Comprehensive Analysis of Biomass from Chlorella sorokiniana Cultivated with Industrial Flue Gas as the Carbon Source
Abstract
1. Introduction
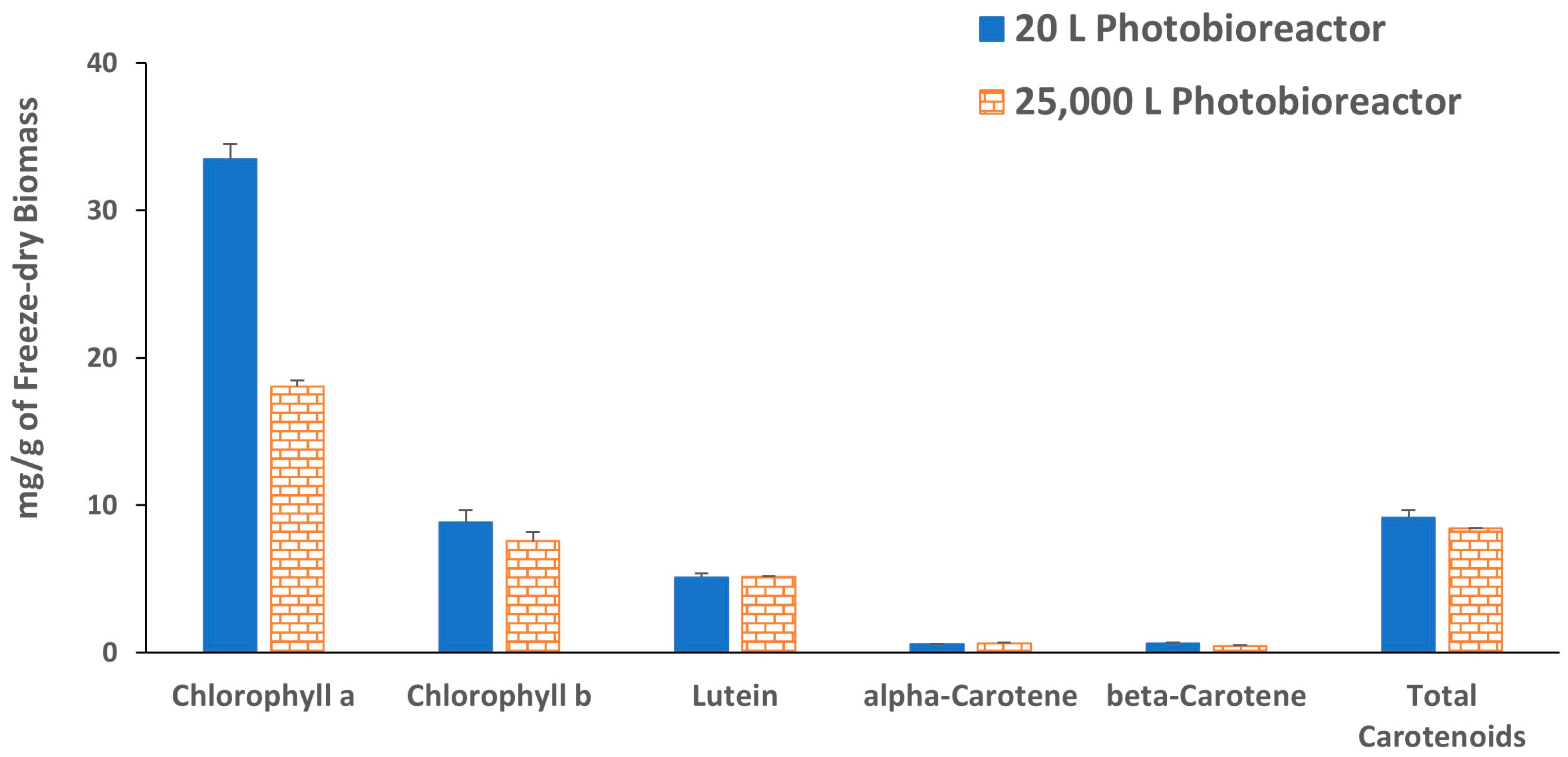
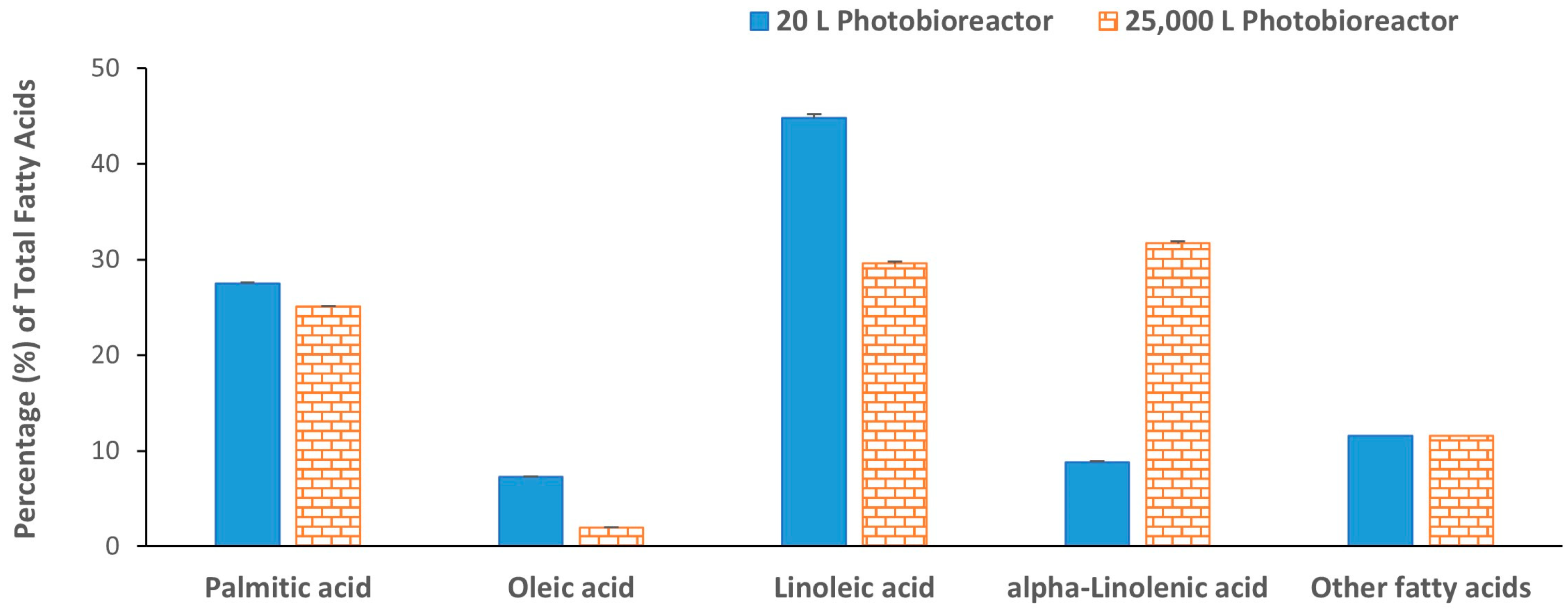
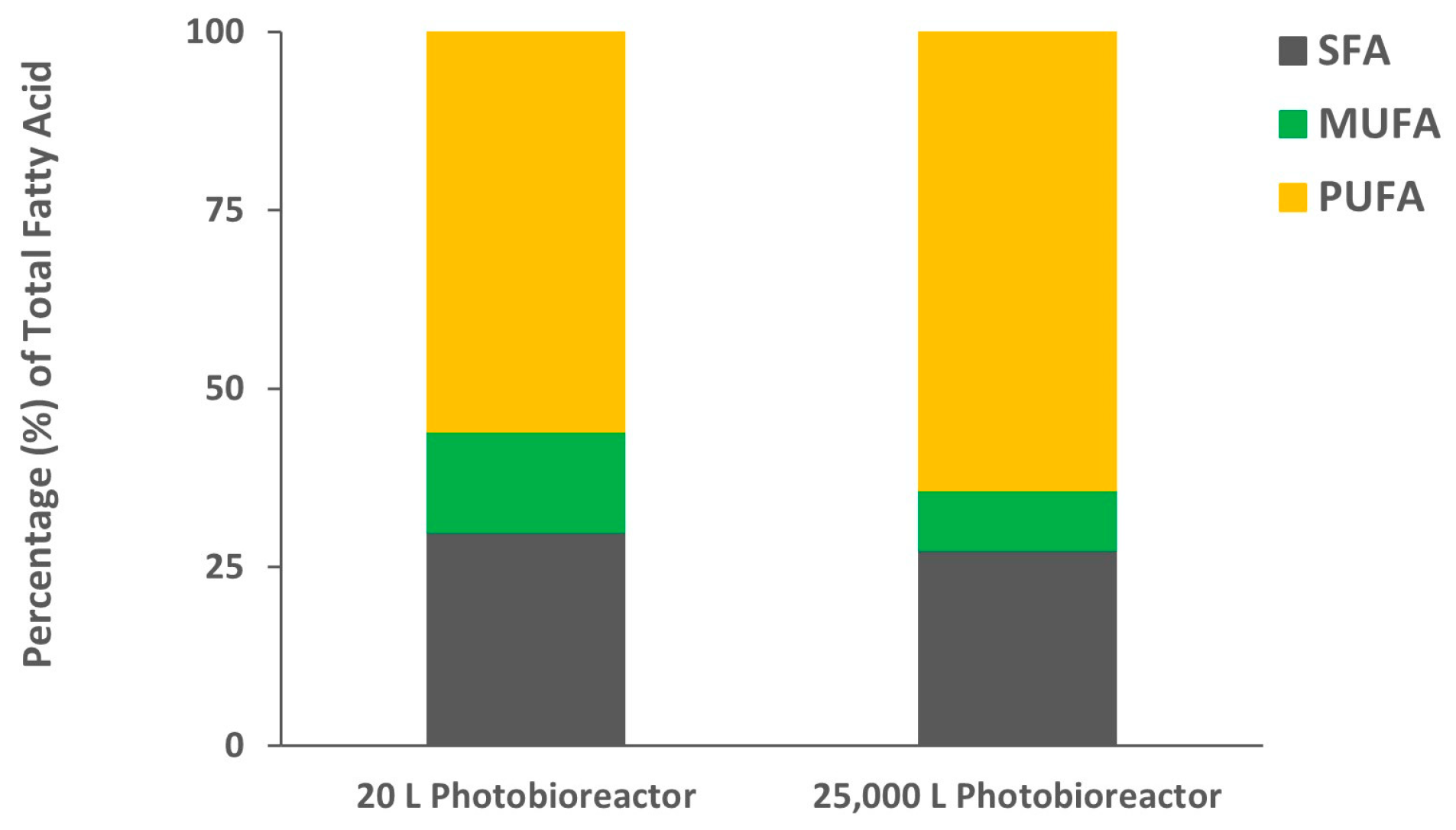
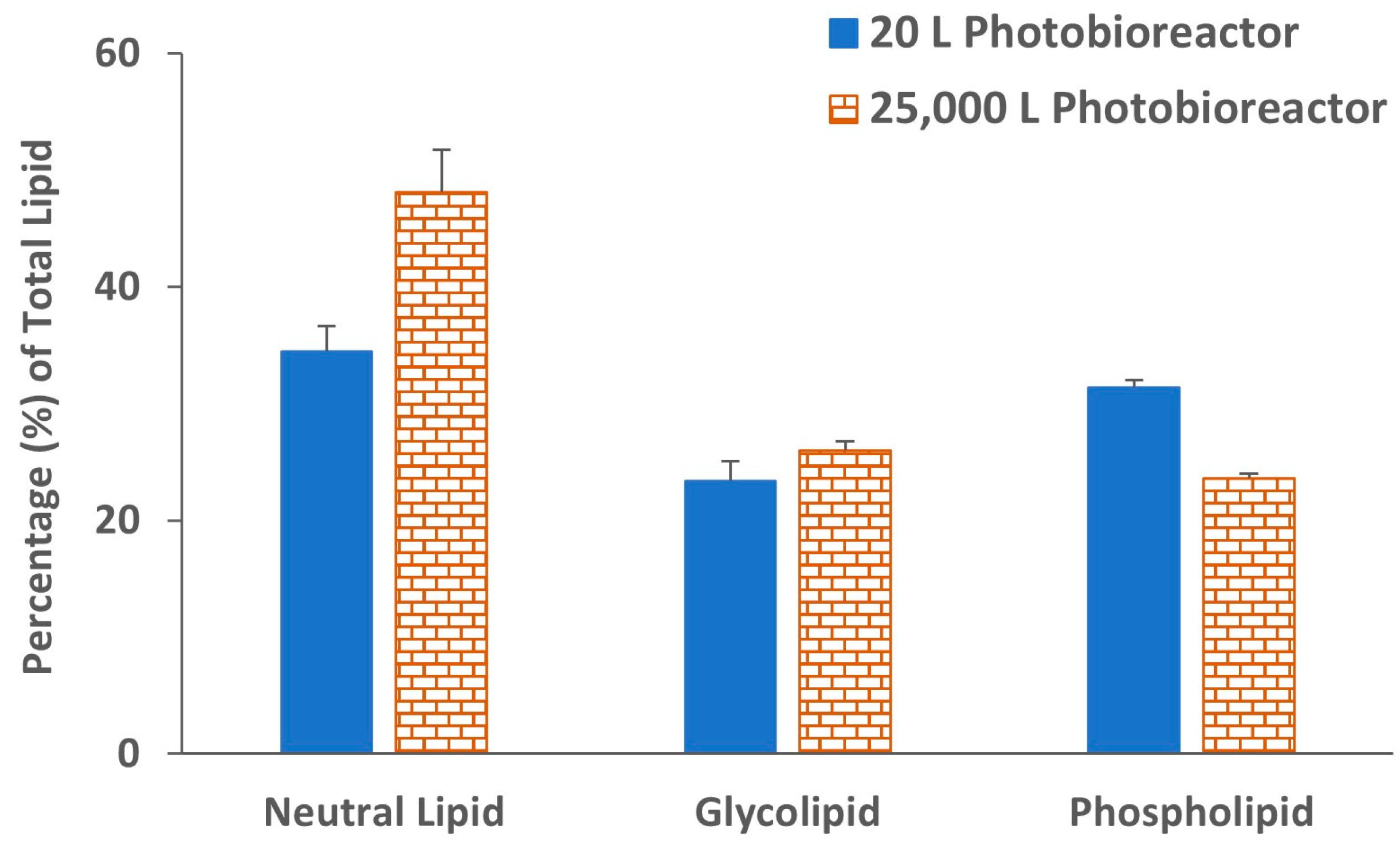
2. Results and Discussion
3. Materials and Methods
3.1. General
3.2. Algal Culture
3.3. Moisture, Ash, Lipid Content and Lipid Class Separation
3.4. Ultra-High-Performance Liquid Chromatography/High-Resolution Mass Spectrometry (UHPLC/HRMS) Analysis
3.5. Fatty Acid Analysis
3.6. Pigment Analysis
3.7. Protein Isolate (PI) and Polysaccharide Extraction
3.8. Amino Acid (AA) Analysis
3.9. Sugar Analysis
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yue, X.-L.; Gao, Q.-X. Contributions of natural systems and human activity to greenhouse gas emissions. Adv. Clim. Chang. Res. 2018, 9, 243–252. [Google Scholar]
- Zhang, S.; Liu, Z. Advances in the biological fixation of carbon dioxide by microalgae. J. Chem. Technol. Biotechnol. 2021, 96, 1475–1495. [Google Scholar] [CrossRef]
- Borowitzka, M.A. High-value products from microalgae-their development and commercialization. J. Appl. Phycol. 2013, 25, 743–756. [Google Scholar] [CrossRef]
- Occhipinti, P.S.; Russo, N.; Foti, P.; Zingale, I.M.; Pino, A.; Romeo, F.V.; Randazzo, C.L.; Caggia, C. Current challenges of microalgae applications: Exploiting the potential of non-conventional microalgae species. J. Sci. Food Agric. 2023, 104, 3823–3833. [Google Scholar] [CrossRef]
- Bito, T.; Okumura, E.; Fujishima, M.; Watanabe, F. Potential of Chlorella as a dietary supplement to promote human health. Nutrients 2020, 12, 2524. [Google Scholar] [CrossRef]
- Andrade, C.J.; Andrade, L.M. An overview on the application of genus Chlorella in biotechnological processes. J. Adv. Biotech. 2017, 2, 1–9. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Lu, J.-C.; Chang, Y.-H.; Chen, J.-H.; Nagarajan, D.; Lee, D.-J.; Chang, J.-S. Optimizing heterotrophic production of Chlorella sorokiniana SU-9 proteins potentially used as s sustainable protein substitute in aquafeed. Bioresour. Technol. 2023, 370, 128538. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, T.; Yu, X.; Bates, P.D.; Dong, T.; Chen, S. High-density fed-batch culture of a thermotolerant microalga Chlorella sorokiniana for biofuel production. Appl. Energy 2013, 108, 281–287. [Google Scholar] [CrossRef]
- Morita, M.; Watanabe, Y.; Saiki, H. High photosynthetic productivity of green microalga Chlorella sorokiniana. Appl. Biochem. Biotechnol. 2000, 87, 203–218. [Google Scholar] [CrossRef]
