Widely Targeted Metabolomic Analysis Reveals the Improvement in Panax notoginseng Triterpenoids Triggered by Arbuscular Mycorrhizal Fungi via UPLC–ESI–MS/MS
Abstract
:1. Introduction
2. Results
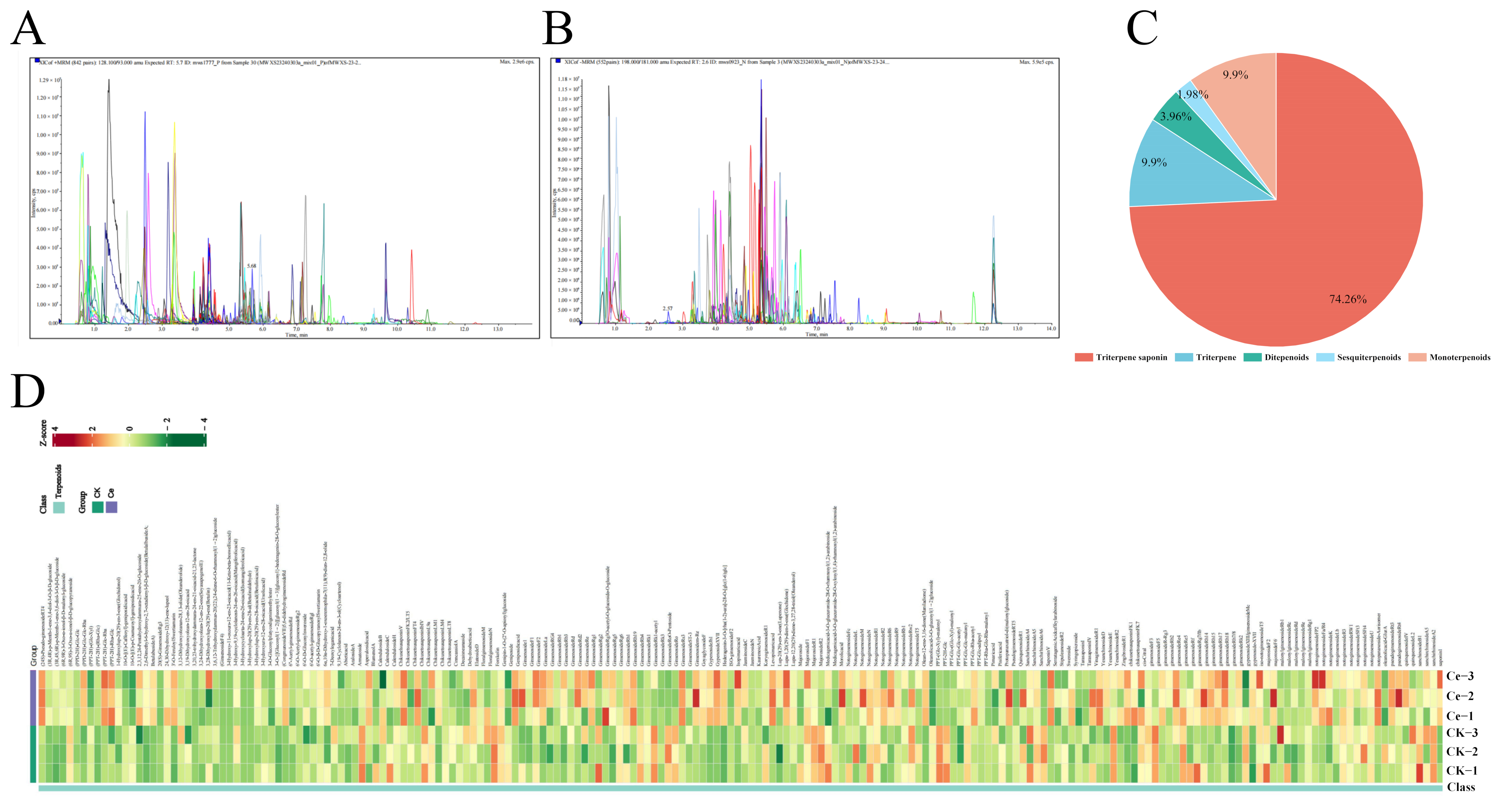
2.1. Terpenoid Profiling of P. notoginseng via UPLC–ESI–MS/MS
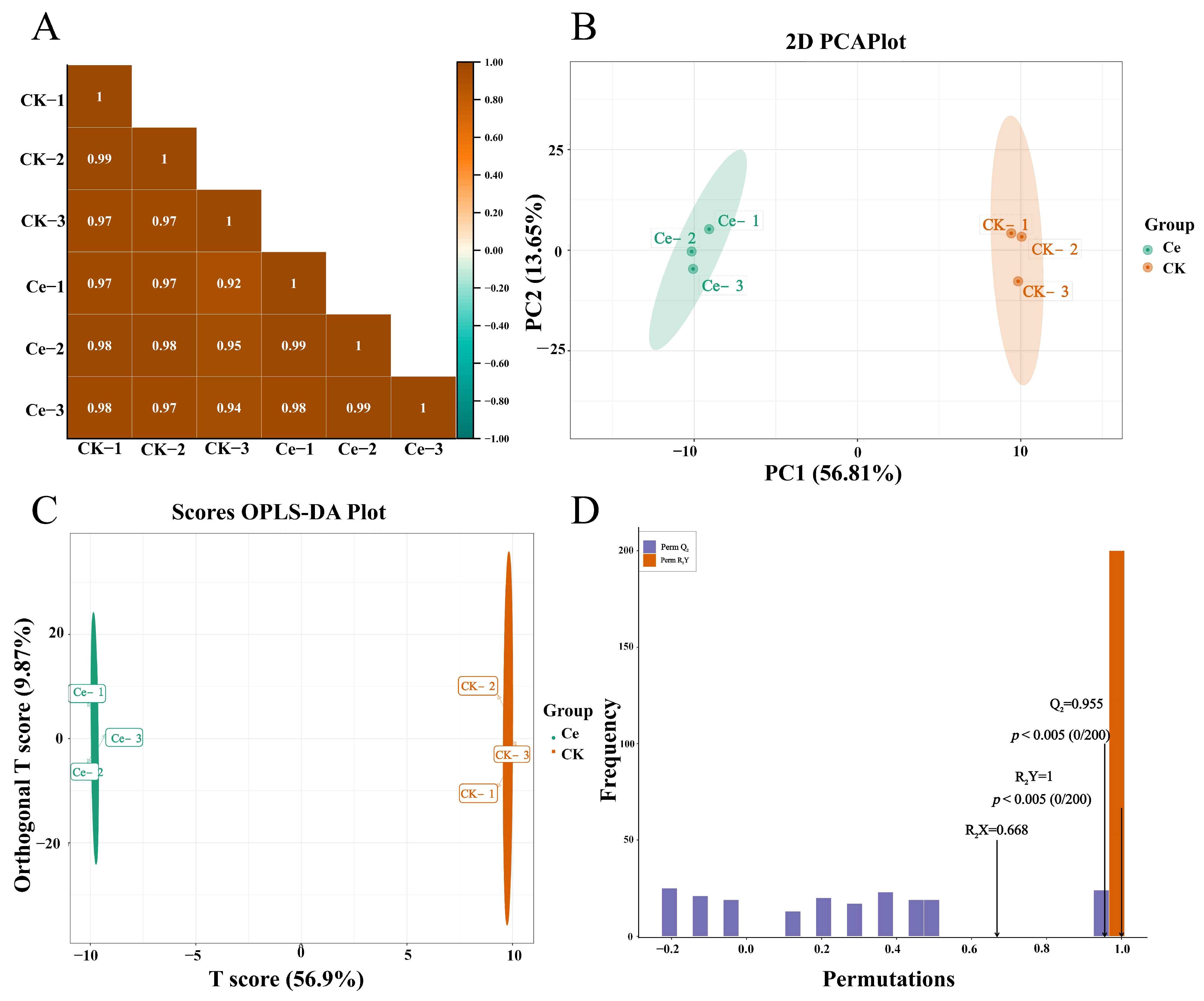
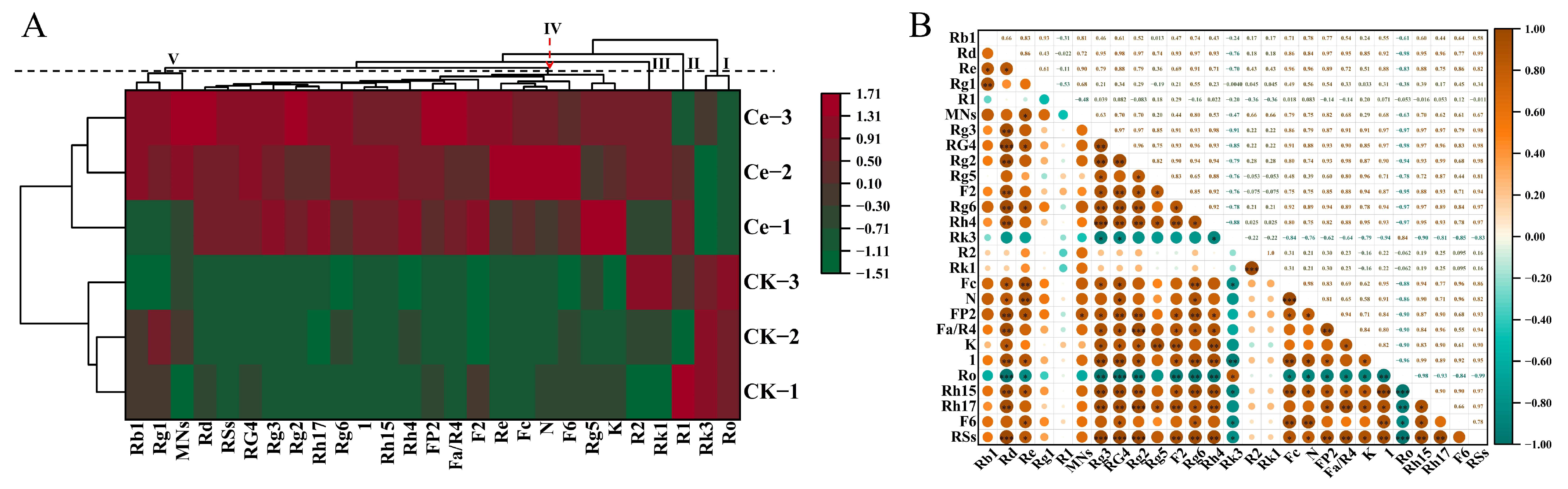
2.2. Multivariate Statistical Analysis
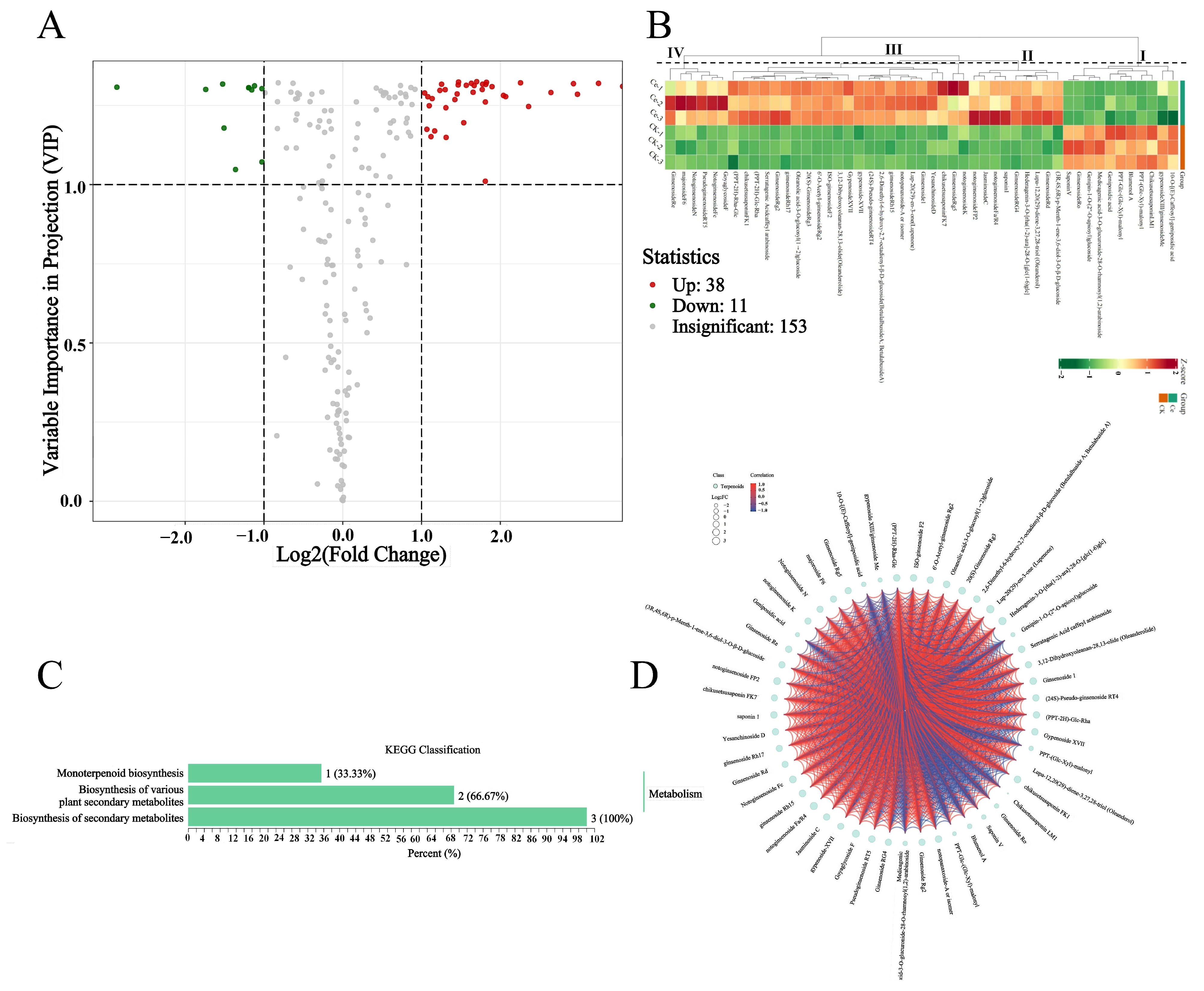
2.3. Differential Terpene Metabolites between the AMF and CK Groups
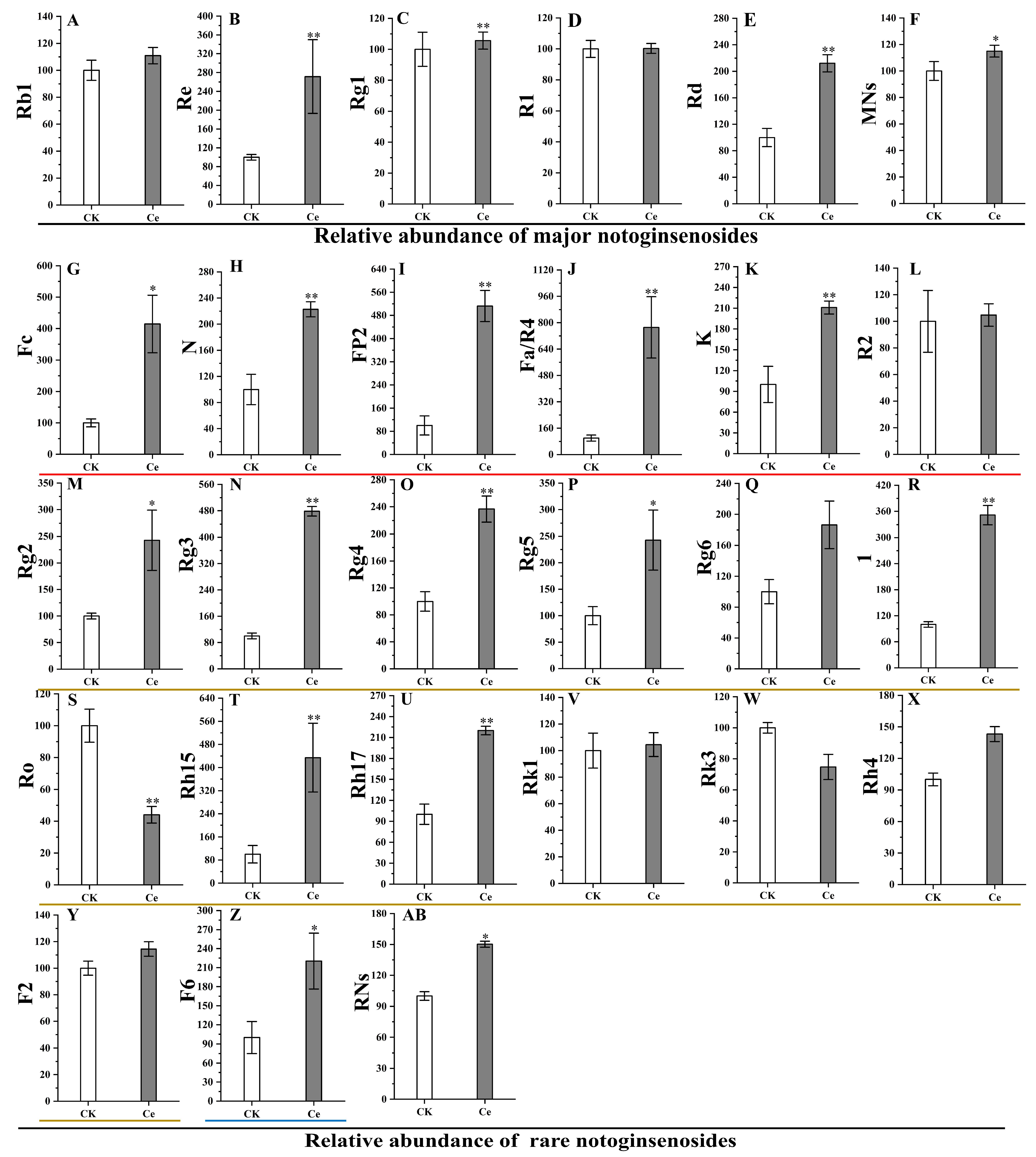
2.4. Accumulation of PNSs Triggered by an AMF
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Pot Experiments
4.2. Sample Preparation and Extraction
4.3. UPLC Conditions
4.4. ESI–Q TRAP–MS/MS
4.5. Qualitative and Quantitative Analyses of Terpene Metabolites
4.6. Principal Component Analysis
4.7. Hierarchical Cluster Analysis and Pearson Correlation Coefficients
4.8. Differentially Abundant Metabolite Selection and KEGG Annotation and Enrichment Analysis
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, T.; Guo, R.; Zhou, G.; Zhou, X.; Kou, Z.; Sui, F.; Li, C.; Tang, L.; Wang, Z. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) FH Chen: A review. J. Ethnopharmacol. 2016, 188, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.H.; Li, Z.D.; Wang, X.F.; Zhang, X.; Zhao, R.H.; Gu, W.; Chen, D.; Yu, J.; He, S. Phosphate transporters, PnPht1; 1 and PnPht1; 2 from Panax notoginseng enhance phosphate and arsenate acquisition. BMC Plant Biol. 2020, 20, 124. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, Q.; Huang, J.; Wang, Z. Antidepressant active ingredients from Chinese traditional herb Panax notoginseng: A pharmacological mechanism review. Front. Pharmacol. 2022, 13, 922337. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Chu, Y.; Chen, Y.; Zhao, Q.; Liao, B.; Zhang, J.; Gao, Y.; Xu, J.; Chen, S. Genome-wide identification and transcriptional profiling analysis of PIN/PILS auxin transporter gene families in Panax ginseng. Chin. Herb. Med. 2022, 14, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, W.; Wang, B.; Zhang, T.; Cui, X.; Pu, Y.; Li, N. Analytical methods and biological activities of Panax notoginseng saponins: Recent trends. J. Ethnopharmacol. 2019, 236, 443–465. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.H.; Wang, X.F.; Li, Z.D.; Zhang, X.; Li, X.G.; Gu, W.; Zhang, F.; Yu, J.; He, S. A Panax notoginseng phosphate transporter, PnPht1; 3, greatly contributes to phosphate and arsenate uptake. Funct. Plant Biol. 2022, 49, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, J.; Diao, M.; Li, J.; Xie, N. Production and pharmaceutical research of minor saponins in Panax notoginseng (Sanqi): Current status and future prospects. Phytochemistry 2024, 223, 114099. