Insights into Transfer of Supramolecular Doxorubicin/Congo Red Aggregates through Phospholipid Membranes
Abstract
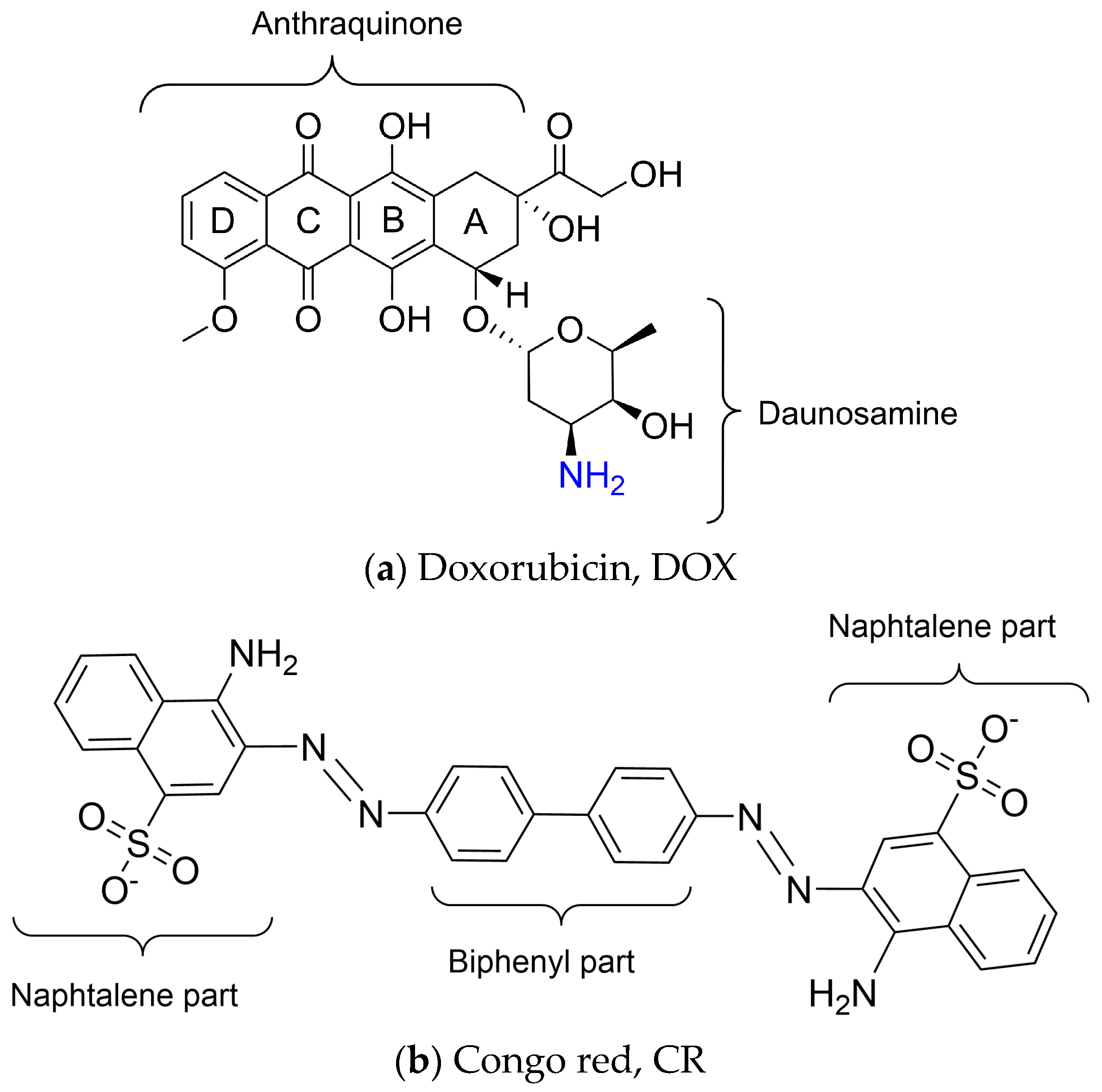
:1. Introduction
2. Results
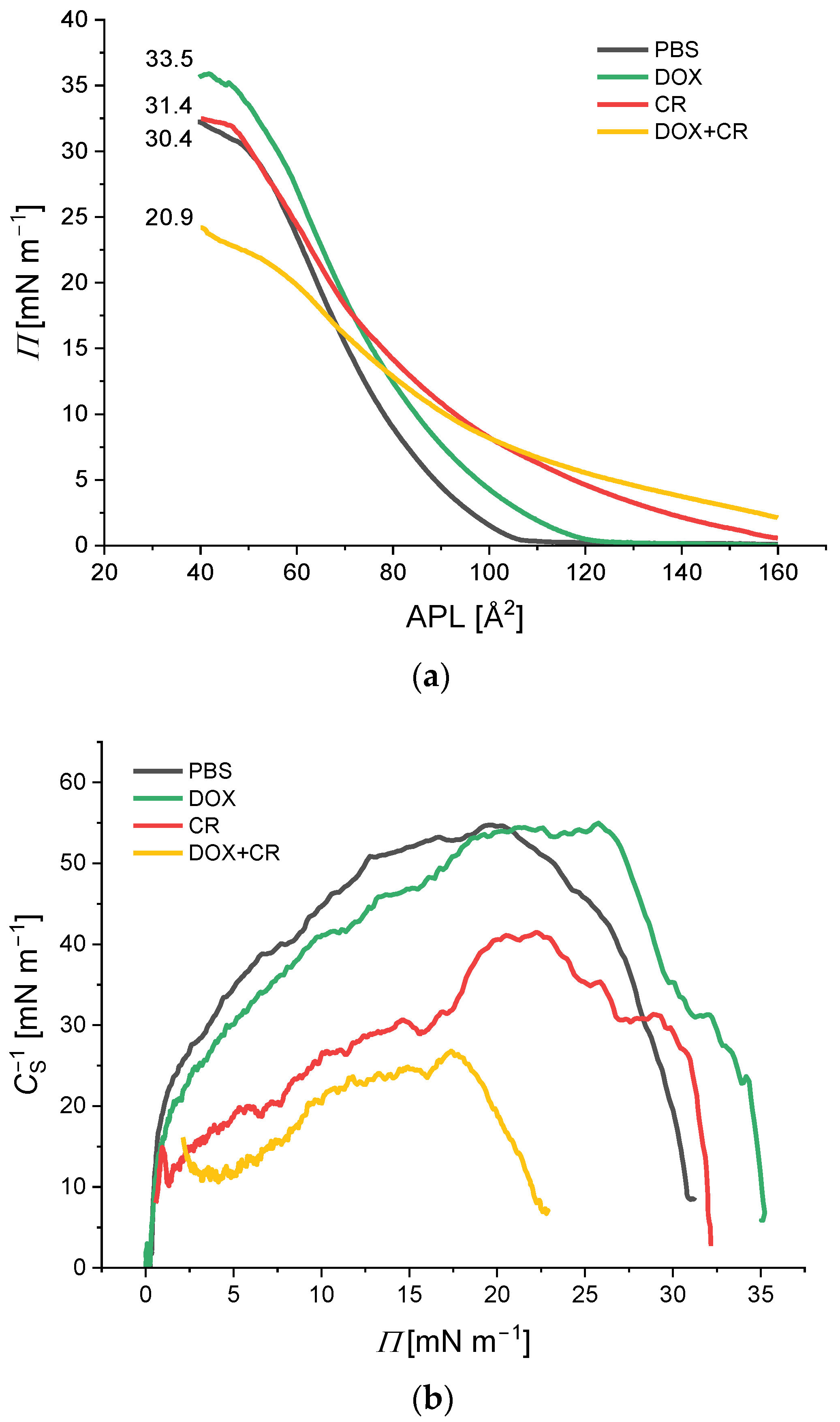
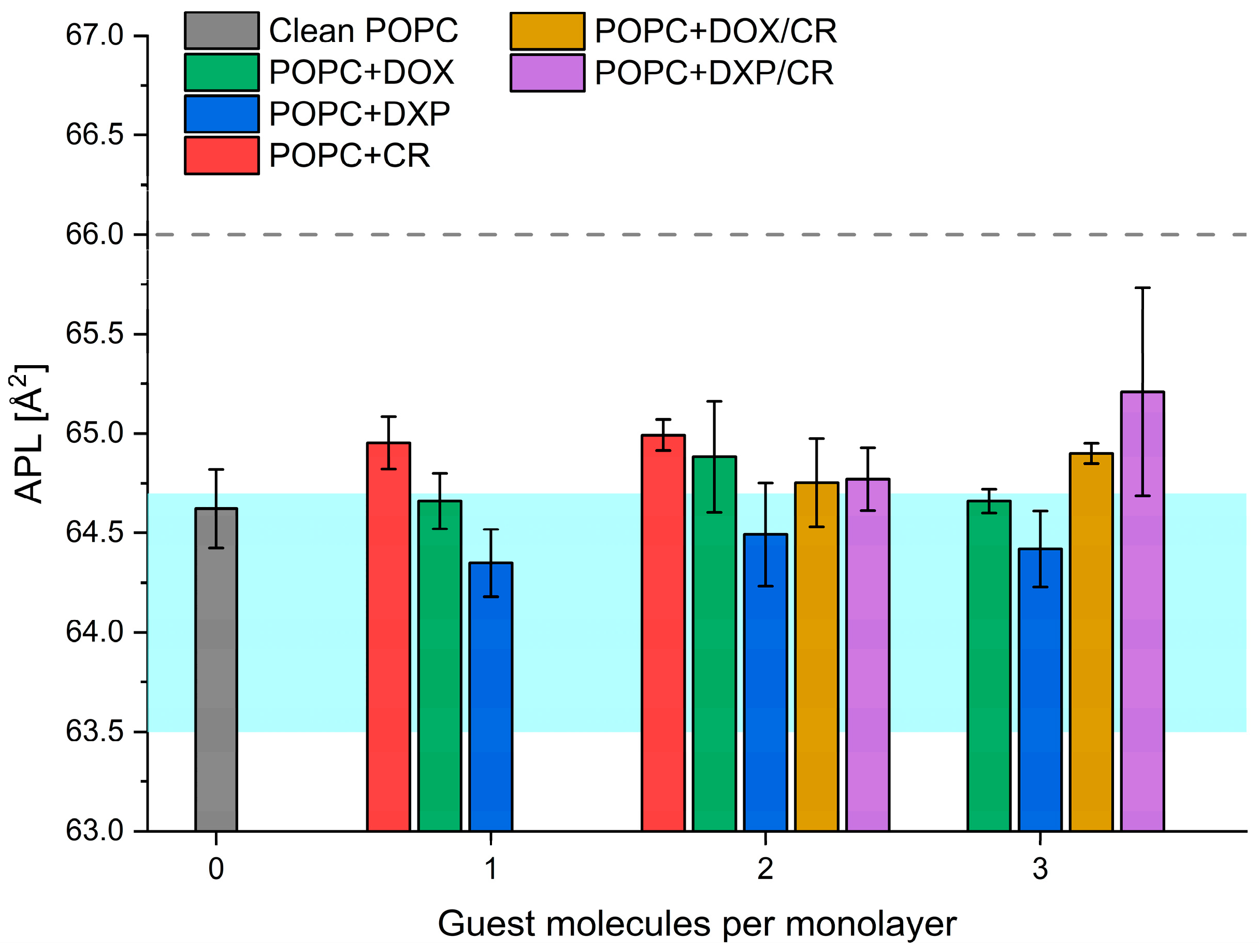
2.1. Langmuir Trough Experiments
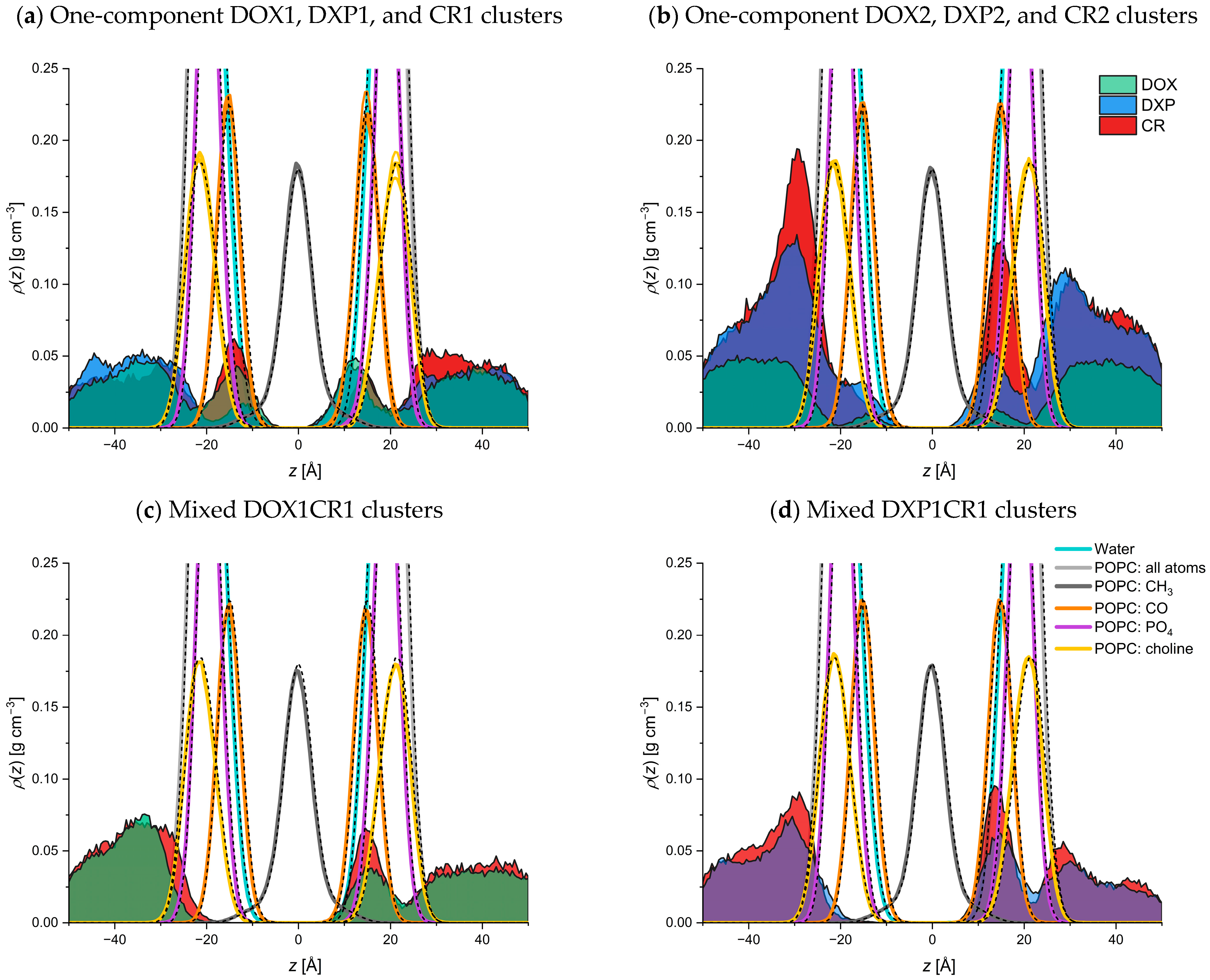
2.2. Molecular Dynamics Simulations
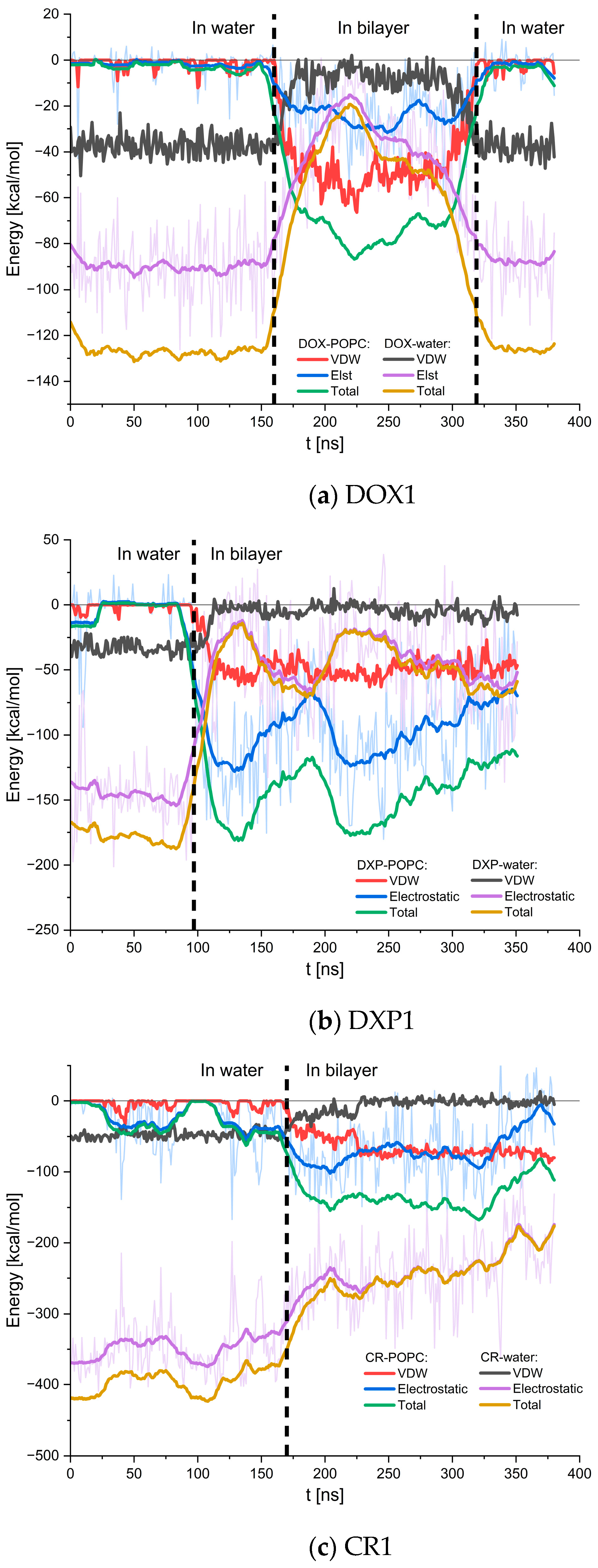
2.2.1. Adsorption of DOX, DXP, CR, and Their Clusters to POPC Bilayer
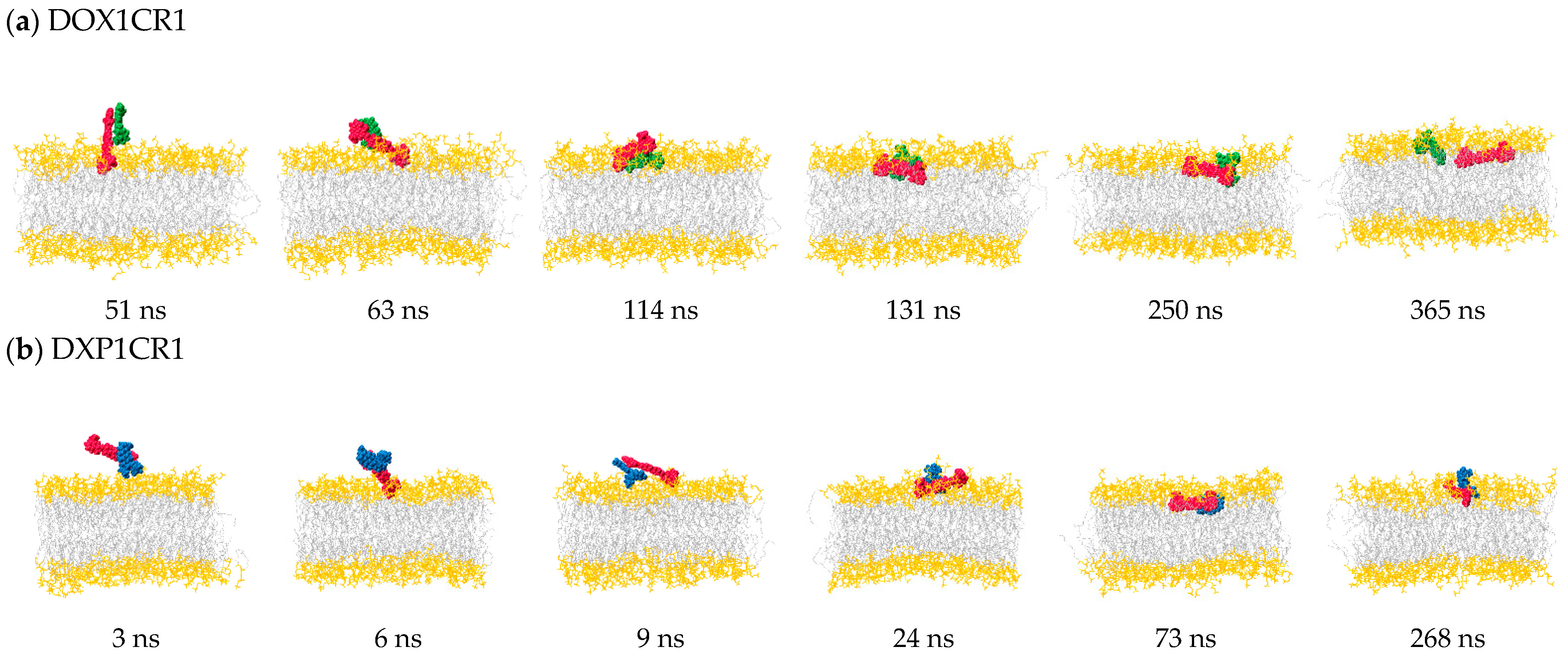
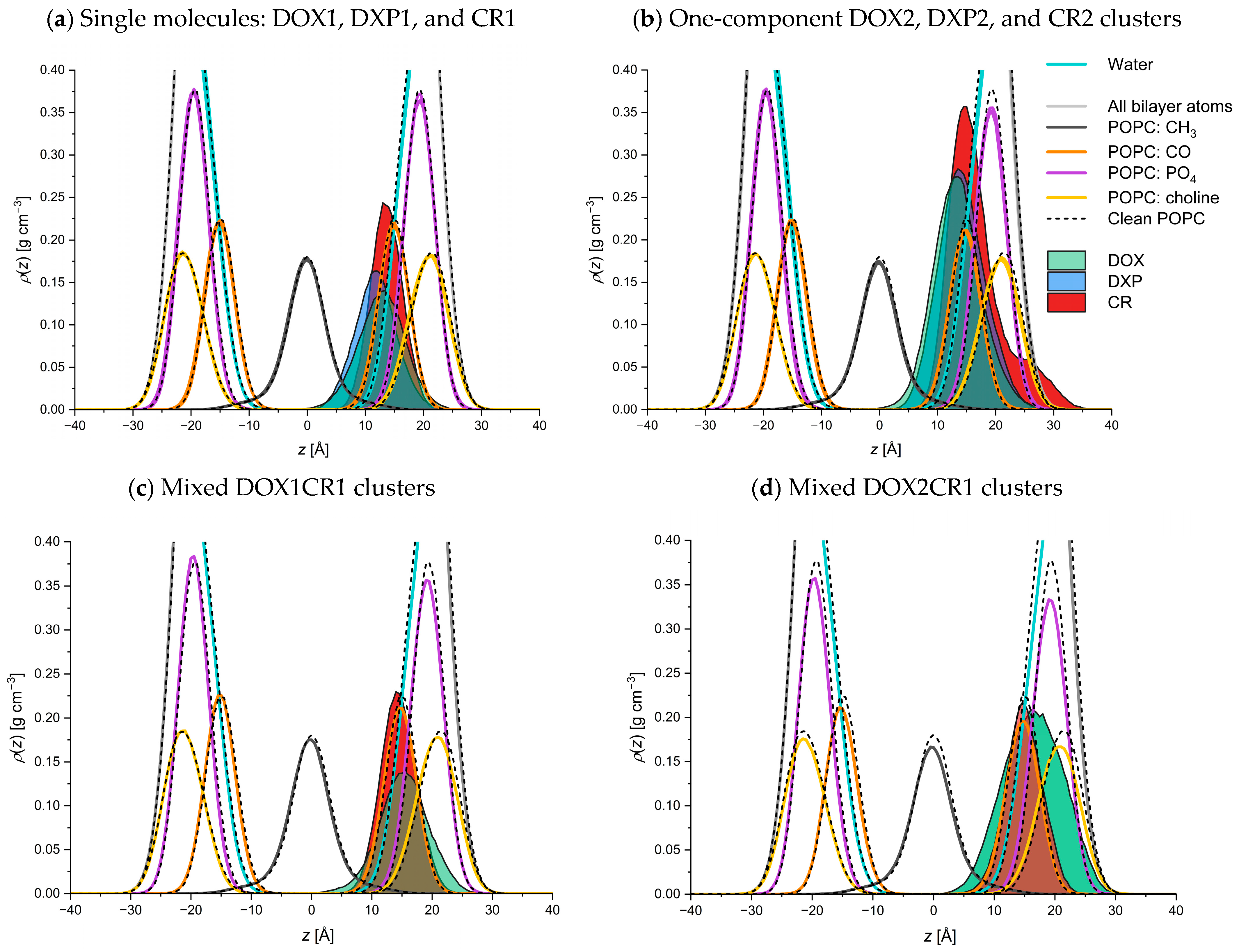
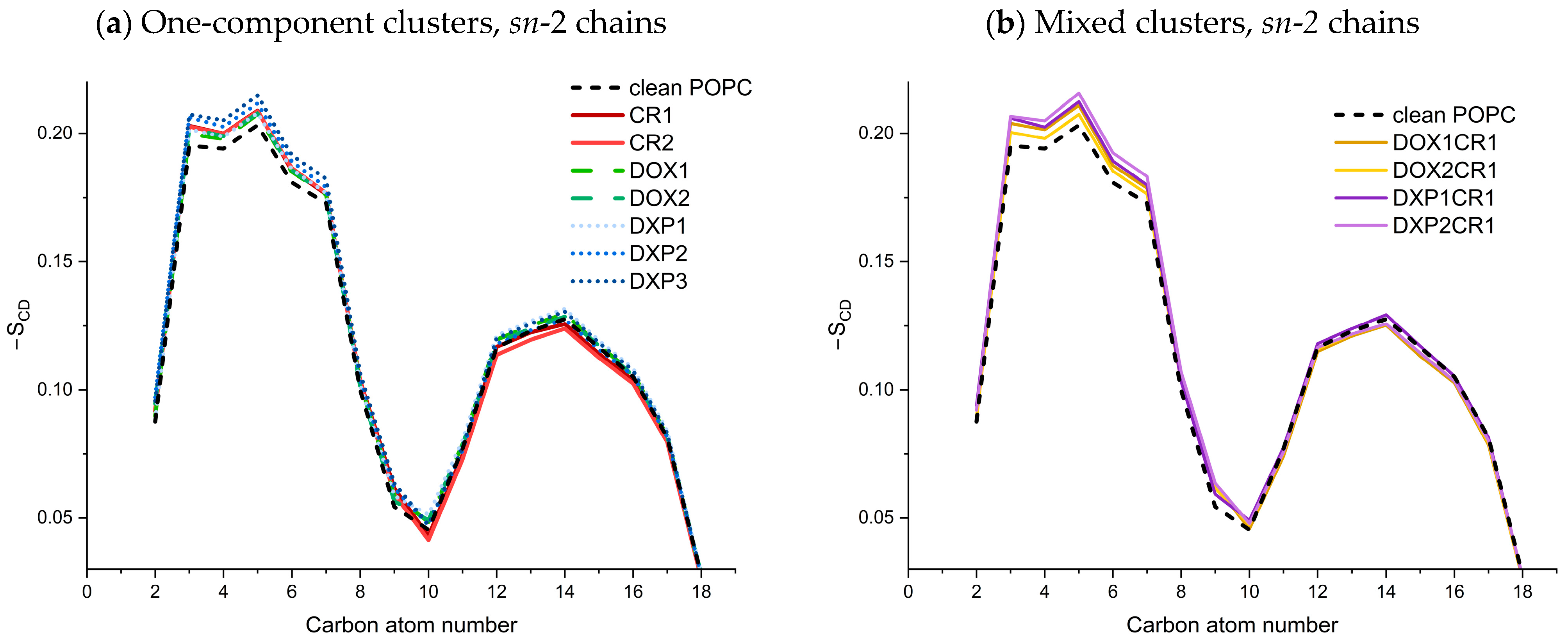
2.2.2. Behavior of Guest Species Inside POPC Bilayer
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Langmuir Trough Experiments
4.3. Computational Details
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tacar, O.; Sriamornsak, P.; Dass, C.R. Doxorubicin: An Update on Anticancer Molecular Action, Toxicity and Novel Drug Delivery Systems. J. Pharm. Pharmacol. 2012, 65, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Bachur, N.R. Anthracyclines. In Encyclopedia of Cancer; Elsevier: Amsterdam, The Netherlands, 2002; pp. 57–61. [Google Scholar]
- Momparler, R.L.; Karon, M.; Siegel, S.E.; Avila, F. Effect of Adriamycin on DNA, RNA, and Protein Synthesis in Cell-Free Systems and Intact Cells. Cancer Res. 1976, 36, 2891–2895. [Google Scholar] [PubMed]
- Jawad, B.; Poudel, L.; Podgornik, R.; Steinmetz, N.F.; Ching, W.-Y. Molecular Mechanism and Binding Free Energy of Doxorubicin Intercalation in DNA. Phys. Chem. Chem. Phys. 2019, 21, 3877–3893. [Google Scholar] [CrossRef] [PubMed]
- Chaires, J.B.; Satyanarayana, S.; Suh, D.; Fokt, I.; Przewloka, T.; Priebe, W. Parsing the Free Energy of Anthracycline Antibiotic Binding to DNA. Biochemistry 1996, 35, 2047–2053. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, B.; Mukherjee, S. Cancer Therapy Using Antibiotics. J. Cancer Ther. 2015, 06, 849–858. [Google Scholar] [CrossRef]
- Hrelia, S.; Fiorentini, D.; Maraldi, T.; Angeloni, C.; Bordoni, A.; Biagi, P.L.; Hakim, G. Doxorubicin Induces Early Lipid Peroxidation Associated with Changes in Glucose Transport in Cultured Cardiomyocytes. Biochim. Et Biophys. Acta BBA Biomembr. 2002, 1567, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D. Risk Factors for Doxorubicin-Lnduced Congestive Heart Failure. Ann. Intern. Med. 1979, 91, 710. [Google Scholar] [CrossRef]