- Lizzul, A.M.; Lekuona-Amundarain, A.; Purton, S.; Campos, L.C. Characterization of Chlorella soroniniana, UTEX 1230. Biology 2018, 7, 25. [Google Scholar] [CrossRef]
- Cordero, B.F.; Obraztsova, I.; Couso, I.; Vargas, M.A.; Rodriguez, H. Enhancement of lutein production in Chlorella sorokiniana (Chorophyta) by improvement of culture conditions and random mutagenesis. Mar. Drugs 2011, 9, 1607–1624. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Gallant, P.; Osborne, J.; Melanson, R.; O’Leary, S.J.B. Nitric oxide inhibitory activity of monogalactosylmonoacylglycerols from freshwater microalgae Chlorella sorokiniana. Nat. Prod. Res. 2013, 27, 1028–1031. [Google Scholar] [CrossRef]
- Banskota, A.H.; Steevensz, A.; Stefanova, R.; Sperker, S.; Melanson, R.; Osborne, J.A.; O’Leary, S.J.B.; Melanson, J. Monogalactosyldiacylglycerols: Lipase inhibitor from the freshwater microalga Chlorella sorokiniana. J. Appl. Phycol. 2016, 28, 169–175. [Google Scholar] [CrossRef][Green Version]
- Huang, G.; Chen, F.; Kuang, Y.; He, H.; Qun, A. Current technologies of growing algae using flue gas from exhaust gas industry: A review. Appl. Biochem. Biotechnol. 2016, 178, 1220–1238. [Google Scholar] [CrossRef]
- Li, F.-F.; Yang, Z.-H.; Zeng, R.; Yang, G.; Chang, J.-B.; Hou, Y.-L. Microalgae capture of CO2 from actual flue gas discharged from a combustion chamber. Ind. Eng. Chem. Res. 2011, 50, 6496–6502. [Google Scholar] [CrossRef]
- Scapini, T.; Woiciechowski, A.L.; Manzoki, M.C.; Molina-Aulestia, D.T.; Martinez-Burgos, W.J.; Fanka, L.S.; Duda, L.J.; da Silva Vale, A.; Carvalho, J.C.; de Soccol, C.R. Microalgae-mediated biofixation as an innovative technology for flue gases towards carbon neutrality: A comprehensive review. J. Environ. Manag. 2024, 363, 121329. [Google Scholar] [CrossRef]
- Chauhan, D.S.; Sahoo, L.; Mohanty, K. Maximize microbial carbon dioxide utilization and lipid productivity by using toxic flue gas compounds as nutrient source. Bioresour. Technol. 2022, 348, 126784. [Google Scholar]
- Nagappan, S.; Tsai, P.-C.; Devendran, S.; Alagarsamy, V.; Ponnusamy, V.K. Enhancement of biofuel production by microalgae using cement flue gas as substrate. Environ. Sci. Poll. Res. 2020, 27, 17571–17586. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Zhu, T. Research progress and application of carbon sequestration in industrial flue gas by microalgae: A review. J. Environ. Sci. 2025, 152, 14–28. [Google Scholar] [CrossRef]
- Ma, X.; Mi, Y.; Zhao, C.; Wei, Q. A comprehensive review on carbon source effect of microalgae lipid accumulation for biofuel production. Sci. Total Environ. 2022, 806, 151387. [Google Scholar] [CrossRef]