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yuan, M.; Li, X.; Li, J.; Xu, M.; Wei, D.; Wu, D.; Wan, J.; Mei, S.; Cui, T.; et al. New dammarane-type triterpenoid saponins from Panax notoginseng saponins. J. Ginseng Res. 2020, 44, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.Y.; Wang, H.C.; Jin, Y.P.; Li, Y.L.; Wang, Y.P. Isolation, identification, and quantification of triterpene saponins in the fresh fruits of Panax notoginseng. Nat. Prod. Res. 2022, 36, 5319–5329. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.M.; Chen, M.H.; Han, F.; Wang, J.W.; Tu, Y.X. Role of bioactive constituents of Panax notoginseng in the modulation of tumorigenesis: A potential review for the treatment of cancer. Front. Pharmacol. 2021, 12, 738914. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Zhou, C.; Zeng, Y.; Zhang, H.; Hossen, M.A.; Dai, J.; Li, S.; Qin, W.; Liu, Y. Structures, physicochemical and bioactive properties of polysaccharides extracted from Panax notoginseng using ultrasonic/microwave-assisted extraction. LWT 2022, 154, 112446. [Google Scholar] [CrossRef]
- Cao, G.H.; Bai, X.; Zhang, C.R.; Li, X.G.; Dai, H.Y.; Bi, Y.; Zhang, X.K.; He, S. Physiological response and transcriptome profiling reveal phosphate-mediated amelioration of arsenic accumulation and toxicity in Panax notoginseng. Environ. Exp. Bot. 2023, 206, 105136. [Google Scholar] [CrossRef]
- Gu, X.; Hao, D.; Xiao, P. Research progress of Chinese herbal medicine compounds and their bioactivities: Fruitful 2020. Chin. Herb. Med. 2022, 14, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Pang, X.; Xu, K.; Wang, M.; Ma, Z.; Tan, H.; Han, L.; Chang, C.; Chen, M.; Huang, Z.; et al. Anaphylactic rare saponins separated from Panax notoginseng saponin and a proteomic approach to their anaphylactic mechanism. Evid. Based Complement Altern. Med. 2022, 2022, 7565177. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Ye, Y.P.; Sun, H.X. Haemolytic activity and adjuvant effect of notoginsenoside K from the roots of Panax notoginseng. Chem. Biodivers. 2006, 3, 1144–1152. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Yi, Y.X.; Zhang, T.; Ding, Y.; Le, J. Stereoisomers of saponins in Panax notoginseng (Sanqi): A review. Front. Pharmacol. 2018, 9, 328597. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lu, X.; Hu, Y.; Fan, X. Chemical constituents of Panax ginseng and Panax notoginseng explain why they differ in therapeutic efficacy. Pharmacol. Res. 2020, 161, 105263. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Zhang, G.; Li, M.; Liu, C.; Wei, F.; Wang, Y.; Huang, Z.; Chen, Z.; Zheng, Y.; Chen, S.; et al. Core rhizosphere microbiome of Panax notoginseng and its associations with belowground biomass and saponin contents. Environ. Microbiol. 2022, 24, 6238–6251. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Wang, S.; Xiong, B.; Gu, H.; Wang, H.; Ji, C.; Jia, W.; Horowitz, A.R.; Zhen, W.; Asher, J.B.; et al. Application of bioorganic fertilizer on Panax notoginseng improves plant growth by altering the rhizosphere microbiome structure and metabolism. Microorganisms 2022, 10, 275. [Google Scholar] [CrossRef]
- Cao, P.; Wei, X.; Wang, G.; Chen, X.; Han, J.; Li, Y. Microbial inoculants and garbage fermentation liquid reduced root-knot nematode disease and As uptake in Panax quinquefolium cultivation by modulating rhizosphere microbiota community. Chin. Herb. Med. 2022, 14, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.H.; He, S.; Chen, D.; Li, T.; Zhao, Z.W. EpABC genes in the adaptive responses of Exophiala pisciphila to metal stress: Functional importance and relation to metal tolerance. Appl. Environ. Microbiol. 2019, 85, e01844-19. [Google Scholar] [CrossRef]
- Khaliq, A.; Perveen, S.; Alamer, K.H.; Zia Ul Haq, M.; Rafique, Z.; Alsudays, I.M.; Althobaiti, A.T.; Saleh, M.A.; Hussain, S.; Attia, H. Arbuscular mycorrhizal fungi symbiosis to Enhance plant-soil interaction. Sustainability 2022, 14, 7840. [Google Scholar] [CrossRef]
- Mandal, S.; Upadhyay, S.; Wajid, S.; Ram, M.; Jain, D.C.; Singh, V.P.; Abdin, M.Z.; Kapoor, R. Arbuscular mycorrhiza increase artemisinin accumulation in Artemisia annua by higher expression of key biosynthesis genes via enhanced jasmonic acid levels. Mycorrhiza 2015, 25, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Domokos, E.; Jakab-Farkas, L.; Darkó, B.; Bíró-Janka, B.; Mara, G.; Albert, C.; Balog, A. Increase in Artemisia annua plant biomass artemisinin content and guaiacol peroxidase activity using the arbuscular mycorrhizal fungus Rhizophagus irregularis. Front. Plant Sci. 2018, 9, 478. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Hao, Z.; Zhou, X.; Jiang, X.; Xu, L.; Wu, S.; Zhao, A.; Zhang, X.; Chen, B. Arbuscular mycorrhiza facilitates the accumulation of glycyrrhizin and liquiritin in Glycyrrhiza uralensis under drought stress. Mycorrhiza 2018, 28, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Hao, Z.; Yu, M.; Wu, Z.; Zhao, A.; Li, J.; Zhang, X.; Chen, B. Improved phosphorus nutrition by arbuscular mycorrhizal symbiosis as a key factor facilitating glycyrrhizin and liquiritin accumulation in Glycyrrhiza uralensis. Plant Soil 2019, 439, 243–257. [Google Scholar] [CrossRef]
