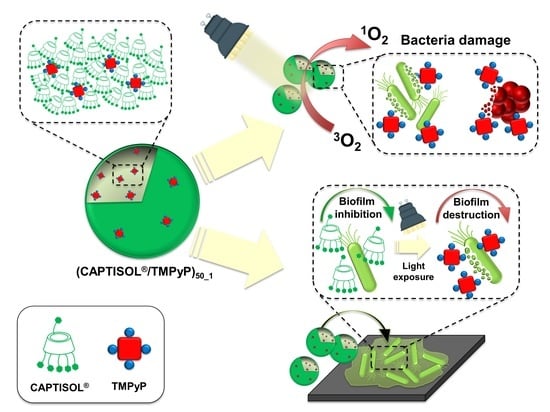
Antimicrobial and Antibiofilm Photodynamic Action of Photosensitizing Nanoassemblies Based on Sulfobutylether-β-Cyclodextrin
Abstract
:1. Introduction
2. Results and Discussion
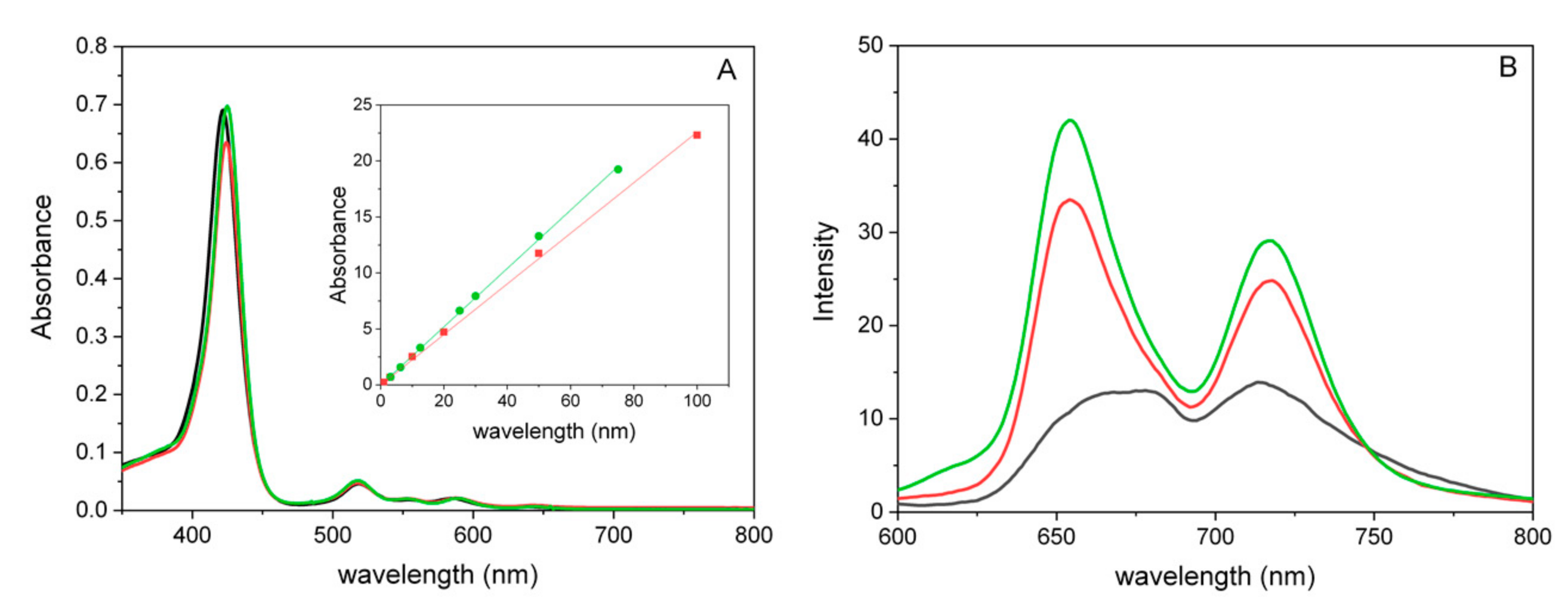
Properties and Spectroscopic Investigation of CAPTISOL®/TMPyP Nanoassemblies
3. Materials and Methods
3.1. Materials
3.2. Nanoassemblies Preparation
3.3. UV/Vis Absorption and Steady-State Fluorescence Spectroscopy