- Zhu, B.; Shen, H.; Li, Y.; Liu, Q.; Jin, G.; Han, J.; Zhao, Y.; Pan, K. Large-scale cultivation of Spirulina for biological CO2 mitigation in open raceway ponds using purified CO2 from a coal chemical flue gas. Front. Bioeng. Biotechnol. 2020, 7, 441. [Google Scholar] [CrossRef]
- Sung, Y.J.; Lee, J.S.; Yoon, H.K.; Ko, H.; Sim, S.J. Outdoor cultivation of microalgae in a coal-fired power plant for conversion of flue gas CO2 into microalgal direct combustion fuels. Syst. Microbiol. Biomanuf. 2021, 1, 90–99. [Google Scholar] [CrossRef]
- Mortensen, L.M.; Gislerod, H.R. The growth of Chlorella sorokiniana as influenced by CO2, light and flue gases. J. Appl. Phycol. 2016, 28, 813–820. [Google Scholar] [CrossRef]
- Kumar, K.; Banerjee, D.; Das, D. Carbon dioxide sequestration from industrial flue gas by Chlorella sorokiniana. Bioresour. Technol. 2014, 152, 225–233. [Google Scholar] [CrossRef]
- Lizzul, A.M.; Hellier, P.; Purton, S.; Baganz, F.; Ladommatos, N.; Campos, L. Combined remediation and lipid production using Chlorella sorokiniana grown on wastewater and exhaust gases. Bioresour. Technol. 2014, 151, 12–18. [Google Scholar] [CrossRef]
- Dickinson, K.E.; Stemmler, K.; Bermarija, T.; Tibbetts, S.M.; MacQuarrie, S.P.; Bhatti, S.; Kozera, C.; O’Leary, S.J.B.; McGinn, P.J. Photosynthetic conversion of carbon dioxide from cement production to microalgae biomass. Appl. Microbio. Biotechnol. 2023, 107, 7375–7390. [Google Scholar] [CrossRef]
- Negi, S.; Barry, A.N.; Friedland, N.; Sudasinghe, N.; Subramanina, S.; Pieris, S.; Holguin, F.O.; Dungan, B.; Schaub, T.; Sayre, R. Impact of nitrogen limitation on biomass, photosynthesis, and lipid accumulation in Chlorella sorokiniana. J. Appl. Phycol. 2016, 28, 803–812. [Google Scholar] [CrossRef]
- Mitra, S.; Rauf, A.; Tareq, A.M.; Jahan, S.; Emran, T.B.; Shahriar, T.G.; Dhama, K.; Alhumaydhi, F.A.; Aljohani, A.S.; Rebezov, M.; et al. Potential health benefits of carotenoid lutein: An updated review. Food Chem. Toxicol. 2021, 154, 112328. [Google Scholar] [CrossRef]
- Hongjin, Q.; Guangce, W. Effect of carbon source on growth and lipid accumulation in Chlorella sorokiniana GXNN01. Chin. J. Oceanol. Limnol. 2009, 27, 762–768. [Google Scholar]
- Qiu, R.; Gao, S.; Lopez, P.A.; Ogden, K.L. Effects of pH on cell growth, lipid production and CO2 addition of microalgae Chlorella sorokiniana. Algal Res. 2017, 28, 192–199. [Google Scholar] [CrossRef]