- Sun, R.T.; Zhang, Z.Z.; Liu, M.Y.; Feng, X.C.; Zhou, N.; Feng, H.D.; Hashem, A.; Abd_Allah, E.F.; Harsonowati, W.; Wu, Q.S. Arbuscular mycorrhizal fungi and phosphorus supply accelerate main medicinal component production of Polygonum cuspidatum. Front. Microbiol. 2022, 13, 1006140. [Google Scholar] [CrossRef]
- Pei, Y.; Yin, M.; Li, Q.H.; Zhang, Y.F.; Zhong, Y.; Chen, X.; Zhang, Y.P.; Huang, B.; Ren, Z. Diversity and community structure of arbuscular mycorrhizal fungi (AMF) in the rhizospheric soil of Panax notoginseng in different ages. Eurasian Soil Sci. 2023, 56, 329–339. [Google Scholar] [CrossRef]
- Cao, G.H.; Zhang, X.; Chen, D.; Bai, X.; Lu, Z.Q.; Zhang, B.; He, S. Colonization of arbuscular mycorrhizal fungi (AMF) and dark septate endophytes (DSE) in the roots of Panax notoginseng and correlation analysis with saponins content. J. Chin. Med. Mater. 2019, 42, 1509–1512. [Google Scholar] [CrossRef]
- Dai, H.Y.; Zhang, X.K.; Bi, Y.; Chen, D.; Long, X.N.; Wu, Y.; Cao, G.H.; He, S. Improvement of Panax notoginseng saponin accumulation triggered by methyl jasmonate under arbuscular mycorrhizal fungi. Front. Plant Sci. 2024, 15, 1360919. [Google Scholar] [CrossRef] [PubMed]
- Bisht, A.; Garg, N. AMF species improve yielding potential of Cd stressed pigeonpea plants by modulating sucrose-starch metabolism, nutrients acquisition and soil microbial enzymatic activities. Plant Growth Regul. 2022, 96, 409–430. [Google Scholar] [CrossRef]
- Saboor, A.; Ali, M.A.; Hussain, S.; El Enshasy, H.A.; Hussain, S.; Ahmed, N.; Gafur, A.; Sayyed, R.Z.; Fahad, S.; Danish, S.; et al. Zinc nutrition and arbuscular mycorrhizal symbiosis effects on maize (Zea mays L.) growth and productivity. Saudi J. Biol. Sci. 2021, 28, 6339–6351. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Suseela, V. Unraveling arbuscular mycorrhiza-induced changes in plant primary and secondary metabolome. Metabolites 2020, 10, 335. [Google Scholar] [CrossRef]
- Iula, G.; Miras-Moreno, B.; Lucini, L.; Trevisan, M. The mycorrhiza and trichoderma-mediated elicitation of secondary metabolism and modulation of phytohormone profile in pomato plants. Horticulturae 2021, 7, 394. [Google Scholar] [CrossRef]
- Ran, Z.; Ding, W.; Cao, S.; Fang, L.; Zhou, J.; Zhang, Y. Arbuscular mycorrhizal fungi: Effects on secondary metabolite accumulation of traditional Chinese medicines. Plant Biol. 2022, 24, 932–938. [Google Scholar] [CrossRef]