- Chatterjee, K.; Zhang, J.; Honbo, N.; Karliner, J.S. Doxorubicin Cardiomyopathy. Cardiology 2010, 115, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Zocchi, E.; Tonetti, M.; Polvani, C.; Guida, L.; Benatti, U.; De Flora, A. Encapsulation of Doxorubicin in Liver-Targeted Erythrocytes Increases the Therapeutic Index of the Drug in a Murine Metastatic Model. Proc. Natl. Acad. Sci. USA 1989, 86, 2040–2044. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer Nanomedicine: Progress, Challenges and Opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.A.; Waterhouse, D.N.; Mayer, L.D.; Cullis, P.R.; Madden, T.D.; Bally, M.B. The Liposomal Formulation of Doxorubicin. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2005; pp. 71–97. [Google Scholar]
- Yaghoubi, A.; Ramazani, A. Anticancer DOX Delivery System Based on CNTs: Functionalization, Targeting and Novel Technologies. J. Control. Release 2020, 327, 198–224. [Google Scholar] [CrossRef] [PubMed]
- Hashemzadeh, H.; Raissi, H. Understanding Loading, Diffusion and Releasing of Doxorubicin and Paclitaxel Dual Delivery in Graphene and Graphene Oxide Carriers as Highly Efficient Drug Delivery Systems. Appl. Surf. Sci. 2020, 500, 144220. [Google Scholar] [CrossRef]
- Ghahari, A.; Raissi, H.; Farzad, F. Design of a New Drug Delivery Platform Based on Surface Functionalization 2D Covalent Organic Frameworks. J. Taiwan. Inst. Chem. Eng. 2021, 125, 15–22. [Google Scholar] [CrossRef]
- Weng, Q.; Wang, B.; Wang, X.; Hanagata, N.; Li, X.; Liu, D.; Wang, X.; Jiang, X.; Bando, Y.; Golberg, D. Highly Water-Soluble, Porous, and Biocompatible Boron Nitrides for Anticancer Drug Delivery. ACS Nano 2014, 8, 6123–6130. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; He, X.; Wang, K.; He, D.; Yang, S.; Qiu, P.; Chen, S. A PH-Responsive Polymer/Mesoporous Silica Nano-Container Linked through an Acid Cleavable Linker for Intracellular Controlled Release and Tumor Therapy in Vivo. J. Mater. Chem. B 2014, 2, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, J.; Wang, F.; Ding, Y.; Wang, T.; Jin, G.; Martz, M.; Gui, Z.; Ouyang, P.; Chen, P. Histidine-Rich Cell-Penetrating Peptide for Cancer Drug Delivery and Its Uptake Mechanism. Langmuir 2019, 35, 3513–3523. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sheng, Y.; Zehtab Yazdi, A.; Sarikhani, K.; Wang, F.; Jiang, Y.; Liu, J.; Zheng, T.; Wang, W.; Ouyang, P.; et al. Surface-Assisted Assembly of a Histidine-Rich Lipidated Peptide for Simultaneous Exfoliation of Graphite and Functionalization of Graphene Nanosheets. Nanoscale 2019, 11, 2999–3012. [Google Scholar] [CrossRef] [PubMed]