- He, Q.; Yang, H.; Wu, L.; Hu, C. Effect of light intensity on physiological changes, carbon allocation and neutral lipid accumulation in oleaginous microalgae. Bioresour. Technol. 2015, 191, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.-S.; Kim, Y.-S.; Yoon, H.-S. Characterization of Chlorella sorokiniana and Chlorella vulgaris fatty acid components under a wide range of light intensity and growth temperature for their use as biological resources. Heliyon 2020, 6, e04447. [Google Scholar] [CrossRef]
- Ryckebosch, E.; Muylaert, K.; Foubert, I. Optimization of an analytical procedure for extraction of lipids from microalgae. J. Am. Oil Chem. Soc. 2012, 89, 189–198. [Google Scholar] [CrossRef]
- Banskota, A.H.; Hui, J.P.M.; Jones, A.; McGinn, P.J. Characterization of neutral lipids of the oleaginous alga Micractinum inermum. Molecules 2024, 29, 359. [Google Scholar] [CrossRef]
- Smith, K.M.; Goff, D.A. The NMR spectra of porphyrins. 27—Proton NMR spectra of chlorophyll-a and pheophytin-a. Org. Magn. Reson. 1984, 22, 779–783. [Google Scholar] [CrossRef]
- Melo, T.; Alves, E.; Azevedo, V.; Martins, A.S.; Neves, B.; Domingues, P.; Calado, R.; Abreu, M.H.; Domingues, M.R. Lipidomics as a new approach for the bioprospecting of marine macroalgae—Unraveling the polar lipid and fatty acid composition of Chondrus crispus. Algal Res. 2015, 8, 181–191. [Google Scholar] [CrossRef]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucl. Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef]
- Xu, J.; Chen, D.; Yan, X.; Chen, J.; Zhou, C. Global characterization of the photosynthetic glycolipids from a marine diatom Stephanodiscus sp. by ultra performance liquid chromatography coupled with electrospray ionization-quadrupole-time of flight mass spectrometry. Anal. Chim. Acta 2010, 663, 60–68. [Google Scholar] [CrossRef]
- Guella, G.; Frassanito, R.; Mancini, I. A new solution for an old problem: The regiochemical distribution of acyl chains in galactolipids can be established by electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 1982–1994. [Google Scholar] [CrossRef] [PubMed]
- Widzgowski, J.; Vogel, A.; Altrogge, L.; Pfaff, J.; Schoof, H.; Usadel, B.; Nedbal, L.; Schurr, U.; Pfaff, C. High light induces species specific changes in the membrane lipid composition of Chlorella. Biochem. J. 2020, 477, 2543–2559. [Google Scholar] [CrossRef]
- Junpeng, J.; Xupeng, C.; Miao, Y.; Song, X. Monogalactosyldiacylglycerols with high PUFA content from microalgae for value-added products. Appl. Biochem. Biotechnol. 2020, 190, 1212–1223. [Google Scholar] [CrossRef]
- Christensen, L.P. Galactolipids as potential health promoting compounds in vegetable foods. Recent Pat. Food Nutr. Agric. 2009, 1, 50–58. [Google Scholar] [CrossRef]