- Commisso, M.; Strazzer, P.; Toffali, K.; Stocchero, M.; Guzzo, F. Untargeted metabolomics: An emerging approach to determine the composition of herbal products. Comput. Struct. Biotechnol. J. 2013, 4, e201301007. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Lu, C.; Zhang, Y.; Zhao, S.; Liu, B.; Xu, X.; Liu, Y. Application of plant metabonomics in quality assessment for large-scale production of traditional Chinese medicine. Planta Med. 2013, 79, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.; Kong, M.; Shen, H.; Zhu, H.; Zhou, S.S.; Li, S.L. LC-MS-based metabolomics in traditional Chinese medicines research: Personal experiences. Chin. Herb. Med. 2017, 9, 14–21. [Google Scholar] [CrossRef]
- Chen, J.; Guo, X.; Song, Y.; Zhao, M.; Tu, P.; Jiang, Y. MRM-based strategy for the homolog-focused detection of minor ginsenosides from notoginseng total saponins by ultra-performance liquid chromatography coupled with hybrid triple quadrupole-linear ion trap mass spectrometry. RSC Adv. 2016, 6, 96376–96388. [Google Scholar] [CrossRef]
- Wu, W.; Jiao, C.; Li, H.; Ma, Y.; Jiao, L.; Liu, S. LC-MS based metabolic and metabonomic studies of Panax ginseng. Phytochem. Anal. 2018, 29, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Liang, K.; Zhou, Z.; Wang, X.; Huang, G. UPLC-ESI-MS/MS-Based widely targeted metabolomics analysis of wood metabolites in Teak (Tectona grandis). Molecules 2020, 25, 2189. [Google Scholar] [CrossRef]
- Devanesan, S.; Elankathirselvan, K.; AlSalhi, A.; AlSalhi, M.S.; Asemi, N.N.; Aldawsari, M.; Jhanani, G.K. UPLC–ESI–MS/MS profiling of active polyphenolics in Morinda coreia leaf extract and in vitro antioxidant and antibacterial activity. Chemosphere 2023, 323, 138179. [Google Scholar] [CrossRef] [PubMed]
- Tarraf, W.; Ruta, C.; Tagarelli, A.; De Cillis, F.; De Mastro, G. Influence of arbuscular mycorrhizae on plant growth, essential oil production and phosphorus uptake of Salvia officinalis L. Ind. Crops Prod. 2017, 102, 144–153. [Google Scholar] [CrossRef]
- de Assis, R.M.; Carneiro, J.J.; Medeiros, A.P.; de Carvalho, A.A.; da Cunha Honorato, A.; Carneiro, M.A.; Bertolucci, S.K.; Pinto, J.E. Arbuscular mycorrhizal fungi and organic manure enhance growth and accumulation of citral, total phenols, and flavonoids in Melissa officinalis L. Ind. Crops Prod. 2020, 158, 112981. [Google Scholar] [CrossRef]
- Wu, Y.H.; Qin, Y.; Cai, Q.Q.; Liu, M.; He, D.M.; Chen, X.; Wang, H.; Yan, Z.Y. Effect the accumulation of bioactive constituents of a medicinal plant (Salvia Miltiorrhiza Bge.) by arbuscular mycorrhizal fungi community. BMC Plant Biol. 2023, 23, 597. [Google Scholar] [CrossRef] [PubMed]