- Haghi, A.; Raissi, H.; Hashemzadeh, H.; Farzad, F. Development of the Poly(L-Histidine) Grafted Carbon Nanotube as a Possible Smart Drug Delivery Vehicle. Comput. Biol. Med. 2022, 143, 105336. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.J.; Cooper, E.A.; Park, T.I.H.; Denny, W.A.; Jose, J. Novel Synthetic Approach for Accessing Drug–Dye Conjugates for Targeted Tumour Therapy. Results Chem. 2022, 4, 100343. [Google Scholar] [CrossRef]
- Ali, Y.; Hamid, S.A.; Rashid, U. Biomedical Applications of Aromatic Azo Compounds. Mini-Rev. Med. Chem. 2018, 18, 1548–1558. [Google Scholar] [CrossRef] [PubMed]
- Jeong, C.; Noh, I.; Rejinold, N.S.; Kim, J.; Jon, S.; Kim, Y.C. Self-Assembled Supramolecular Bilayer Nanoparticles Composed of Near-Infrared Dye as a Theranostic Nanoplatform to Encapsulate Hydrophilic Drugs Effectively. ACS Biomater. Sci. Eng. 2020, 6, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Król, M.; Roterman, I.; Piekarska, B.; Konieczny, L.; Rybarska, J.; Stopa, B.; Spólnik, P.; Szneler, E. An Approach to Understand the Complexation of Supramolecular Dye Congo Red with Immunoglobulin L Chain λ. Biopolymers 2005, 77, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Kwiecińska, K.; Stachowicz-Kuśnierz, A.; Jagusiak, A.; Roterman, I.; Korchowiec, J. Impact of Doxorubicin on Self-Organization of Congo Red: Quantum Chemical Calculations and Molecular Dynamics Simulations. ACS Omega 2020, 5, 19377–19384. [Google Scholar] [CrossRef] [PubMed]
- Jagusiak, A.; Pańczyk, T. Interaction of Congo Red, Evans Blue and Titan Yellow with Doxorubicin in Aqueous Solutions. A Molecular Dynamics Study. J. Mol. Liq. 2019, 279, 640–648. [Google Scholar] [CrossRef]
- Jagusiak, A.; Chłopaś, K.; Zemanek, G.; Kościk, I.; Roterman, I. Interaction of Supramolecular Congo Red and Congo Red-Doxorubicin Complexes with Proteins for Drug Carrier Design. Pharmaceutics 2021, 13, 2027. [Google Scholar] [CrossRef] [PubMed]
