Enantioselective Labeling of Zebrafish for D-Phenylalanine Based on Graphene-Based Nanoplatform
Abstract
1. Introduction
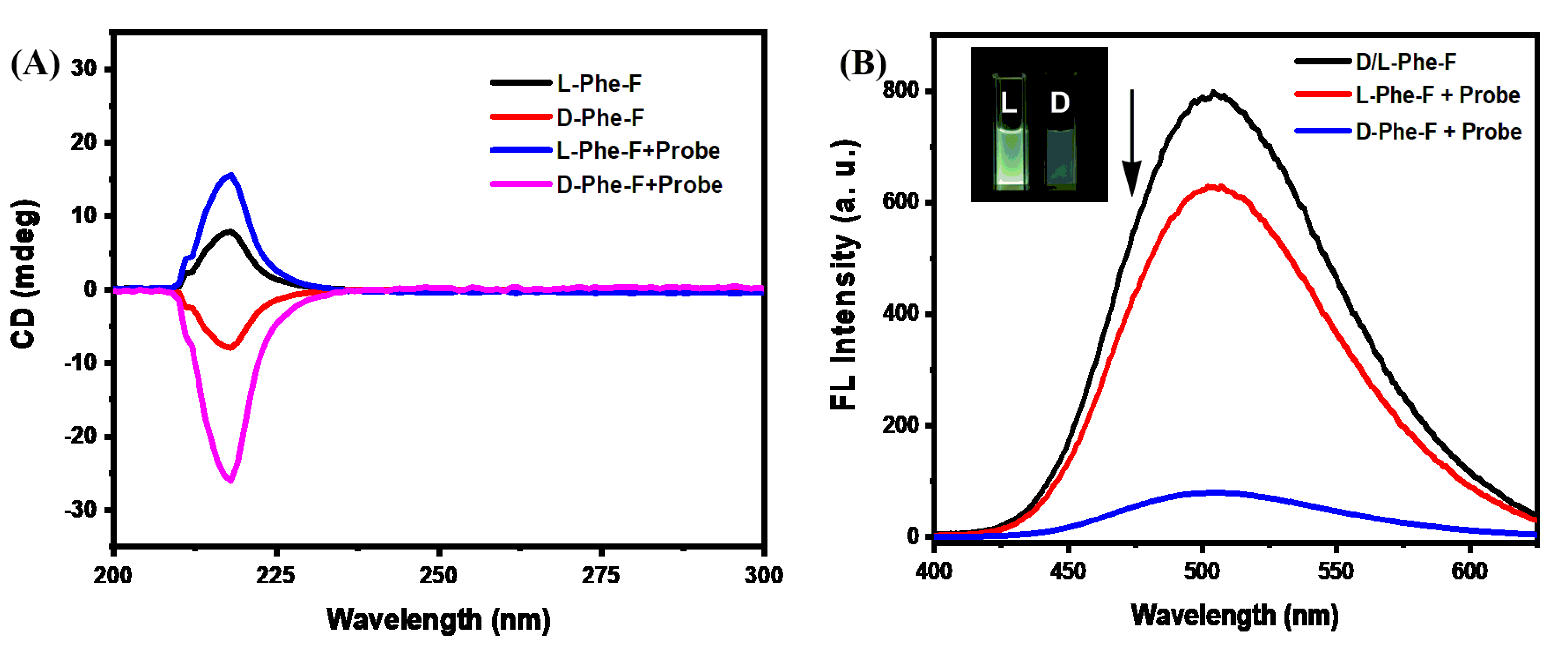
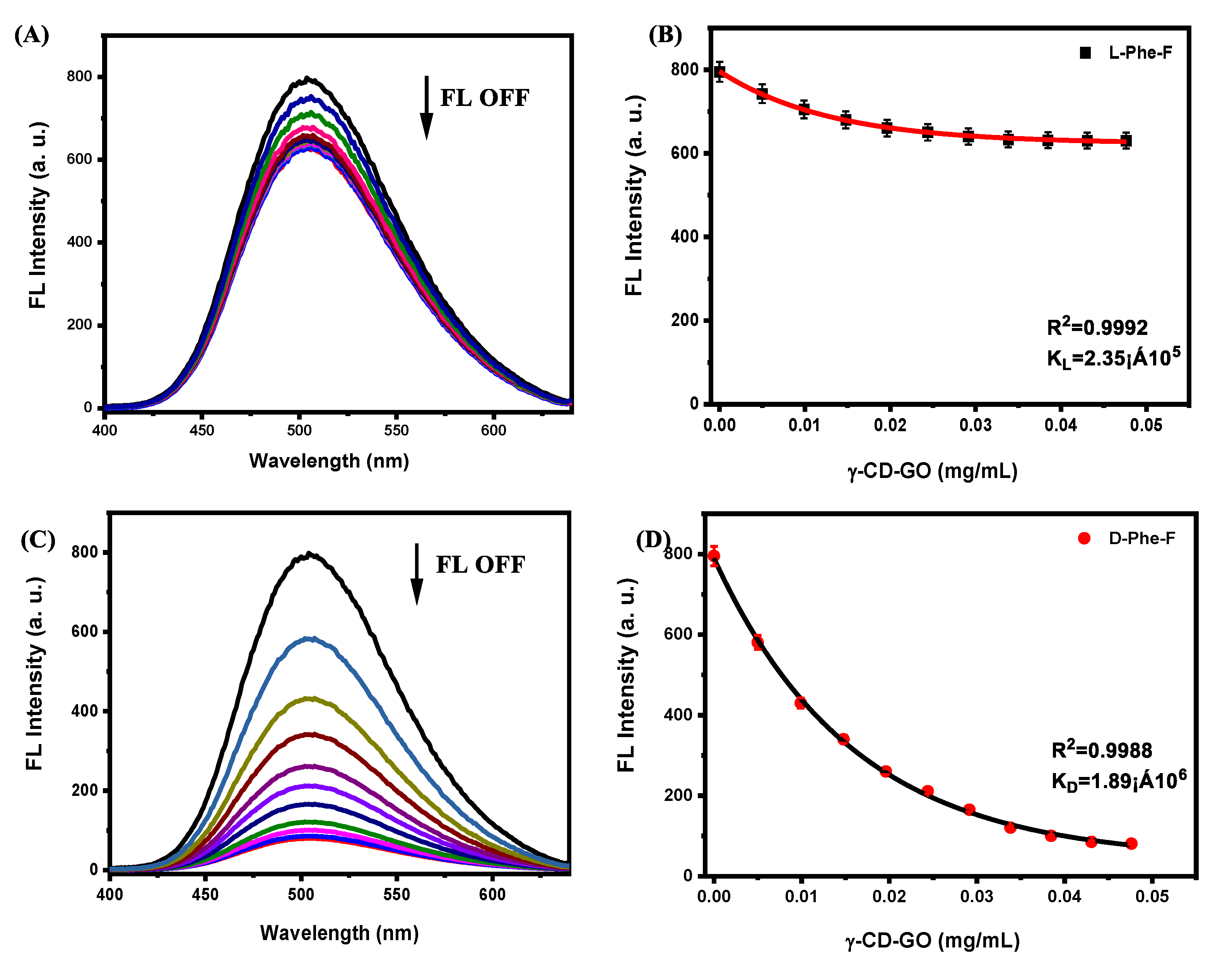
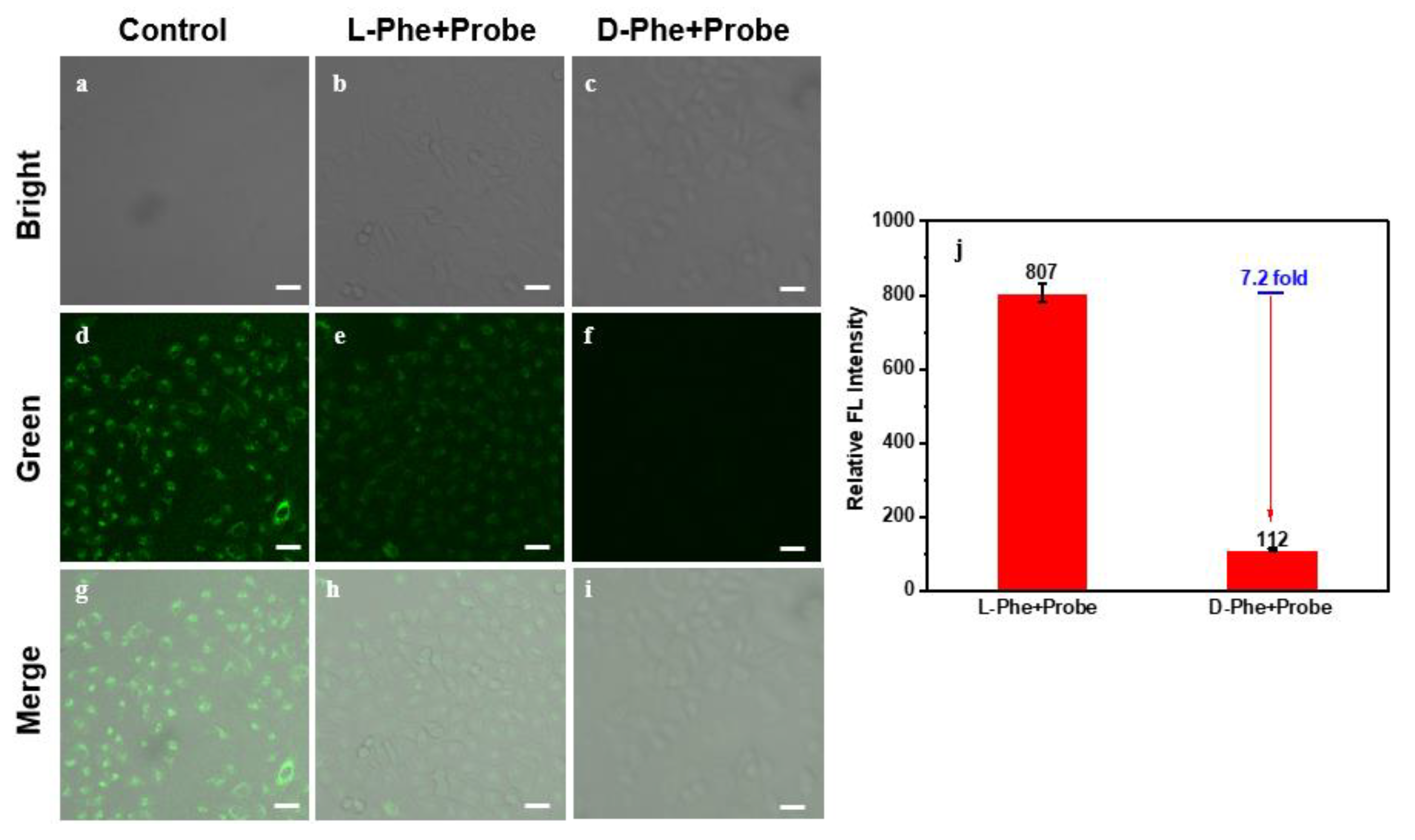
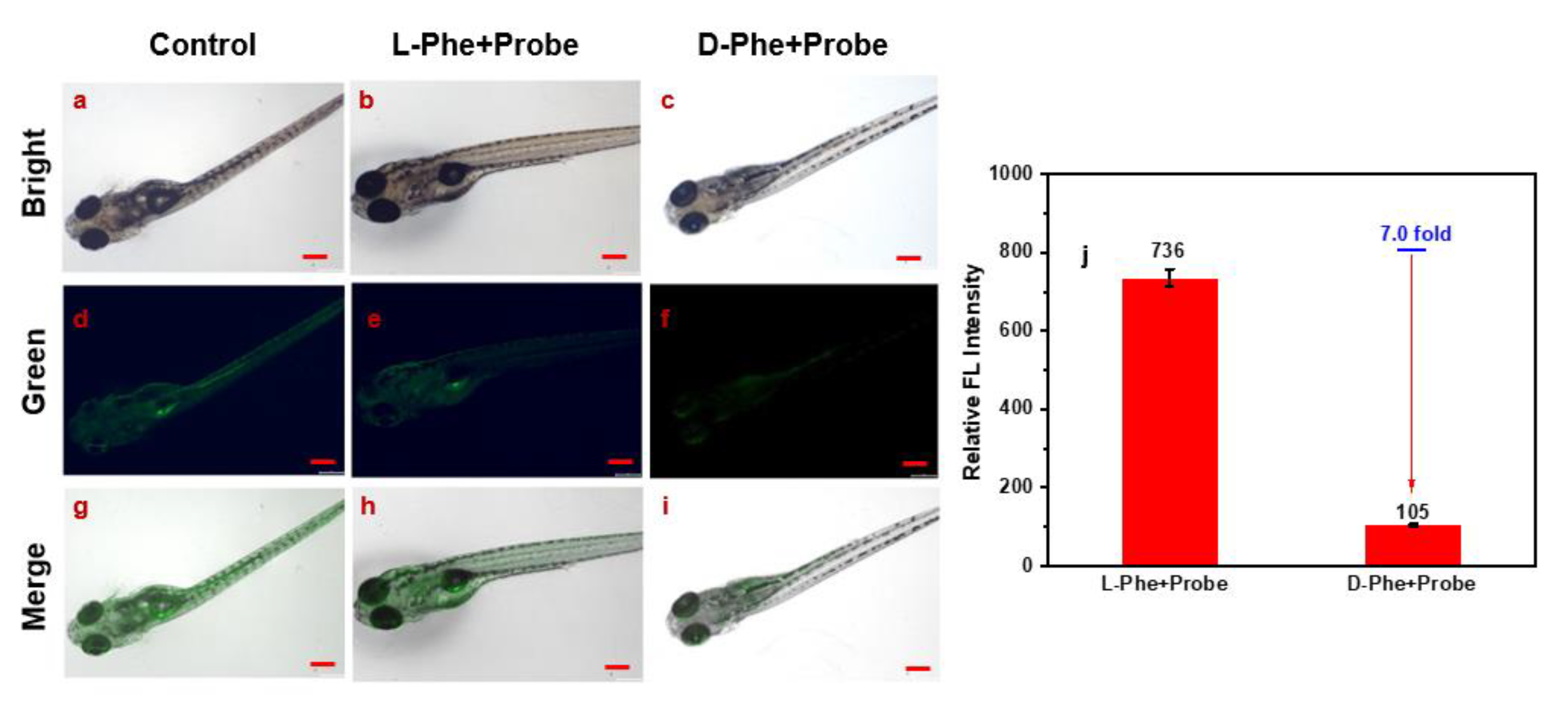
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Apparatus
3.3. Preparation of γ-CD-GO
3.4. Preparation of F-D/L-Phe
3.5. Cell Culture and Fluorescence Microscopy
3.6. Chiral Imaging in Zebrafish
3.7. Data Analysis and Fitting
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, Y.Y.; Luo, H.; Ge, Q.M.; Liu, M.; Tao, Z.; Cong, H. An ultrasensitive photoelectrochemical sensor with layer-by-layer assembly of chiral multifarene [3,2,1] and g-C3N4 quantum dots for enantiorecognition towards thyroxine. Sens. Actuat. B-Chem. 2021, 336, 129750. [Google Scholar] [CrossRef]
- Aboul-Enein, H.Y.; Bounoua, N.; Rebizi, M.; Wagdy, H. Application of nanoparticles in chiral analysis and chiral separation. Chirality 2021, 33, 196–208. [Google Scholar] [CrossRef]
- Nakano, T.; Okamoto, Y. Synthetic Helical Polymers: Conformation and function. Chem. Rev. 2001, 101, 4013–4038. [Google Scholar] [CrossRef]
- Schneider, J.H. Binding mechanisms in supramolecular complexes. Angew. Chem. Int. Ed. 2009, 48, 3924–3977. [Google Scholar] [CrossRef]
- Will, J.M.; Behrens, A.; Macke, M.; Quarles, C.D.; Karst, U. Automated chiral analysis of amino acids based on chiral derivatization and trapped ion mobility-mass spectrometry. Anal. Chem. 2021, 23, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Sang, M.X.; Wang, M.L.; Han, L.B.; Gong, Z.L.; Tang, X.H.; Long, X.Z.; Xiong, H.; Peng, H.L. Rapid detection of d-limonene emanating from citrus infestation by Bactrocera dorsalis (Hendel) using a developed gas-sensing system based on QCM sensors coated with ethyl cellulose. Sens. Actuat. B-Chem. 2021, 328, 129048. [Google Scholar] [CrossRef]