- Muniz, B.C.; Falcão, E.L.; de Paula Monteiro, R.; dos Santos, E.L.; Bastos Filho, C.J.A.; da Silva, F.S.B. Acaulospora longula Spain & NC Schenck: A low-cost bioinsumption to optimize phenolics and saponins production in Passiflora alata Curtis. Ind. Crops Prod. 2021, 167, 113498. [Google Scholar]
- Yilmaz, A.; Karik, Ü. AMF and PGPR enhance yield and secondary metabolite profile of basil (Ocimum basilicum L.). Ind. Crops Prod. 2022, 176, 114327. [Google Scholar] [CrossRef]
- Arpanahi, A.A.; Feizian, M.; Mehdipourian, G.; Khojasteh, D.N. Arbuscular mycorrhizal fungi inoculation improve essential oil and physiological parameters and nutritional values of Thymus daenensis Celak and Thymus vulgaris L. under normal and drought stress conditions. Eur. J. Soil Biol. 2020, 100, 103217. [Google Scholar] [CrossRef]
- Cao, G.H.; Li, X.G.; Zhang, C.R.; Xiong, Y.R.; Li, X.; Li, T.; He, S.; Cui, Z.; Yu, J. Physiological response mechanism of heavy metal-resistant endophytic fungi isolated from the roots of Polygonatum kingianum. Environ. Microbiol. Rep. 2023, 15, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.; Shin, W.-C.; Oh, S.-M.; Choi, B.-R.; Lee, D.Y. Integration of multiplatform metabolomics and multivariate analysis for geographical origin discrimination of Panax ginseng. Food Res. Int. 2022, 159, 111610. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wu, D.; Sun, Y.; Zhao, H.; Wang, Y.; Zhang, W.; Su, F.; Yang, B.; Wang, Q.; Kuang, H. Comprehensive analysis of Eleutherococcus senticosus (Rupr. & Maxim.) Maxim. leaves based on UPLC-MS/MS: Separation and rapid qualitative and quantitative analysis. Front. Pharmacol. 2022, 13, 865586. [Google Scholar]
- Sun, Y.; Liu, X.; Fu, X.; Xu, W.; Guo, Q.; Zhang, Y. Discrepancy study of the chemical constituents of Panax Ginseng from different growth environments with UPLC-MS-based metabolomics strategy. Molecules 2023, 28, 2928. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.H.; Wang, H.; Liu, M.; Li, B.; Chen, X.; Ma, Y.T.; Yan, Z.Y. Effects of native arbuscular mycorrhizae isolated on root biomass and secondary metabolites of Salvia miltiorrhiza Bge. Front. Plant Sci. 2021, 12, 617892. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Peñuelas, J.; Chen, Y.; Sardans, J.; Yu, J.; Xu, Z.; Cui, Q.; Liu, J.; Cui, Y.; Zhao, S.; et al. Arbuscular mycorrhizal fungal interactions bridge the support of root-associated microbiota for slope multifunctionality in an erosion-prone ecosystem. iMeta 2024, 3, e187. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Zhang, T.; Wang, Z.; Li, G.; Zhao, W.; Lv, X.; Zhuang, L. Succession of endophytic fungi and arbuscular mycorrhizal fungi associated with the growth of plant and their correlation with secondary metabolites in the roots of plants. BMC Plant Biol. 2021, 21, 165. [Google Scholar] [CrossRef]