- Ptak-Kaczor, M.; Kwiecińska, K.; Korchowiec, J.; Chłopaś, K.; Banach, M.; Roterman, I.; Jagusiak, A. Structure and Location of Protein Sites Binding Self-Associated Congo Red Molecules with Intercalated Drugs as Compact Ligands—Theoretical Studies. Biomolecules 2021, 11, 501. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.T.; Kuhlmann, M.; Hvam, M.L.; Howard, K.A. Albumin-Based Drug Delivery: Harnessing Nature to Cure Disease. Mol. Cell Ther. 2016, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Bahrami, S.; Ravari, S.B.; Zangabad, P.S.; Mirshekari, H.; Bozorgomid, M.; Shahreza, S.; Sori, M.; Hamblin, M.R. Albumin Nanostructures as Advanced Drug Delivery Systems. Expert Opin. Drug Deliv. 2016, 13, 1609–1623. [Google Scholar] [CrossRef] [PubMed]
- Jagusiak, A.; Chłopaś, K.; Zemanek, G.; Kościk, I.; Skorek, P.; Stopa, B. Albumin Binds Doxorubicin via Self–Assembling Dyes as Specific Polymolecular Ligands. Int. J. Mol. Sci. 2022, 23, 5033. [Google Scholar] [CrossRef] [PubMed]
- Lohcharoenkal, W.; Wang, L.; Chen, Y.C.; Rojanasakul, Y. Protein Nanoparticles as Drug Delivery Carriers for Cancer Therapy. Biomed. Res. Int. 2014, 2014, 180549. [Google Scholar] [CrossRef] [PubMed]
- Jagusiak, A.; Chlopas, K.; Zemanek, G.; Wolski, P.; Panczyk, T. Controlled Release of Doxorubicin from the Drug Delivery Formulation Composed of Single-Walled Carbon Nanotubes and Congo Red: A Molecular Dynamics Study and Dynamic Light Scattering Analysis. Pharmaceutics 2020, 12, 622. [Google Scholar] [CrossRef] [PubMed]
- Bosio, V.E.; López, A.G.; Mukherjee, A.; Mechetti, M.; Castro, G.R. Tailoring Doxorubicin Sustainable Release from Biopolymeric Smart Matrix Using Congo Red as Molecular Helper. J. Mater. Chem. B 2014, 2, 5178. [Google Scholar] [CrossRef]
- Kwiecińska, K.; Stachowicz-Kusnierz, A.; Korchowiec, B.; Roman, M.; Kwiatek, W.M.; Jagusiak, A.; Roterman, I.; Korchowiec, J. Congo Red as a Supramolecular Carrier System for Doxorubicin: An Approach to Understanding the Mechanism of Action. Int. J. Mol. Sci. 2022, 23, 8935. [Google Scholar] [CrossRef] [PubMed]
- Fülöp, Z.; Gref, R.; Loftsson, T. A Permeation Method for Detection of Self-Aggregation of Doxorubicin in Aqueous Environment. Int. J. Pharm. 2013, 454, 559–561. [Google Scholar] [CrossRef] [PubMed]