3.4. Emission Quantum Yield
3.5. Size and ζ Potential of Nanoassemblies
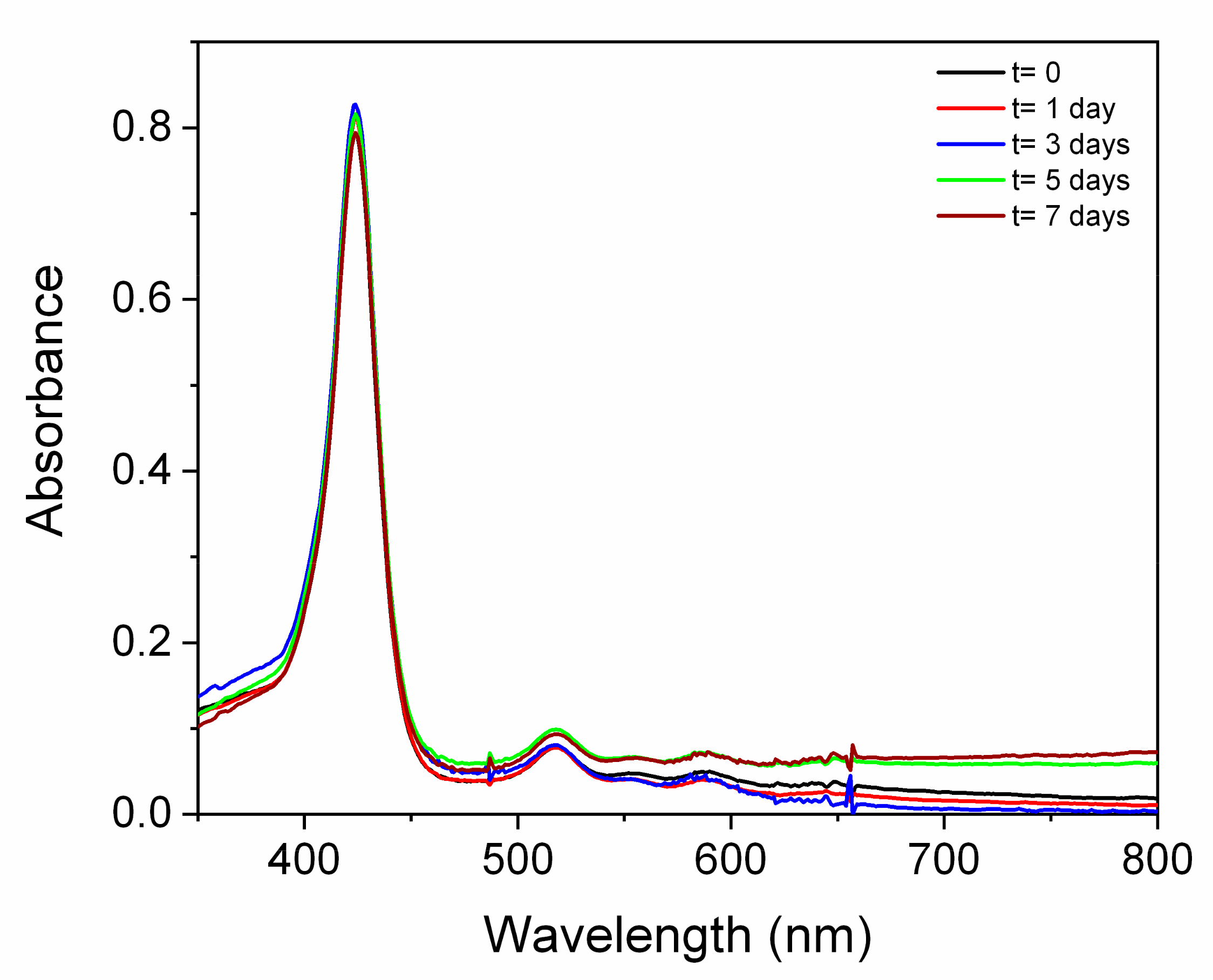
3.6. Stability Studies
3.7. Bacteria Strain, Media and Growth Conditions
3.8. Antibacterial Assay
3.9. Antibiofilm Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- 35 000 Annual Deaths from Antimicrobial Resistance in the EU/EEA. Available online: https://www.ecdc.europa.eu/en/news-events/eaad-2022-launch (accessed on 20 November 2022).