- Kong, H.J.; Sun, X.P.; Yang, L.; Liu, X.L.; Yang, H.F.; Jin, R.H. Chirality detection by raman spectroscopy: The case of enantioselective interactions between amino acids and polymer-modified chiral silica. Anal. Chem. 2020, 92, 14292–14296. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Wang, W.Q.; Zhang, F.; Dhinakaran, M.K.; Wang, Y.Q.; Wang, R.; Cheng, J.; Toimil-Molares, M.E.; Trautmannbc, C.; Li, H. Selective transmembrane transport of Aβ protein regulated by tryptophan enantiomers. Chem. Commn. 2021, 57, 215–218. [Google Scholar] [CrossRef]
- Shen, K.; Wang, L.; He, Q.; Jin, Z.; Chen, W.Y.; Sun, C.R.; Pan, Y.J. Sensitive bromine-labeled probe D-BPBr for simultaneous identification and quantification of chiral amino acids and amino-containing metabolites profiling in human biofluid by HPLC/MS. Anal. Chem. 2020, 92, 1763–1769. [Google Scholar] [CrossRef]
- Lu, H.J.; Wang, Z.M.; Fan, X.Y.; Wang, H.Y.; Zhang, Q.; Fu, M.X.; Ning, G.Y.; Zhang, Y.F.; Wang, H. Electrochemical chiral amino acid biosensor based on dopamine-localized gold nanoparticles@left-handed spiral chiral carbon nanotubes. Anal. Methods 2020, 12, 3901–3908. [Google Scholar] [CrossRef] [PubMed]
- Barron, L.D. Chirality and Life. Space Sci. Rev. 2008, 135, 187–201. [Google Scholar]
- Zhang, J.; Wang, Z.J.; Lv, S.X.; Zeng, X.F.; Sun, Y.; Li, H.B.; Zhang, R.P. The chiral interfaces fabricated by D/L-alaninepillar [5] arenes for selectively adsorbing ctDNA. Chem. Commn. 2019, 55, 778. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Yang, Y.X.; Gu, Y.L.; Zhang, J.; Cheng, J.; Wang, J.H.; Sun, K.P.; Li, H.B. Chiral galactose responsive S-phenethylamine calix [4] arene-based sensing surface. Sens. Actuat. B-Chem. 2019, 297, 126662. [Google Scholar] [CrossRef]