- Li, L.; Ni, J.; Li, M.; Chen, J.; Han, L.; Zhu, Y.; Kong, D.; Mao, J.; Wang, Y.; Zhang, B.; et al. Ginsenoside Rg3 micelles mitigate doxorubicin-induced cardiotoxicity and enhance its anticancer efficacy. Drug Deliv. 2017, 24, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Kee, J.Y.; Hong, S.H. Ginsenoside Rg3 suppresses mast cell-mediated allergic inflammation via mitogen-activated protein kinase signaling pathway. J. Ginseng Res. 2019, 43, 282–290. [Google Scholar] [CrossRef]
- Duan, Z.; Wei, B.; Deng, J.; Mi, Y.; Dong, Y.; Zhu, C.; Fu, R.; Qu, L.; Fan, D. The anti-tumor effect of ginsenoside Rh4 in MCF-7 breast cancer cells in vitro and in vivo. Biochem. Biophys. Res. Commun. 2018, 499, 482–487. [Google Scholar] [CrossRef]
- Wu, Q.; Deng, J.; Fan, D.; Duan, Z.; Zhu, C.; Fu, R.; Wang, S. Ginsenoside Rh4 induces apoptosis and autophagic cell death through activation of the ROS/JNK/p53 pathway in colorectal cancer cells. Biochem. Pharmacol. 2018, 148, 64–74. [Google Scholar] [CrossRef]
- To, K.I.; Zhu, Z.X.; Wang, Y.N.; Li, G.A.; Sun, Y.M.; Li, Y.; Jin, Y.H. Integrative network pharmacology and experimental verification to reveal the anti-inflammatory mechanism of ginsenoside Rh4. Front. Pharmacol. 2022, 13, 953871. [Google Scholar] [CrossRef]
- Murugesan, M.; Mathiyalagan, R.; Boopathi, V.; Kong, B.M.; Choi, S.-K.; Lee, C.-S.; Yang, D.C.; Kang, S.C.; Thambi, T. Production of minor ginsenoside CK from major ginsenosides by biotransformation and its advances in targeted delivery to tumor tissues using nanoformulations. Nanomaterials 2022, 12, 3427. [Google Scholar] [CrossRef] [PubMed]
- Karmazyn, M.; Gan, X.T. Chemical components of ginseng, their biotransformation products and their potential as treatment of hypertension. Mol. Cell Biochem. 2021, 476, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, L.; Fang, X.; Zhao, L. Highly efficient biotransformation of notoginsenoside R1 into ginsenoside Rg1 by Dictyoglomus thermophilum β-xylosidase Xln-DT. J. Microbiol. Biotechnol. 2022, 32, 447. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lin, D.; Li, F.; Cui, X.; Lou, D.; Yang, X. Production of rare ginsenosides by biotransformation of Panax notoginseng saponins using Aspergillus fumigatus. Bioresour. Bioprocess. 2024. [Google Scholar] [CrossRef]