- Godzien, J.; Ciborowski, M.; Martínez-Alcázar, M.P.; Samczuk, P.; Kretowski, A.; Barbas, C. Rapid and reliable identification of phospholipids for untargeted metabolomics with LC-ESI-QTOF-MS/MS. J. Proteome Res. 2015, 14, 3204–3216. [Google Scholar] [CrossRef]
- Pi, J.; Wu, X.; Feng, Y. Fragmentation patterns of five types of phospholipids by ultra-high-performance liquid chromatography electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Anal. Methods 2016, 8, 1319–1332. [Google Scholar] [CrossRef]
- Han, X.; Gross, R.W. Structural determination of lysophospholipid regioisomers by electrospray ionization Tandem mass spectrometry. J. Am. Chem. Soc. 1996, 118, 451–457. [Google Scholar] [CrossRef]
- Sun, N.; Chen, J.; Wang, D.; Lin, S. Advance in food-derived phospholipids: Sources, molecular species and structure as well as their biological activities. Trends Food Sci. Technol. 2018, 80, 199–211. [Google Scholar] [CrossRef]
- Barone, R.S.C.; Sonoda, D.Y.; Lorenz, E.K.; Cyrino, E.E.P. Digestivility and pricing of Chlorella sorokiniana meal for use in tilapia feeds. Sci. Agric. 2018, 75, 184–190. [Google Scholar] [CrossRef]
- Ballesteros-Torres, J.M.; Samaniego-Moreno, L.; Gomez-Flores, R.; Tamez-Guerra, R.S.; Rodriguez-Padilla, C.; Tamez-Guerra, P. Amino acids and acylcarnitine production by Chlorella vulgaris and Chlorella sorokiniana microalgae from wastewater culture. Peer J. 2019, 7, e7977. [Google Scholar] [CrossRef]
- Cao, M.; Kang, J.; Cao, Y.; Wang, X.; Pan, X.; Liu, P. Optimization of cultivation conditions for enhancing biomass, polysaccharide and protein yields of Chlorella sorokiniana by response surface methodology. Aquac. Res. 2020, 51, 2456–2471. [Google Scholar] [CrossRef]
- Chakraborty, M.; Miao, C.; McDonald, A.; Chen, S. Concomitant extraction of bio-oil and value added polysaccharides from Chlorella sorokiniana using a unique sequential hydrothermal extraction technology. Fuel 2012, 95, 63–70. [Google Scholar] [CrossRef]
- Chakraborty, M.; McDonald, A.G.; Nindo, C.; Chen, S. An a-glucan isolated as a co-product of biofuel by hydrothermal liquefaction of Chlorella sorokinniana biomass. Algal Res. 2013, 2, 230–236. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.; Jones, A.; Hui, J.P.M.; Stefanova, R.; Burton, J. Analysis of polar lipids in hemp (Cannabis sativa L.) by-products by ultra-high performance liquid chromatography and high-resolution mass spectrometry. Molecules 2022, 27, 5856. [Google Scholar] [CrossRef] [PubMed]
- AOAC 991.39-1995; Fatty Acids in Encapsulated Fish Oils and Fish Oil Methyl and Ethyl Esters. Gas Chromatographic Method. AOAC International: Rockville, MD, USA, 1995.