- Liu, J.Y.; Fu, B.H.; Zhang, Z.H. Ionic current rectification triggered photoelectrochemical chiral sensing platform for recognition of amino acid enantiomers on self-standing nanochannel arrays. Anal. Chem. 2020, 92, 8670–8674. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.J.; Cao, Z.Z.; Hao, J.J.; Zhou, J.H.; Yang, Z.J.; Yang, Y.Z.; Wei, J.J. Controlled synthesis of Au chiral propellers from seeded growth of Au nanoplates for chiral differentiation of biomolecules. J. Phys. Chem. C 2020, 124, 24306–24314. [Google Scholar] [CrossRef]
- Zaidi, S.A. Facile and efficient electrochemical enantiomer recognition of phenylalanine using β-Cyclodextrin immobilized on reduced graphene oxide. Biosens. Bioelectron. 2017, 94, 714–718. [Google Scholar] [CrossRef]
- Niu, X.H.; Mo, Z.l.; Yang, X.; Shuai, C.; Liu, N.J.; Guo, R.B. Graphene-ferrocene functionalized cyclodextrin composite with high electrochemical recognition capability for phenylalanine enantiomers. Bioelectrochemistry 2019, 128, 74–82. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Zhang, S.Y.; Yan, H.W.; Quan, J.X.; Yang, L.; Chen, X.; Toimil-Molares, M.E.; Trautmann, C.; Li, H.B. Efficient chiral nanosenor based on Tip-modified nanochannels. Anal. Chem. 2021, 93, 6145–6150. [Google Scholar] [CrossRef]
- Kuang, H.; Xu, C.L.; Tang, Z.Y. Emerging chiral materials. Adv. Mat. 2020, 32, e2005110. [Google Scholar] [CrossRef] [PubMed]
- Zor, E.; Bingol, H.; Ersoz, M. Chiral sensors. Trac-Trend Anal. Chem. 2019, 121, 115662. [Google Scholar] [CrossRef]