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Navidinia, M. The Clinical Importance of Emerging ESKAPE Pathogens in Nosocomial Infections. Arch. Adv. Biosci. 2016, 7, 43–57. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyaue, S.; Suzuki, E.; Komiyama, Y.; Kondo, Y.; Morikawa, M.; Maeda, S. Bacterial Memory of Persisters: Bacterial Persister Cells Can Retain Their Phenotype for Days or Weeks After Withdrawal From Colony–Biofilm Culture. Front. Microbiol. 2018, 9, 1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, T.K. Strategies for Combating Persister Cell and Biofilm Infections. Microb. Biotechnol. 2017, 10, 1054–1056. [Google Scholar] [CrossRef] [Green Version]
- Douafer, H.; Andrieu, V.; Phanstiel, O.I.; Brunel, J.M. Antibiotic Adjuvants: Make Antibiotics Great Again! J. Med. Chem. 2019, 62, 8665–8681. [Google Scholar] [CrossRef]
- Wright, G.D. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends Microbiol. 2016, 24, 862–871. [Google Scholar] [CrossRef]
- De Plano, L.M.; Fazio, E.; Rizzo, M.G.; Franco, D.; Carnazza, S.; Trusso, S.; Neri, F.; Guglielmino, S.P.P. Phage-Based Assay for Rapid Detection of Bacterial Pathogens in Blood by Raman Spectroscopy. J. Immunol. Methods 2019, 465, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Romero-Calle, D.; Guimarães Benevides, R.; Góes-Neto, A.; Billington, C. Bacteriophages as Alternatives to Antibiotics in Clinical Care. Antibiotics 2019, 8, 138. [Google Scholar] [CrossRef] [Green Version]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial Peptides and Their Therapeutic Potential for Bacterial Skin Infections and Wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Mumtaz, S.; Li, C.-H.; Hussain, I.; Rotello, V.M. Combatting Antibiotic-Resistant Bacteria Using Nanomaterials. Chem. Soc. Rev. 2019, 48, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.; Calabrese, G.; Petralia, S.; Neri, G.; Corsaro, C.; Forte, L.; Squarzoni, S.; Guglielmino, S.; Traina, F.; Fazio, E.; et al. Antimicrobial Effect and Cytotoxic Evaluation of Mg-Doped Hydroxyapatite Functionalized with Au-Nano Rods. Molecules 2021, 26, 1099. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Manuguri, S.; Proietti, G.; Romson, J.; Fu, Y.; Inge, A.K.; Wu, B.; Zhang, Y.; Häll, D.; Ramström, O.; et al. Design and Synthesis of Theranostic Antibiotic Nanodrugs That Display Enhanced Antibacterial Activity and Luminescence. Proc. Natl. Acad. Sci. USA 2017, 114, 8464–8469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torrisi, L.; Guglielmino, S.; Silipigni, L.; De Plano, L.M.; Kovacik, L.; Lavrentiev, V.; Torrisi, A.; Fazio, M.; Fazio, B.; Di Marco, G. Study of Gold Nanoparticle Transport by M13 Phages towards Disease Tissues as Targeting Procedure for Radiotherapy Applications. Gold Bull. 2019, 52, 135–144. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and Promise of Bacterial Quorum Sensing Research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saurav, K.; Costantino, V.; Venturi, V.; Steindler, L. Quorum Sensing Inhibitors from the Sea Discovered Using Bacterial N-Acyl-Homoserine Lactone-Based Biosensors. Mar. Drugs 2017, 15, 53. [Google Scholar] [CrossRef] [Green Version]
- Hawver, L.A.; Jung, S.A.; Ng, W.-L. Specificity and Complexity in Bacterial Quorum-Sensing Systems. FEMS Microbiol. Rev. 2016, 40, 738–752. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.; Zhang, X.-H. Quorum Quenching Agents: Resources for Antivirulence Therapy. Mar. Drugs 2014, 12, 3245–3282. [Google Scholar] [CrossRef] [Green Version]
- Cieplik, F.; Deng, D.; Crielaard, W.; Buchalla, W.; Hellwig, E.; Al-Ahmad, A.; Maisch, T. Antimicrobial Photodynamic Therapy—What We Know and What We Don’t. Crit. Rev. Microbiol. 2018, 44, 571–589. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Jiang, W.; Xu, Q.; Zheng, Y. Progress in Antibacterial Hydrogel Dressing. Gels 2022, 8, 503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; Zou, Q.; Xing, R.; Jiao, T.; Yan, X. An Injectable Dipeptide–Fullerene Supramolecular Hydrogel for Photodynamic Antibacterial Therapy. J. Mater. Chem. B 2018, 6, 7335–7342. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sampedro, A.; Tabero, A.; Mahamed, I.; Acedo, P. Multimodal Use of the Porphyrin TMPyP: From Cancer Therapy to Antimicrobial Applications. J. Porphyr. Phthalocyanines 2019, 23, 11–27. [Google Scholar] [CrossRef] [Green Version]
- Hanakova, A.; Bogdanova, K.; Tomankova, K.; Pizova, K.; Malohlava, J.; Binder, S.; Bajgar, R.; Langova, K.; Kolar, M.; Mosinger, J.; et al. The Application of Antimicrobial Photodynamic Therapy on S. Aureus and E. Coli Using Porphyrin Photosensitizers Bound to Cyclodextrin. Microbiol. Res. 2014, 169, 163–170. [Google Scholar] [CrossRef]