- White, J.A.; Hart, R.J.; Fry, J.C. An evaluation of the Waters Pico-Tag system for the amino-acid analysis of food materials. J. Autom. Chem. 1986, 8, 170–177. [Google Scholar] [CrossRef] [PubMed]
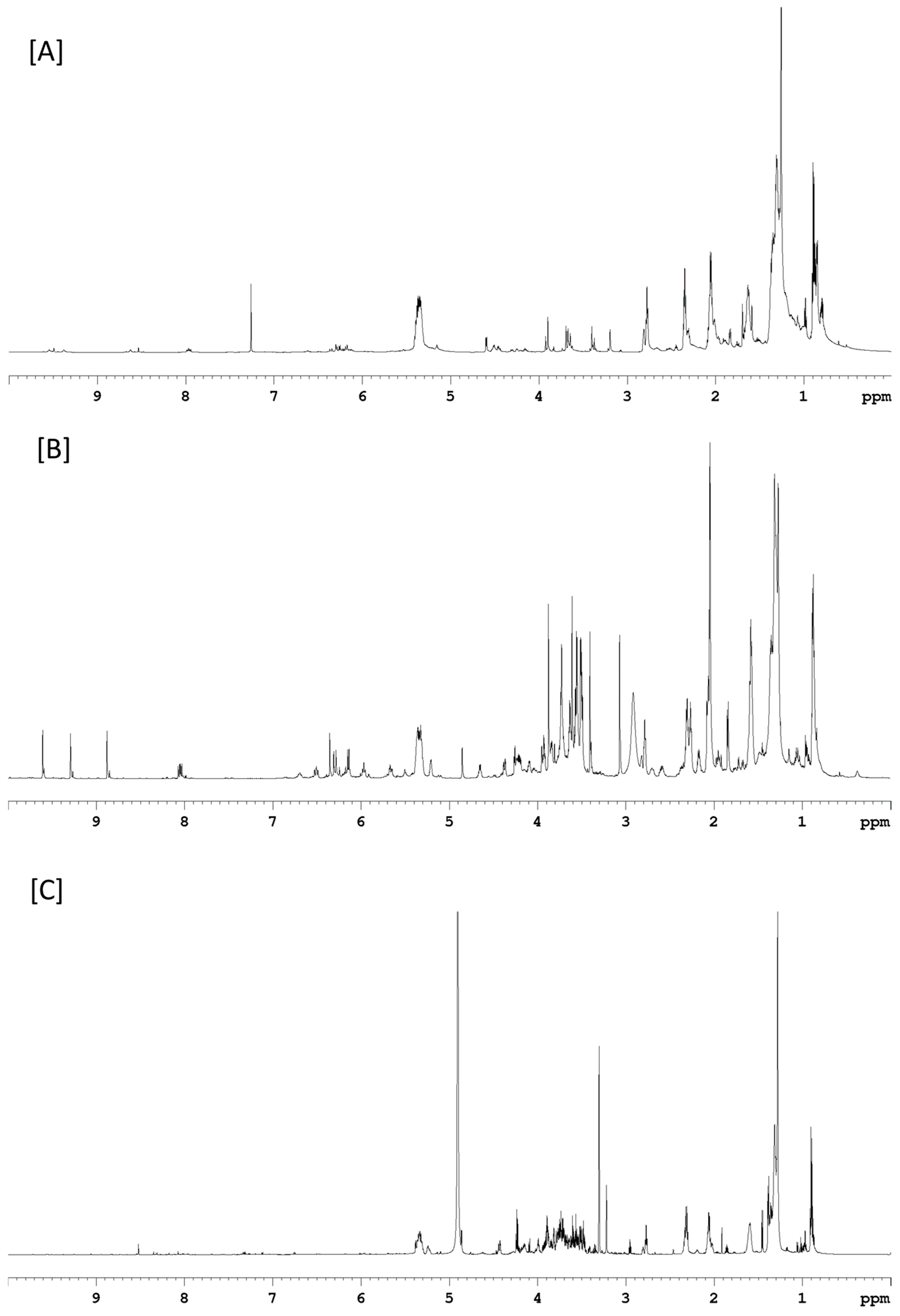
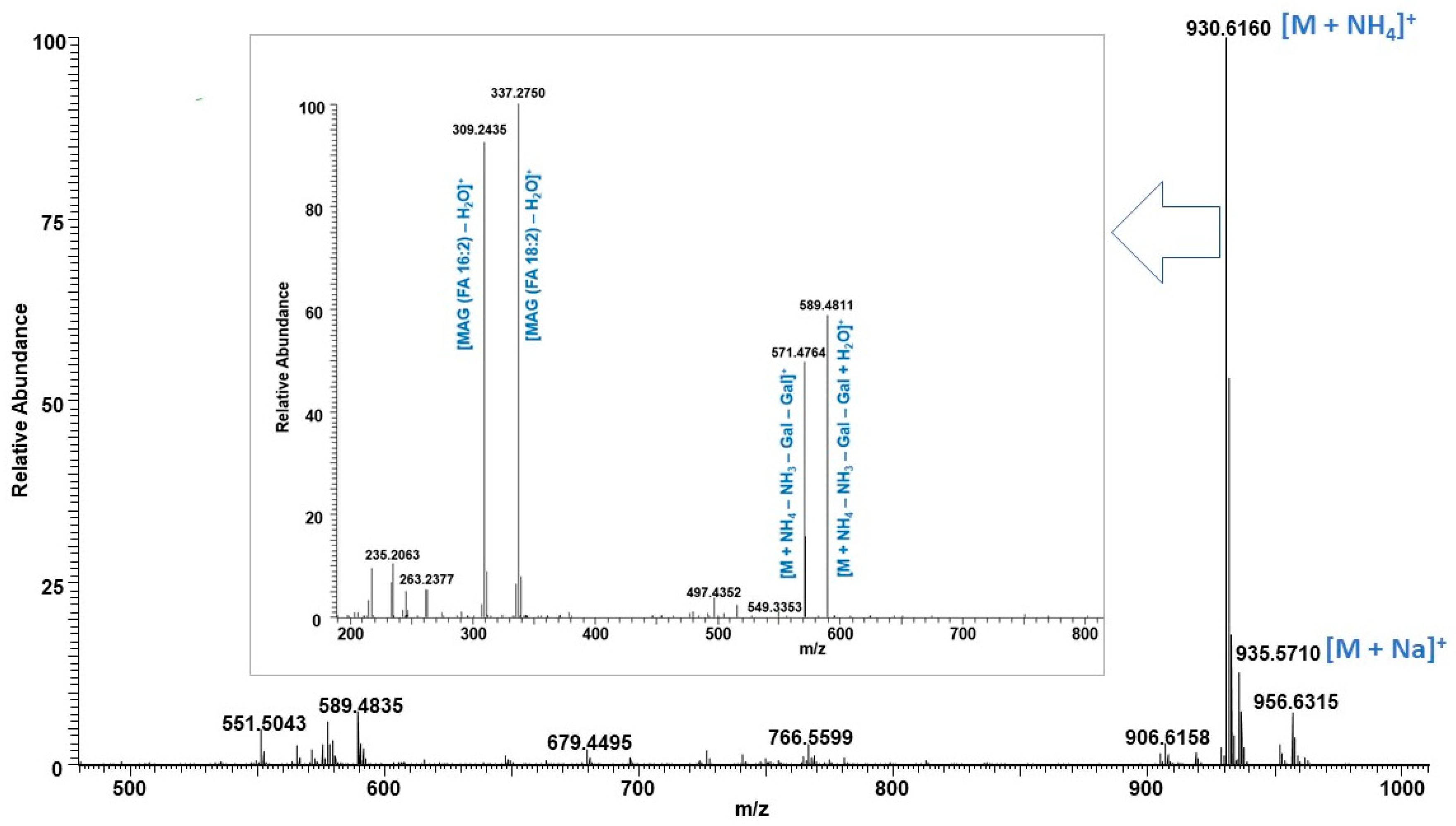


| RT (min) | Measured ms (m/z) | Exact ms (m/z) | Error ppm * | TAG C:DB | TAG Identity | RA 20 L | RA 25,000 L |
|---|---|---|---|---|---|---|---|
| 1.55 | 838.6931 | 838.6919 | −1.4 | 50:7 | 18:3/16:3/16:1; 18:3/18:2/16:2; 18:2/16:3/16:2 | 3.2 | 0.0 |
| 1.55 | 864.7107 | 864.7076 | −3.6 | 52:8 | 18:3/18:1/16:4; 18:3/18:2/16:3 | 1.2 | 0.0 |
| 1.84 | 840.7073 | 840.7076 | 0.4 | 50:6 | 18:3/16:3/16:0; 18:3/16:2/16:1; 18:2/16:3/16:1; 18:2/16:4/16:0 | 7.0 | 6.9 |
| 1.84 | 866.7231 | 866.7232 | 0.1 | 52:7 | 18:3/18:1/16:3; 18:2/18:1/16:4 | 4.1 | 8.1 |
| 1.95 | 894.7540 | 894.7545 | 0.6 | 54:7 | 18:3/18:2/18:3; 18:3/18:3/18:1 | 1.2 | 3.0 |
| 2.10 | 816.7070 | 816.7076 | 0.7 | 48:4 | 16:4/16:0/16:0; 16:3:16:1/16:0; 16:2/16:2/16:0 | 4.0 | 1.5 |
| 2.10 | 842.7226 | 842.7232 | 0.7 | 50:5 | 18:3/16:2/16:0; 18:3/16:1/16:1; 18:2/16:3/16:0; 18:2/16:2/16:1; 18:1/16:3/16:2; 18:1/16:4/16:0 | 8.0 | 7.5 |
| 2.10 | 868.7383 | 868.7389 | 0.7 | 52:6 | 18:3/18:2/16:1; 18:3/18:3/16:0; 18:1/18:0/16:5; 18:3/18:1/16:2; 18:1/18:1/16:4; 18:2/18:1/16:3 | 5.5 | 7.7 |
| 2.35 | 818.7233 | 818.7232 | −0.1 | 48:3 | 16:3/16:0/16:0; 16:2/16:1/16:0 | 2.7 | 1.9 |
| 2.35 | 844.7388 | 844.7389 | 0.1 | 50:4 | 18:1/16:3/16:0; 18:1/16:2/16:1; 18:2/16:2/16:0; 18:2/16:1/16:1; 18:3/16:1/16:0 | 7.1 | 5.4 |