- Castro-Neto, A.H.; Guinea, F.; Peres, N.M.R.; Novoselov, K.S. Lifshitz transition including many-body effects in bi-layer graphene and change in stacking order. Rev. Mod. Phys. 2009, 81, 109–162. [Google Scholar]
- Chhowalla, M.; Shin, H.S.; Eda, G.; Li, L.J.; Loh, K.P.; Zhang, H. The chemistry of two-dimensional layered transition metal dichalcogenide nanosheets. Nat. Chem. 2013, 5, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Geim, A.K. Graphene: Status and prospects. Science 2009, 324, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Brakat, A.; Zhu, H.W. Nanocellulose-Graphene Hybrids: Advanced functional materials as multifunctional sensing platform. Nanomicro Lett. 2021, 13, 94. [Google Scholar] [CrossRef]
- Sainz-Urruela, C.; Vera-Lopez, S.; San-Andres, M.P.; Diez-Pascual, A.M. Graphene-based sensors for the detection of bioactive compounds: A review. Int. J. Mol. Sci. 2021, 22, 3316. [Google Scholar] [CrossRef]
- Zhu, Y.W.; Murali, S.; Cai, W.W.; Li, X.S.; Suk, J.W.; Potts, J.R.; Ruoff, R.S. Population pharmacokinetics of ustekinumab in patients with active psoriatic arthritis. Adv. Mater. 2010, 22, 3906–3924. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.L.; Cao, X.H.; Wu, X.J.; He, Q.Y.; Yang, J.; Zhang, X.; Chen, J.Z.; Zhao, W.; Han, S.K.; Nam, G.H.; et al. Recent advances in ultrathin two-dimensional nanomaterials. Chem. Rev. 2017, 117, 6225–6331. [Google Scholar] [CrossRef]
- Wei, W.L.; Qu, K.G.; Ren, J.S.; Qu, X.G. Chiral detection using reusable fluorescent amylose-functionalized graphene. Chem. Sci. 2011, 2, 2050. [Google Scholar] [CrossRef]