- Gallois, L.; Fiallo, M.; Laigle, A.; Priebe, W.; Garnier-Suillerot, A. The Overall Partitioning of Anthracyclines into Phosphatidyl-Containing Model Membranes Depends Neither on the Drug Charge nor the Presence of Anionic Phospholipids. Eur. J. Biochem. 1996, 241, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Hyslop, P.A.; Morel, B.; Sauerheber, R.D. Organization and Interaction of Cholesterol and Phosphatidylcholine in Model Bilayer Membranes. Biochemistry 1990, 29, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Shahane, G.; Ding, W.; Palaiokostas, M.; Orsi, M. Physical Properties of Model Biological Lipid Bilayers: Insights from All-Atom Molecular Dynamics Simulations. J. Mol. Model. 2019, 25, 76. [Google Scholar] [CrossRef] [PubMed]
- Paloncýová, M.; Devane, R.; Murch, B.; Berka, K.; Otyepka, M. Amphiphilic Drug-like Molecules Accumulate in a Membrane below the Head Group Region. J. Phys. Chem. B 2014, 118, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Speelmans, G.; Staffhorst, R.W.H.M.; Steenbergen, H.G.; de Kruijff, B. Transport of the Anti-Cancer Drug Doxorubicin across Cytoplasmic Membranes and Membranes Composed of Phospholipids Derived from Escherichia Coli Occurs via a Similar Mechanism. Biochim. Et Biophys. Acta BBA Biomembr. 1996, 1284, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Frézard, F.; Garnier-Suillerot, A. Permeability of Lipid Bilayer to Anthracycline Derivatives. Role of the Bilayer Composition and of the Temperature. Biochim. Et Biophys. Acta BBA Lipids Lipid Metab. 1998, 1389, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Kotecka, K.; Krysinski, P. Effect of Tetracycline Antibiotic on the Monolayers of Phosphatidylcholines at the Air–Water Interface. Colloids Surf. A Physicochem. Eng. Asp. 2015, 482, 678–686. [Google Scholar] [CrossRef]
- Heywang, C.; Chazalet, M.S.P.; Masson, M.; Bolard, J. Orientation of Anthracyclines in Lipid Monolayers and Planar Asymmetrical Bilayers: A Surface-Enhanced Resonance Raman Scattering Study. Biophys. J. 1998, 75, 2368–2381. [Google Scholar] [CrossRef]
- Peetla, C.; Bhave, R.; Vijayaraghavalu, S.; Stine, A.; Kooijman, E.; Labhasetwar, V. Drug Resistance in Breast Cancer Cells: Biophysical Characterization of and Doxorubicin Interactions with Membrane Lipids. Mol. Pharm. 2010, 7, 2334–2348. [Google Scholar] [CrossRef] [PubMed]
- Ramu, A.; Glaubiger, D.; Magrath, I.T.; Joshi, A. Plasma Membrane Lipid Structural Order in Doxorubicin-Sensitive and -Resistant P388 Cells. Cancer Res. 1983, 43, 5533–5537. [Google Scholar] [PubMed]