- Santoro, A.M.; Lo Giudice, M.C.; D’Urso, A.; Lauceri, R.; Purrello, R.; Milardi, D. Cationic Porphyrins Are Reversible Proteasome Inhibitors. J. Am. Chem. Soc. 2012, 134, 10451–10457. [Google Scholar] [CrossRef] [Green Version]
- Jurczak, A.; Szramka, B.; Grinholc, M.; Legendziewicz, J.; Bielawski, K.P. Photodynamic Effect of Lanthanide Derivatives of Meso-Tetra(N-Methyl-4-Pyridyl)Porphine against Staphylococcus Aureus. Acta Biochim. Pol. 2008, 55, 581–585. [Google Scholar] [CrossRef]
- Collins, T.L.; Markus, E.A.; Hassett, D.J.; Robinson, J.B. The Effect of a Cationic Porphyrin on Pseudomonas Aeruginosa Biofilms. Curr. Microbiol. 2010, 61, 411–416. [Google Scholar] [CrossRef]
- Cieplik, F.; Tabenski, L.; Buchalla, W.; Maisch, T. Antimicrobial Photodynamic Therapy for Inactivation of Biofilms Formed by Oral Key Pathogens. Front. Microbiol. 2014, 5, 405. [Google Scholar] [CrossRef] [Green Version]
- Ghorbani, J.; Rahban, D.; Aghamiri, S.; Teymouri, A.; Bahador, A. Photosensitizers in Antibacterial Photodynamic Therapy: An Overview. Laser. Ther. 2018, 27, 293–302. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Xu, F.-J.; Yu, B. Smart Polymeric Delivery System for Antitumor and Antimicrobial Photodynamic Therapy. Front. Bioeng. Biotechnol. 2021, 9, 783354. [Google Scholar] [CrossRef] [PubMed]
- Youf, R.; Nasir, A.; Müller, M.; Thétiot, F.; Haute, T.; Ghanem, R.; Jonas, U.; Schönherr, H.; Lemercier, G.; Montier, T.; et al. Ruthenium(II) Polypyridyl Complexes for Antimicrobial Photodynamic Therapy: Prospects for Application in Cystic Fibrosis Lung Airways. Pharmaceutics 2022, 14, 1664. [Google Scholar] [CrossRef] [PubMed]
- Păduraru, D.N.; Niculescu, A.-G.; Bolocan, A.; Andronic, O.; Grumezescu, A.M.; Bîrlă, R. An Updated Overview of Cyclodextrin-Based Drug Delivery Systems for Cancer Therapy. Pharmaceutics 2022, 14, 1748. [Google Scholar] [CrossRef] [PubMed]
- González-Delgado, J.A.; Castro, P.M.; Machado, A.; Araújo, F.; Rodrigues, F.; Korsak, B.; Ferreira, M.; Tomé, J.P.C.; Sarmento, B. Hydrogels Containing Porphyrin-Loaded Nanoparticles for Topical Photodynamic Applications. Int. J. Pharm. 2016, 510, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Crini, G. Review: A History of Cyclodextrins. Chem. Rev. 2014, 114, 10940–10975. [Google Scholar] [CrossRef] [PubMed]
- Do, T.T.; Van Hooghten, R.; Van den Mooter, G. A Study of the Aggregation of Cyclodextrins: Determination of the Critical Aggregation Concentration, Size of Aggregates and Thermodynamics Using Isodesmic and K2–K Models. Int. J. Pharm. 2017, 521, 318–326. [Google Scholar] [CrossRef]