- Mao, X.W.; Li, H.B. Chiral imaging in living cells with functionalized graphene oxide. J. Mater. Chem. B 2013, 1, 4267–4272. [Google Scholar] [CrossRef]
- Mohamadhoseini, M.; Mohamadnia, Z. Supramolecular self-healing materials via host-guest strategy between cyclodextrin and specific types of guest molecules. Coord. Chem. Rev. 2021, 432, 213711. [Google Scholar] [CrossRef]
- Xu, W.J.; Li, X.M.; Wang, L.; Li, S.Y.; Chu, S.N.; Wang, J.C.; Li, Y.J.; Hou, J.X.; Luo, Q.A.; Liu, J.Q. Design of cyclodextrin-based functional systems for biomedical applications. Front. Chem. 2021, 9, 635507. [Google Scholar] [CrossRef] [PubMed]
- Harada, A.; Takashima, Y.; Nakahata, M. Supramolecular polymeric materials via cyclodextrin-guest interactions. Accounts Chem. Res. 2014, 47, 2128–2140. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.D.; Tang, G.P.; Chu, P.K. Cyclodextrin-based host-guest supramolecular nanoparticles for delivery: From design to applications. Acc. Chem. Res. 2014, 47, 2017–2025. [Google Scholar] [CrossRef] [PubMed]
- Skidmore, M.W. Handbook of Derivatives for Chromatography; Blau, K., Halket, J.M., Eds.; Wiley-VCH: Weinheim, Germany, 1993; Volume 3, pp. 215–252. [Google Scholar]
- Singh, V.; Joung, D.; Zhai, L.; Das, S.; Khondaker, S.I.; Seal, S. Graphene based materials: Past, present and future. Prog. Mater. Sci. 2011, 56, 1178–1271. [Google Scholar] [CrossRef]
- Allen, M.J.; Tung, V.C.; Kaner, R.B. Honeycomb carbon: A review of graphene. Chem. Rev. 2010, 110, 132–145. [Google Scholar] [CrossRef]
- Craciun, M.F.; Russo, S.; Yamamoto, M.; Tarucha, S. Tuneable electronic properties in graphene. Nano. Today 2011, 6, 42–60. [Google Scholar] [CrossRef]
- Hummers, W.S.; Offeman, R.E. Preparation of graphitic oxide. J. Am. Chem. Soc. 1958, 80, 1339. [Google Scholar] [CrossRef]