| 2.35 | 870.7543 | 870.7545 | 0.2 | 52:5 | 18:3/18:2/16:0; 18:2/18:2/16:1; 18:3/18:1/16:1; 18:2/18:1/16:2; 18:1/18:1/16:3; 18:0/16:2/16:3 | 4.4 | 7.8 |
| 2.35 | 896.7702 | 896.7702 | 0.0 | 54:6 | 18:3/18:2/18:1 | 1.4 | 3.2 |
| 2.60 | 820.7392 | 820.7389 | −0.4 | 48:2 | 16:2/16:0/16:0; 16:1/16:1/16:0; 18:2/16:0/14:0; 18:1/16:0/14:1; 18:1/16:1/14:0 | 5.3 | 1.8 |
| 2.85 | 898.7859 | 898.7858 | −0.1 | 54:5 | 18:3/18:1/18:1; 18:3/18:2/18:0; 18:2/18:2/18:1 | 1.4 | 2.9 |
| 2.85 | 846.7547 | 846.7545 | −0.2 | 50:3 | 18:3/16:0/16:0; 18:2/16:1/16:0; 18:1/16:1/16:1; 18:0/16:2/16:1 | 6.5 | 4.4 |
| 2.85 | 872.7702 | 872.7702 | 0.0 | 52:4 | 18:3/18:1/16:0; 18:2/18:2/16:0 (major); 18:3/18:0/16:0 | 9.6 | 9.2 |
| 3.10 | 900.8013 | 900.8015 | 0.2 | 54:4 | 18:2/18:2/18:0 (major); 18:3/18:1/18:0 | 1.2 | 2.7 |
| 3.10 | 874.7855 | 874.7858 | 0.3 | 52:3 | 18:2/18:1/16:0; 18:2/18:0/16:1 | 4.6 | 6.9 |
| 3.10 | 848.7704 | 848.7702 | −0.2 | 50:2 | 18:2/16:0/16:0 (major); 18:1/16:1/16:0 | 12.3 | 6.0 |
| 3.10 | 862.7859 | 862.7858 | −0.1 | 51:2 | 18:1/17:1:16:0; 18:2/17:0:16:0; | 3.0 | 1.5 |
| 3.10 | 888.8016 | 888.8015 | −0.1 | 53:3 | 18:2:/18:1/17:0 | 0.7 | 0.9 |
| 3.70 | 850.7860 | 850.7858 | −0.2 | 50:1 | 18:1/16:0/16:0 | 1.9 | 2.8 |
| 3.70 | 876.8016 | 876.8015 | −0.1 | 52:2 | 18:1/18:1/16:0 | 2.7 | 4.5 |
| 3.70 | 902.8174 | 902.8171 | −0.3 | 54:3 | 18:1/18:1/18:1 (major); 18:2/18:1/18:0 (minor) | 0.5 | 2.0 |
| 3.90 | 878.8168 | 878.8171 | 0.3 | 52:1 | 18:1/18:0/16:0 | 0.5 | 1.5 |
 high abundance.