- Aminipour, Z.; Khorshid, M.; Keshvari, H.; Bonakdar, S.; Wagner, P.; Van der Bruggen, B. Passive Permeability Assay of Doxorubicin through Model Cell Membranes under Cancerous and Normal Membrane Potential Conditions. Eur. J. Pharm. Biopharm. 2020, 146, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Speelmans, G.; Staffhorst, R.W.H.M.; de Kruijff, B.; de Wolf, F.A. Transport Studies of Doxorubicin in Model Membranes Indicate a Difference in Passive Diffusion across and Binding at the Outer and Inner Leaflet of the Plasma Membrane. Biochemistry 1994, 33, 13761–13768. [Google Scholar] [CrossRef] [PubMed]
- de Wolf, F.A.; Staffhorst, R.W.H.M.; Smits, H.P.; Onwezen, M.F.; de Kruijff, B. Role of Anionic Phospholipids in the Interaction of Doxorubicin and Plasma Membrane Vesicles: Drug Binding and Structural Consequences in Bacterial Systems. Biochemistry 1993, 32, 6688–6695. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.C.; Magarkar, A.; Horta, M.; Lima, J.L.F.C.; Bunker, A.; Nunes, C.; Reis, S. Influence of Doxorubicin on Model Cell Membrane Properties: Insights from in Vitro and in Silico Studies. Sci. Rep. 2017, 7, 6343. [Google Scholar] [CrossRef]
- Siani, P.; Donadoni, E.; Ferraro, L.; Re, F.; Di Valentin, C. Molecular Dynamics Simulations of Doxorubicin in Sphingomyelin-Based Lipid Membranes. Biochim. Et Biophys. Acta BBA Biomembr. 2022, 1864, 183763. [Google Scholar] [CrossRef]
- Toroz, D.; Gould, I.R. A Computational Study of Anthracyclines Interacting with Lipid Bilayers: Correlation of Membrane Insertion Rates, Orientation Effects and Localisation with Cytotoxicity. Sci. Rep. 2019, 9, 2155. [Google Scholar] [CrossRef]
- Ahmed, Y.; Elkhodary, K.I.; Youssef, M. Molecular Assessment of Drug-Phospholipid Interactions Consequent to Cancer Treatment: A Study of Anthracycline-Induced Cardiotoxicity. Sci. Rep. 2023, 13, 22155. [Google Scholar] [CrossRef] [PubMed]
- Yacoub, T.J.; Reddy, A.S.; Szleifer, I. Structural Effects and Translocation of Doxorubicin in a DPPC/Chol Bilayer: The Role of Cholesterol. Biophys. J. 2011, 101, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Shirazi-Fard, S.; Zolghadr, A.R.; Klein, A. How Does Aggregation of Doxorubicin Molecules Affect Its Solvation and Membrane Penetration? New J. Chem. 2023, 47, 22063–22077. [Google Scholar] [CrossRef]