- He, Y.; Su, Y.; Yang, X.; Kang, Z.; Xu, T.; Zhang, R.; Fan, C.; Lee, S.T. Photo and pH stable, highly-luminescent silicon nanospheres and their bioconjugates for immunofluorescent cell imaging. J. Am. Chem. Soc. 2009, 131, 4434–4438. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Z.; Welsher, K.; Robinson, J.T.; Goodwin, A.; Zaric, S.; Dai, H. Nano-graphene oxide for cellular imaging and drug delivery. Nano Res. 2008, 1, 203–212. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.H.; Hu, D.H.; Lin, C.T.; Li, J.H.; Lin, Y.H. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J. Am. Chem. Soc. 2010, 132, 9274–9276. [Google Scholar] [CrossRef]
- Hembury, G.; Rekharsky, M.; Nakamura, A.; Inoue, Y. Direct correlation between complex conformation and chiral discrimination upon inclusion of amino acid derivatives by beta- and gamma-cyclodextrins. Org. Lett. 2000, 2, 3257–3260. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Blomberg, L.G. Chiral separation of amino acids and peptides by capillary electrophoresis. J. Chromatogr. A 2000, 875, 43–88. [Google Scholar] [CrossRef] [PubMed]
- Zeynep, A. Spectrofluorometric and HPLC determinations of ribavirin in capsules based on fluorescence derivatization of the sugar moiety. Anal. Sci. 2011, 27, 277–282. [Google Scholar]
- Healy, B.; Yu, T.; Alves, D.C.D.; Okeke, C.; Breslin, C.B. Cyclodextrins as supramolecular recognition systems: Applications in the fabrication of electrochemical sensors. Mater. 2021, 14, 1668. [Google Scholar] [CrossRef]
- Shahgaldian, P.; Pieles, U. Cyclodextrin derivatives as chiral supramolecular receptors for enantioselective sensing. Sensors 2006, 6, 593–615. [Google Scholar] [CrossRef]