- Zagami, R.; Rapozzi, V.; Piperno, A.; Scala, A.; Triolo, C.; Trapani, M.; Xodo, L.E.; Monsù Scolaro, L.; Mazzaglia, A. Folate-Decorated Amphiphilic Cyclodextrins as Cell-Targeted Nanophototherapeutics. Biomacromolecules 2019, 20, 2530–2544. [Google Scholar] [CrossRef]
- Zagami, R.; Sortino, G.; Caruso, E.; Malacarne, M.C.; Banfi, S.; Patanè, S.; Monsù Scolaro, L.; Mazzaglia, A. Tailored-BODIPY/Amphiphilic Cyclodextrin Nanoassemblies with PDT Effectiveness. Langmuir 2018, 34, 8639–8651. [Google Scholar] [CrossRef]
- Conte, C.; Scala, A.; Siracusano, G.; Leone, N.; Patanè, S.; Ungaro, F.; Miro, A.; Sciortino, M.T.; Quaglia, F.; Mazzaglia, A. Nanoassembly of an Amphiphilic Cyclodextrin and Zn(II)-Phthalocyanine with the Potential for Photodynamic Therapy of Cancer. RSC Adv. 2014, 4, 43903–43911. [Google Scholar] [CrossRef]
- Zagami, R.; Franco, D.; Pipkin, J.D.; Antle, V.; De Plano, L.; Patanè, S.; Guglielmino, S.; Monsù Scolaro, L.; Mazzaglia, A. Sulfobutylether-β-Cyclodextrin/5,10,15,20-Tetrakis(1-Methylpyridinium-4-Yl)Porphine Nanoassemblies with Sustained Antimicrobial Phototherapeutic Action. Int. J. Pharm. 2020, 585, 119487. [Google Scholar] [CrossRef]
- Brancaccio, D.; Pizzo, E.; Cafaro, V.; Notomista, E.; De Lise, F.; Bosso, A.; Gaglione, R.; Merlino, F.; Novellino, E.; Ungaro, F.; et al. Antimicrobial Peptide Temporin-L Complexed with Anionic Cyclodextrins Results in a Potent and Safe Agent against Sessile Bacteria. Int. J. Pharm. 2020, 584, 119437. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Cui, H.; Shi, R.; Guo, J.; Wang, B.; Xu, Y.; Ding, Y.; Mao, H.; Yan, F. Antimicrobial Anionic Polymers: The Effect of Cations. Eur. Polym. J. 2018, 107, 181–188. [Google Scholar] [CrossRef]
- Mohanty, J.; Bhasikuttan, A.C.; Choudhury, S.D.; Pal, H. Noncovalent Interaction of 5,10,15,20-Tetrakis(4-N-Methylpyridyl)Porphyrin with Cucurbit[7]Uril: A Supramolecular Architecture. J. Phys. Chem. B 2008, 112, 10782–10785. [Google Scholar] [CrossRef] [PubMed]
- Vergeldt, F.J.; Koehorst, R.B.M.; van Hoek, A.; Schaafsma, T.J. Intramolecular Interactions in the Ground and Excited States of Tetrakis(N-Methylpyridyl)Porphyrins. J. Phys. Chem. 1995, 99, 4397–4405. [Google Scholar] [CrossRef]