- Regev, R.; Eytan, G.D. Flip-Flop of Doxorubicin across Erythrocyte and Lipid Membranes. Biochem. Pharmacol. 1997, 54, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.T.; Rideal, E.K. Interfacial Phenomena; Academic Press: Cambridge, MA, USA, 1963; ISBN 9780323161664. [Google Scholar]
- Czapla, K.; Korchowiec, B.; Orlof, M.; Magnieto, J.R.; Rogalska, E. Enzymatic Probing of Model Lipid Membranes: Phospholipase A2 Activity toward Monolayers Modified by Oxicam NSAIDs. J. Phys. Chem. B 2011, 115, 9290–9298. [Google Scholar] [CrossRef] [PubMed]
- Corvis, Y.; Korchowiec, B.; Brezesinski, G.; Follot, S.; Rogalska, E. Impact of Aluminum on the Oxidation of Lipids and Enzymatic Lipolysis in Monomolecular Films at the Air/Water Interface. Langmuir 2007, 23, 3338–3348. [Google Scholar] [CrossRef] [PubMed]
- Korchowiec, B.; Gorczyca, M.; Rogalska, E.; Regnouf-de-Vains, J.-B.; Mourer, M.; Korchowiec, J. The Selective Interactions of Cationic Tetra-p-Guanidinoethylcalix[4]Arene with Lipid Membranes: Theoretical and Experimental Model Studies. Soft Matter 2016, 12, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Gravier, J.; Korchowiec, B.; Schneider, R.; Rogalska, E. Interaction of Amphiphilic Chlorin-Based Photosensitizers with 1,2-Dipalmitoyl-Sn-Glycero-3-Phosphocholine Monolayers. Chem. Phys. Lipids 2009, 158, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Patel, D.S.; Ståhle, J.; Park, S.-J.; Kern, N.R.; Kim, S.; Lee, J.; Cheng, X.; Valvano, M.A.; Holst, O.; et al. CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans. J. Chem. Theory Comput. 2019, 15, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, J.; Jo, S.; Brooks, C.L.; Lee, H.S.; Im, W. CHARMM-GUI Ligand Reader and Modeler for CHARMM Force Field Generation of Small Molecules. J. Comput. Chem. 2017, 38, 1879–1886. [Google Scholar] [CrossRef]
- Besler, B.H.; Merz, K.M.; Kollman, P.A. Atomic Charges Derived from Semiempirical Methods. J. Comput. Chem. 1990, 11, 431–439. [Google Scholar] [CrossRef]
- Mayne, C.G.; Saam, J.; Schulten, K.; Tajkhorshid, E.; Gumbart, J.C. Rapid Parameterization of Small Molecules Using the Force Field Toolkit. J. Comput. Chem. 2013, 34, 2757–2770. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Fiorin, G.; Klein, M.L.; Hénin, J. Using Collective Variables to Drive Molecular Dynamics Simulations. Mol. Phys. 2013, 111, 3345–3362. [Google Scholar] [CrossRef]