- Song, L.X.; Bai, L.; Xu, X.M.; He, J.; Pana, S.Z. Inclusion complexation, encapsulation interaction and inclusion number in cyclodextrin chemistry. Coord. Chem. Rev. 2009, 253, 1276–1284. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Wang, Y.; Zhang, P.; Lu, Z.M.; Xiao, Y. Recent advances of molecularly imprinted polymers based on cyclodextrin. Macromol. Rapid Commun. 2021, 42, e2100004. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.Y.; Wang, Y.; Bai, Y.X.; Li, X.X.; Qiu, L.Z.; Jin, Z.Y. Application of cyclodextrinase in non-complexant production of gamma-cyclodextrin. Biotechnol. Progr. 2020, 36, e2930. [Google Scholar] [CrossRef]
- Hu, Z.M.; Li, S.N.; Wang, S.K.; Zhang, B.; Huang, Q. Encapsulation of menthol into cyclodextrin metal-organic frameworks: Preparation, structure characterization and evaluation of complexing capacity. Food Chem. 2021, 338, 127839. [Google Scholar] [CrossRef]
- Dembereldorj, U.; Kim, M.; Kim, S.; Ganbold, E.O.; Lee, S.Y.; Joo, S.W. A spatiotemporal anticancer drug release platform of PEGylated graphene oxide triggered by glutathione in vitro and in vivo. J. Mater. Chem. 2012, 22, 23845–23851. [Google Scholar] [CrossRef]
- Mao, X.W.; Zhao, H.Y.; Luo, L.; Tian, D.M.; Li, H.B. Highly sensitive chiral recognition of amino propanol in serum with R-mandelic acid-linked calix [4] arene modified graphene. J. Mater. Chem. C 2015, 3, 1325–1329. [Google Scholar] [CrossRef]
- Hoenes, G.; Hauser, M.; Pfleiderer, G. Dynamic Total Fluorescence and anisotropy decay study of the dansyl fluorophor in model compounds and enzymes. Photochem. Photobiol. 1986, 43, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Nakamura, M.; Ise, N.; Oguma, N.; Nakamura, A.; Ikeda, T.; Toda, F.; Ueno, A. Fluorescent cyclodextrins for molecule sensing: Fluorescent properties, NMR characterization, and inclusion phenomena of N-dansylleucine-modified cyclodextrins. J. Am. Chem. Soc. 1996, 118, 10980–10988. [Google Scholar] [CrossRef]