- Pasternack, R.F.; Huber, P.R.; Boyd, P.; Engasser, G.; Francesconi, L.; Gibbs, E.; Fasella, P.; Venturo, G.C.; Hinds, L.d.C. On the Aggregation of Meso-Substituted Water-Soluble Porphyrins. J. Am. Chem. Soc. 1972, 94, 4511–4517. [Google Scholar] [CrossRef]
- Davis, J.L. Chapter 2—Pharmacologic Principles. In Equine Internal Medicine, 4th ed.; Reed, S.M., Bayly, W.M., Sellon, D.C., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2018; pp. 79–137. ISBN 978-0-323-44329-6. [Google Scholar]
- Le Guern, F.; Sol, V.; Ouk, C.; Arnoux, P.; Frochot, C.; Ouk, T.-S. Enhanced Photobactericidal and Targeting Properties of a Cationic Porphyrin Following the Attachment of Polymyxin B. Bioconjug. Chem. 2017, 28, 2493–2506. [Google Scholar] [CrossRef]
- Pierson, L.S.; Pierson, E.A. Metabolism and Function of Phenazines in Bacteria: Impacts on the Behavior of Bacteria in the Environment and Biotechnological Processes. Appl. Microbiol. Biotechnol. 2010, 86, 1659–1670. [Google Scholar] [CrossRef] [Green Version]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Tasca Ribeiro, V.S. Pathogenesis of the Pseudomonas Aeruginosa Biofilm: A Review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef]
- Rodriguez-Urretavizcaya, B.; Pascual, N.; Pastells, C.; Martin-Gomez, M.T.; Vilaplana, L.; Marco, M.-P. Diagnosis and Stratification of Pseudomonas Aeruginosa Infected Patients by Immunochemical Quantitative Determination of Pyocyanin From Clinical Bacterial Isolates. Front. Cell Infect. Microbiol. 2021, 11, 786929. [Google Scholar] [CrossRef]
- Costa, K.C.; Glasser, N.R.; Conway, S.J.; Newman, D.K. Pyocyanin Degradation by a Tautomerizing Demethylase Inhibits Pseudomonas Aeruginosa Biofilms. Science 2017, 355, 170–173. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.M.; Wang, D.; Pierson, L.S.; Pierson, E.A. Effect of Producing Different Phenazines on Bacterial Fitness and Biological Control in Pseudomonas Chlororaphis 30–84. Plant Pathol. J. 2018, 34, 44–58. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory Mechanisms and Promising Applications of Quorum Sensing-Inhibiting Agents in Control of Bacterial Biofilm Formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Inoue, Y.; Suehiro, A.; Ikeshoji, H.; Ishida, T.; Takiguchi, N.; Kuroda, A.; Kato, J.; Ohtake, H. The Effects of Cyclodextrins on Autoinducer Activities of Quorum Sensing in Pseudomonas Aeruginosa. J. Incl. Phenom. 2002, 44, 381–382. [Google Scholar] [CrossRef]