- Piggot, T.J.; Allison, J.R.; Sessions, R.B.; Essex, J.W. On the Calculation of Acyl Chain Order Parameters from Lipid Simulations. J. Chem. Theory Comput. 2017, 13, 5683–5696. [Google Scholar] [CrossRef] [PubMed]



| System | No. of Lipids per Leaflet | No. of Guest Molecules per System | Fraction of Penetrating Guest Molecules | |
|---|---|---|---|---|
| DOX1 | 100 | 2 | 50% | Disintegrated in water, penetrated the bilayer as single molecules |
| DOX2 | 100 | 4 | 50% | |
| DOX3 | 100 | 6 | 0% | |
| DOX3 | 225 | 6 | 17% | |
| DXP1 | 100 | 2 | 33% | Remained aggregated in water, penetrated the bilayer in one piece |
| DXP2 | 100 | 4 | 33% | |
| CR1 | 100 | 2 | 50% | Remained aggregated in water, penetrated the bilayer in one piece |
| CR2 | 100 | 4 | 17% | |
| CR3 | 100 | 6 | 17% | |
| CR3 | 225 | 6 | 17% | |
| DOX1CR1 | 100 | 4 | 17% | Remained aggregated in water, DOX1CR1 penetrated the bilayer in one piece, disintegrated in the membrane |
| DOX2CR2 | 100 | 8 | 0% | |
| DOX4CR2 | 100 | 12 | 0% | |
| DOX4CR2 | 225 | 12 | 0% | |
| DXP1CR1 | 100 | 4 | 33% | Remained aggregated in water, DXP1CR1 penetrated the bilayer in one piece |
| DXP2CR1 | 100 | 6 | 0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stachowicz-Kuśnierz, A.; Rychlik, P.; Korchowiec, J.; Korchowiec, B. Insights into Transfer of Supramolecular Doxorubicin/Congo Red Aggregates through Phospholipid Membranes. Molecules 2024, 29, 2567. https://doi.org/10.3390/molecules29112567
Stachowicz-Kuśnierz A, Rychlik P, Korchowiec J, Korchowiec B. Insights into Transfer of Supramolecular Doxorubicin/Congo Red Aggregates through Phospholipid Membranes. Molecules. 2024; 29(11):2567. https://doi.org/10.3390/molecules29112567
Chicago/Turabian StyleStachowicz-Kuśnierz, Anna, Paulina Rychlik, Jacek Korchowiec, and Beata Korchowiec. 2024. "Insights into Transfer of Supramolecular Doxorubicin/Congo Red Aggregates through Phospholipid Membranes" Molecules 29, no. 11: 2567. https://doi.org/10.3390/molecules29112567
APA StyleStachowicz-Kuśnierz, A., Rychlik, P., Korchowiec, J., & Korchowiec, B. (2024). Insights into Transfer of Supramolecular Doxorubicin/Congo Red Aggregates through Phospholipid Membranes. Molecules, 29(11), 2567. https://doi.org/10.3390/molecules29112567