- Dong, J.; Yin, Z.; Su, L.; Yu, M.; Wang, M.; Li, L.; Mao, C.; Lu, T. Comparative pharmacokinetic analysis of raw and steamed Panax notoginseng roots in rats by UPLC-MS/MS for simultaneously quantifying seven saponins. Pharm. Biol. 2021, 59, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Nanda, B.; Nandy, S.; Mukherjee, A.; Pandey, D.K.; Dey, A. Neoteric trends in medicinal plant-AMF association and elicited accumulation of phytochemicals. In Recent Trends in Mycological Research: Volume 2: Environmental and Industrial Perspective; Springer: Cham, Switzerland, 2021; pp. 359–389. [Google Scholar]
- Ran, Z.; Yang, X.; Zhang, Y.; Zhou, J.; Guo, L. Effects of arbuscular mycorrhizal fungi on photosynthesis and biosynthesis of ginsenoside in Panax quinquefolius L. Theor. Exp. Plant Physiol. 2021, 33, 235–248. [Google Scholar] [CrossRef]
- Ran, Z.; Chen, X.; Li, R.; Duan, W.; Zhang, Y.; Fang, L.; Guo, L.; Zhou, J. Transcriptomics and metabolomics reveal the changes induced by arbuscular mycorrhizal fungi in Panax quinquefolius L. J. Sci. Food Agric. 2023, 103, 4919–4933. [Google Scholar] [CrossRef]
- Singh, N.; Sharma, V.; Kalita, R.D. Interaction between plants and endophytes: Evolutionary significance and its role in plants development. In Plant Endophytes and Secondary Metabolites; Academic Press: New York, NY, USA, 2024; pp. 295–312. [Google Scholar]
- Wang, F.; Chen, L.; Chen, S.; Chen, H.; Liu, Y. Characterization of two closely related citrus cultivars using UPLC-ESI-MS/MS-based widely targeted metabolomics. PLoS ONE 2021, 16, e0254759. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, Y.; Duan, Y.; Zhao, Y.; Zhang, D.; Zang, L.; Ya, H. Widely Targeted Metabolomics Analysis of Different Parts of Salsola collina Pall. Molecules 2021, 26, 1126. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E.M. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.-K.; Wu, Y.; Long, X.-N.; You, X.-X.; Chen, D.; Bi, Y.; He, S.; Cao, G.-H. Widely Targeted Metabolomic Analysis Reveals the Improvement in Panax notoginseng Triterpenoids Triggered by Arbuscular Mycorrhizal Fungi via UPLC–ESI–MS/MS. Molecules 2024, 29, 3235. https://doi.org/10.3390/molecules29133235
Zhang X-K, Wu Y, Long X-N, You X-X, Chen D, Bi Y, He S, Cao G-H. Widely Targeted Metabolomic Analysis Reveals the Improvement in Panax notoginseng Triterpenoids Triggered by Arbuscular Mycorrhizal Fungi via UPLC–ESI–MS/MS. Molecules. 2024; 29(13):3235. https://doi.org/10.3390/molecules29133235
Chicago/Turabian StyleZhang, Xing-Kai, Yue Wu, Xian-Nv Long, Xiao-Xu You, Di Chen, Yue Bi, Sen He, and Guan-Hua Cao. 2024. "Widely Targeted Metabolomic Analysis Reveals the Improvement in Panax notoginseng Triterpenoids Triggered by Arbuscular Mycorrhizal Fungi via UPLC–ESI–MS/MS" Molecules 29, no. 13: 3235. https://doi.org/10.3390/molecules29133235