- Kruk, N.N. Fluorescent Properties and Symmetry of the Monodeprotonated Form of 5,10,15,20-Tetrakis-(4-N-Methylpyridyl)-Porphyrin. J. Appl. Spectrosc. 2006, 73, 686–693. [Google Scholar] [CrossRef]
- Coffey, B.M.; Anderson, G.G. Biofilm Formation in the 96-Well Microtiter Plate. In Pseudomonas Methods and Protocols; Filloux, A., Ramos, J.-L., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2014; pp. 631–641. ISBN 978-1-4939-0473-0. [Google Scholar]

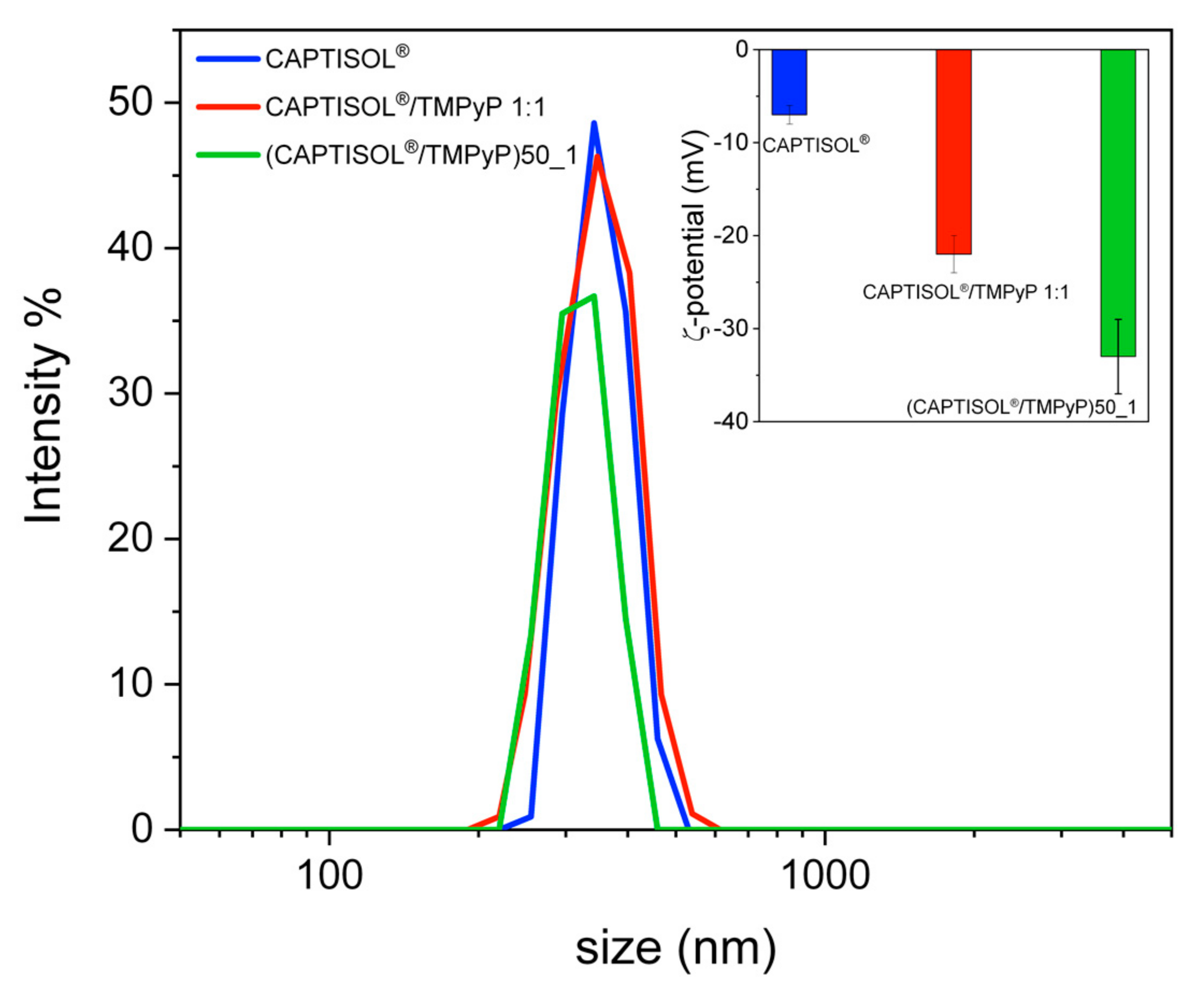

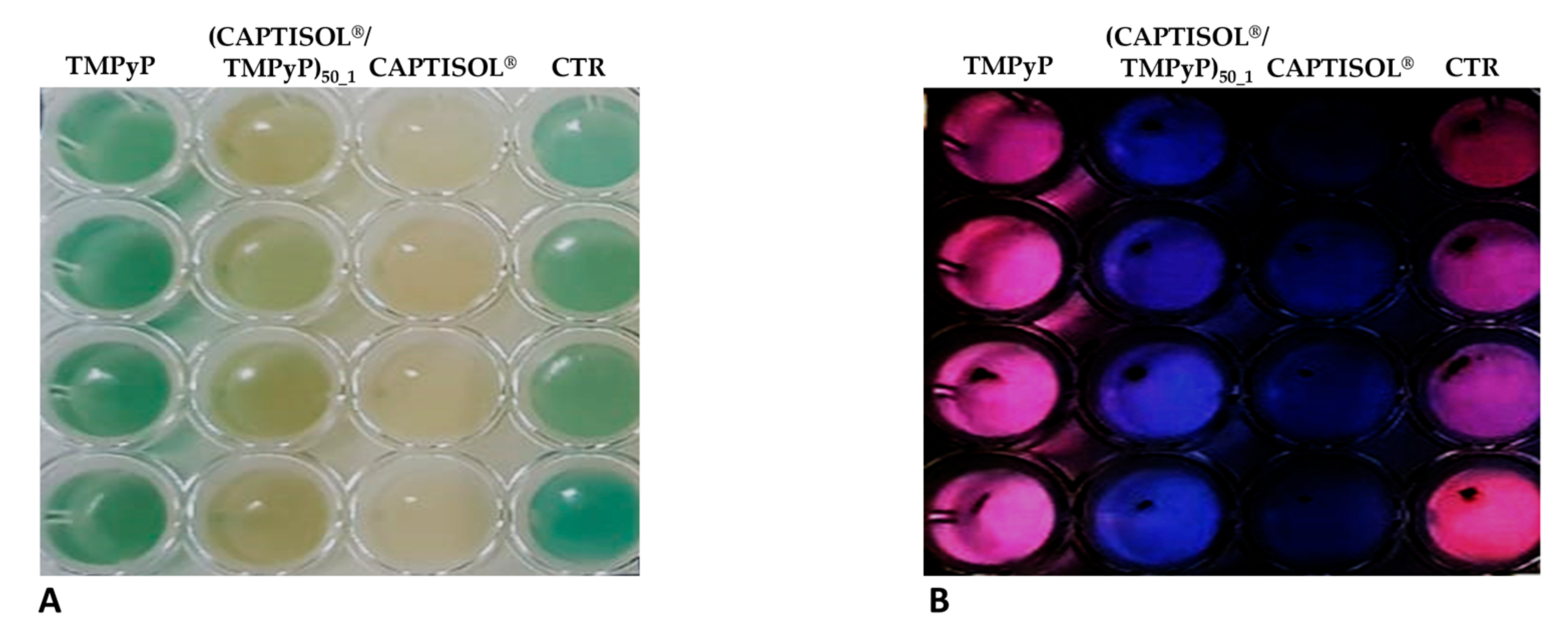
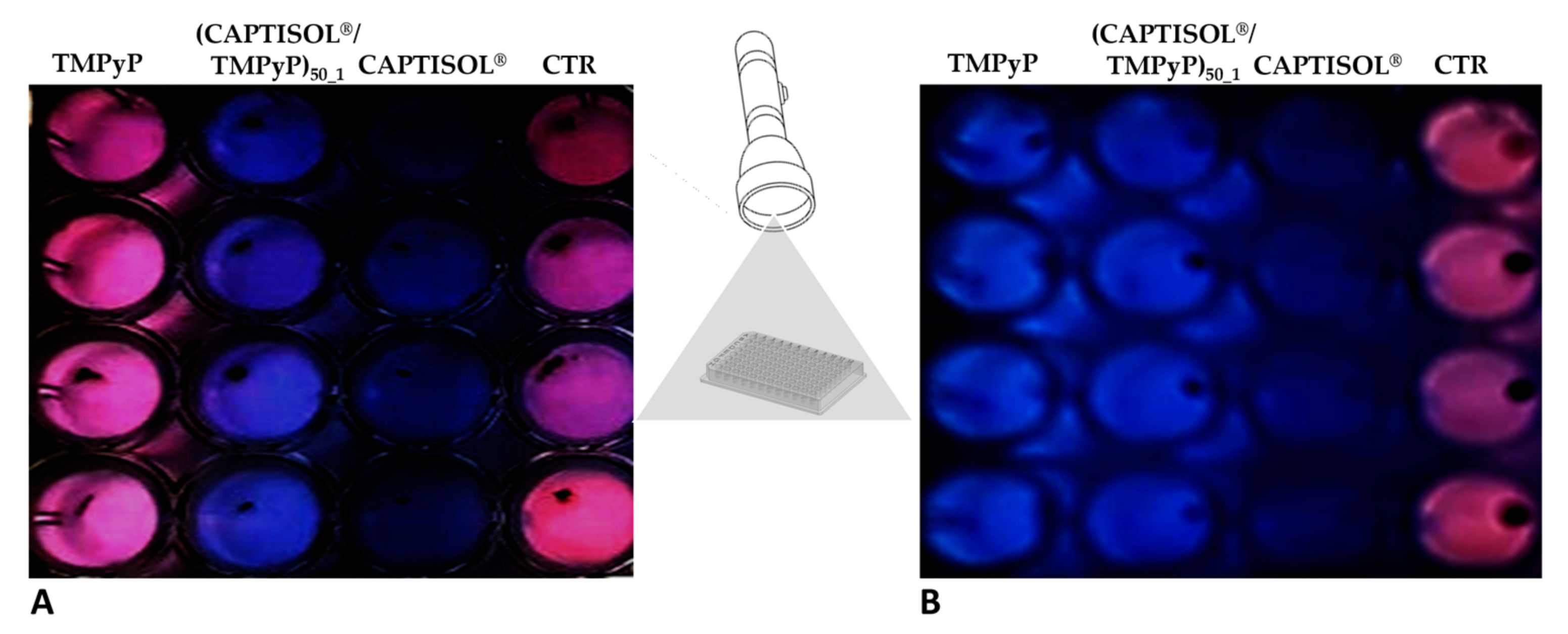
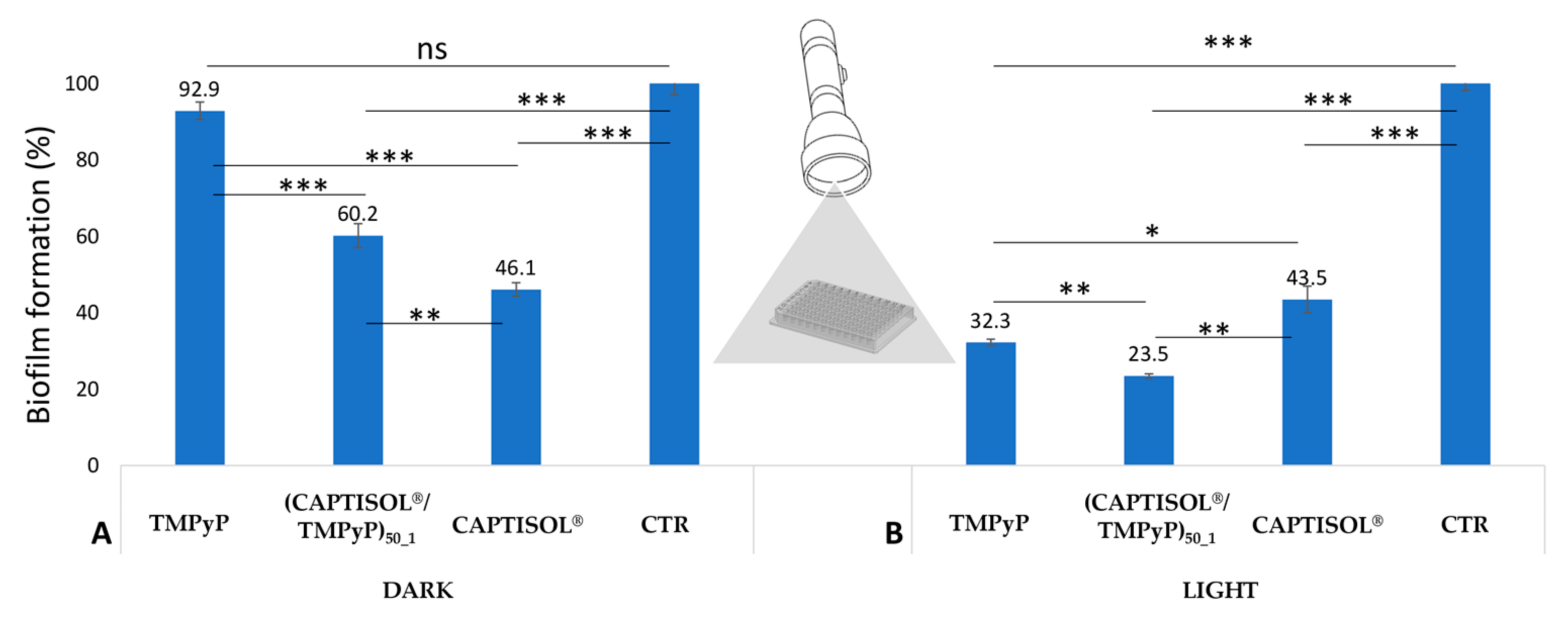
| a System | Mean DH (nm ± SD) | PDI | ζ (mV ± SD) |
|---|---|---|---|
| CAPTISOL® | 362 ± 60 | 0.3 | −7 ± 1 |
| a (CAPTISOL®/TMPyP)50_1 | 322 ± 42 | 0.3 | −33 ± 4 |
| b CAPTISOL®/TMPyP | 360 ± 40 | 0.2 | −22 ± 2 |
| a System | S. aureus | P. aeruginosa | ||||||
|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | |||||
| Light | Dark | Light | Dark | Light | Dark | Light | Dark | |
| TMPyP | 1.5 | - | 3 | - | 3 | - | 6 | - |
| (CAPTISOL®/TMPyP)50_1 | 3 | - | 6 | - | 6 | - | 12 | - |
| CAPTISOL® | - | - | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco, D.; Zagami, R.; De Plano, L.M.; Burduja, N.; Guglielmino, S.P.P.; Scolaro, L.M.; Mazzaglia, A. Antimicrobial and Antibiofilm Photodynamic Action of Photosensitizing Nanoassemblies Based on Sulfobutylether-β-Cyclodextrin. Molecules 2023, 28, 2493. https://doi.org/10.3390/molecules28062493
Franco D, Zagami R, De Plano LM, Burduja N, Guglielmino SPP, Scolaro LM, Mazzaglia A. Antimicrobial and Antibiofilm Photodynamic Action of Photosensitizing Nanoassemblies Based on Sulfobutylether-β-Cyclodextrin. Molecules. 2023; 28(6):2493. https://doi.org/10.3390/molecules28062493
Chicago/Turabian StyleFranco, Domenico, Roberto Zagami, Laura Maria De Plano, Nina Burduja, Salvatore Pietro Paolo Guglielmino, Luigi Monsù Scolaro, and Antonino Mazzaglia. 2023. "Antimicrobial and Antibiofilm Photodynamic Action of Photosensitizing Nanoassemblies Based on Sulfobutylether-β-Cyclodextrin" Molecules 28, no. 6: 2493. https://doi.org/10.3390/molecules28062493
APA StyleFranco, D., Zagami, R., De Plano, L. M., Burduja, N., Guglielmino, S. P. P., Scolaro, L. M., & Mazzaglia, A. (2023). Antimicrobial and Antibiofilm Photodynamic Action of Photosensitizing Nanoassemblies Based on Sulfobutylether-β-Cyclodextrin. Molecules, 28(6), 2493. https://doi.org/10.3390/molecules28062493