- Peng, C.; Hu, W.B.; Zhou, Y.T.; Fan, C.H.; Huang, Q. Intracellular Imaging with a Graphene-Based Fluorescent Probe. Small 2010, 6, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.B.; Peng, C.; Luo, W.J.; Lv, M.; Li, X.M.; Li, D.; Huang, Q.; Fan, C.H. Graphene-based antibacterial paper. ACS Nano 2010, 4, 4317–4323. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Ye, Z.; Zhu, F.; Qiu, T.; Dai, X.; Xie, Y.; Zou, S.; Dong, Q.; Zhang, W.; Ma, J.; et al. Enantioselective Labeling of Zebrafish for D-Phenylalanine Based on Graphene-Based Nanoplatform. Molecules 2023, 28, 3700. https://doi.org/10.3390/molecules28093700
He Y, Ye Z, Zhu F, Qiu T, Dai X, Xie Y, Zou S, Dong Q, Zhang W, Ma J, et al. Enantioselective Labeling of Zebrafish for D-Phenylalanine Based on Graphene-Based Nanoplatform. Molecules. 2023; 28(9):3700. https://doi.org/10.3390/molecules28093700
Chicago/Turabian StyleHe, Yuqing, Ziqi Ye, Fei Zhu, Tianxiang Qiu, Xiyan Dai, Yue Xie, Shibiao Zou, Qingjian Dong, Weiying Zhang, Junkai Ma, and et al. 2023. "Enantioselective Labeling of Zebrafish for D-Phenylalanine Based on Graphene-Based Nanoplatform" Molecules 28, no. 9: 3700. https://doi.org/10.3390/molecules28093700
APA StyleHe, Y., Ye, Z., Zhu, F., Qiu, T., Dai, X., Xie, Y., Zou, S., Dong, Q., Zhang, W., Ma, J., & Mao, X. (2023). Enantioselective Labeling of Zebrafish for D-Phenylalanine Based on Graphene-Based Nanoplatform. Molecules, 28(9), 3700. https://doi.org/10.3390/molecules28093700






