. 2022, 57, e4819. [Google Scholar] [CrossRef]
- Goyon, A.; Scott, B.; Kurita, K.; Crittenden, C.M.; Shaw, D.; Lin, A.; Yehl, P.; Zhang, K. Full Sequencing of CRISPR/Cas9 Single Guide RNA (SgRNA) via Parallel Ribonuclease Digestions and Hydrophilic Interaction Liquid Chromatography–High-Resolution Mass Spectrometry Analysis. Anal. Chem. 2021, 93, 14792–14801. [Google Scholar] [CrossRef] [PubMed]
- Vanhinsbergh, C.J.; Criscuolo, A.; Sutton, J.N.; Murphy, K.; Williamson, A.J.K.; Cook, K.; Dickman, M.J. Characterization and Sequence Mapping of Large RNA and MRNA Therapeutics Using Mass Spectrometry. Anal. Chem. 2022, 94, 7339–7349. [Google Scholar] [CrossRef]
- Wu, J.; McLuckey, S.A. Gas-Phase Fragmentation of Oligonucleotide Ions. Int. J. Mass Spectrom. 2004, 237, 197–241. [Google Scholar] [CrossRef]
- Bahr, U.; Aygün, H.; Karas, M. Sequencing of Single and Double Stranded RNA Oligonucleotides by Acid Hydrolysis and MALDI Mass Spectrometry. Anal. Chem. 2009, 81, 3173–3179. [Google Scholar] [CrossRef]
- Ickert, S.; Schwaar, T.; Springer, A.; Grabarics, M.; Riedel, J.; Beck, S.; Pagel, K.; Linscheid, M.W. Comparison of the Fragmentation Behavior of DNA and LNA Single Strands and Duplexes. J. Mass Spectrom. 2019, 54, 402–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaucage, S.L.; Caruthers, M.H. Deoxynucleoside Phosphoramidites—A New Class of Key Intermediates for Deoxypolynucleotide Synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. [Google Scholar] [CrossRef]
| Oligonucleotide | Sequence | m/Da |
|---|---|---|
| 15 nt | TTT TTT TTT TTT TTT | 4578.70 |
| 20 nt | TTT TTT TTT TTT TTT TTT | 6098.93 |
| 25 nt | TTT TTT TTT TTT TTT TTT TTT | 7619.16 |
| 30 nt | TTT TTT TTT TTT TTT TTT TTT TTT | 9139.39 |
| 35 nt | TTT TTT TTT TTT TTT TTT TTT TTT TTT | 10,659.62 |
| 24 nt | T*C*G *T*C*G *T*T*T *T*G*T *C*G*T *T*T*T *G*T*C *G*T*T | 7696.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gawlig, C.; Rühl, M. Investigation of the Influence of Charge State and Collision Energy on Oligonucleotide Fragmentation by Tandem Mass Spectrometry. Molecules 2023, 28, 1169. https://doi.org/10.3390/molecules28031169
Gawlig C, Rühl M. Investigation of the Influence of Charge State and Collision Energy on Oligonucleotide Fragmentation by Tandem Mass Spectrometry. Molecules. 2023; 28(3):1169. https://doi.org/10.3390/molecules28031169
Chicago/Turabian StyleGawlig, Christopher, and Michael Rühl. 2023. "Investigation of the Influence of Charge State and Collision Energy on Oligonucleotide Fragmentation by Tandem Mass Spectrometry" Molecules 28, no. 3: 1169. https://doi.org/10.3390/molecules28031169
APA StyleGawlig, C., & Rühl, M. (2023). Investigation of the Influence of Charge State and Collision Energy on Oligonucleotide Fragmentation by Tandem Mass Spectrometry. Molecules, 28(3), 1169. https://doi.org/10.3390/molecules28031169