high abundance.| RT (min) | Measured ms (m/z) | Exact ms (m/z) | Error ppm * | Lipid Identity | Fatty Acid Position R1/R2 | RA 20 L | RA 25,000 L |
|---|---|---|---|---|---|---|---|
| Free Fatty Acids | |||||||
| 1.25 | 249.1850 | 249.1849 | −0.4 | FA 16:4 | ND | ND | |
| 1.55 | 251.2005 | 251.2006 | 0.4 | FA 16:3 | ND | ND | |
| 1.95 | 277.2162 | 277.2162 | 0.0 | FA 18:4 | ND | ND | |
| 2.40 | 279.2318 | 279.2319 | 0.4 | FA 18:3 | ND | ND | |
| MGDGs and DGDGs | |||||||
| 5.83 | 738.5156 | 738.5151 | −0.7 | MGDG 32:5 | 16:2/16:3 | 0.9 | 6.9 |
| 5.99 | 740.5312 | 740.5307 | −0.7 | MGDG 32:4 | 16:2/16:2 | 3.4 | 6.2 |
| 6.07 | 766.5477 | 766.5464 | −1.7 | MGDG 34:5 | 18:3/16:2 | 4.8 | 13.3 |
| 6.23 | 768.5622 | 768.5620 | −0.3 | MGDG 34:4 | 18:2/16:2 | 15.1 | 12.5 |
| 6.38 | 770.5777 | 770.5777 | 0.0 | MGDG 34:3 | 18:2/16:1 # | 3.0 | 3.1 |
| 6.51 | 772.5936 | 772.5933 | −0.4 | MGDG 34:2 | 18:2/16:0 # | 1.7 | 1.2 |
| 6.60 | 764.5437 | 764.5307 | 17.0 | MGDG 34:6 | 18:3/16:3 | 0.0 | 0.0 |
| 6.68 | 774.6092 | 774.6090 | −0.3 | MGDG 34:1 | NI | 0.9 | 0.6 |
| 6.82 | 776.6246 | 776.6246 | 0.0 | MGDG 34:0 | 18:0/16:0 # | 1.1 | 0.6 |
| 5.85 | 926.5837 | 926.5835 | −0.2 | DGDG 34:6 | 18:3/16:3 # | 1.2 | 11.6 |
| 902.5841 | 902.5835 | 0.7 | DGDG 32:4 | 16:2/16:2, 16:3/16:1 # | 1.2 | 1.3 | |
| 5.94 | 928.5995 | 928.5992 | −0.3 | DGDG 34:5 | 18:3/16:2; 18:2/16:3 | 7.4 | 11.7 |
| 6.10 | 930.6147 | 930.6148 | 0.1 | DGDG 34:4 | 18:2/16:2 | 34.4 | 18.9 |
| 6.10 | 932.6299 | 932.6305 | 0.6 | DGDG 34:3 | 18:3/16:0; 18:2/16:1; 18:1/16:2 # | 7.9 | 3.5 |
| 6.26 | 958.6458 | 958.6461 | 0.3 | DGDG 36:4 | 18:2/18:2 | 4.3 | 2.5 |
| 6.45 | 934.6463 | 934.6461 | −0.2 | DGDG 34:2 | 18:2/16:0 | 9.2 | 4.8 |
| 6.45 | 936.6618 | 936.6618 | 0.0 | DGDG 34:1 | 18:1/16:0 # | 3.6 | 1.4 |
| SQDG | |||||||
| 5.94 | 812.5552 | 812.5552 | SQDG 32:0 | 16:0/16:0 | |||
 high abundance.
high abundance.| RT (min) | Observed Mass (m/z) | Exact Mass (m/z) | Error ppm * | Lipid Identity | Fatty Acid Position R1/R2 | RA 20 L | RA 25,000 L |
|---|---|---|---|---|---|---|---|
| LPCs and PCs | |||||||
| 2.55 | 490.2929 | 490.2928 | −0.2 | LPC 16:3 | 16:3/0:0 | 0.1 | 3.4 |
| 3.10 | 492.3086 | 492.3085 | −0.2 | LPC 16:2 | 16:2/0:0 | 1.0 | 2.9 |
| 3.10 | 494.3252 | 494.3241 | −2.2 | LPC 16:1 | 16:1/0:0 | 0.2 | 0.9 |
| 3.45 | 518.3244 | 518.3241 | −0.6 | LPC 18:3 | 18:3/0:0 | 0.4 | 9.0 |
| 3.96 | 520.3402 | 520.3398 | −0.8 | LPC 18:2 | 18:2/0:0 | 6.2 | 7.3 |
| 4.25 | 496.3400 | 496.3398 | −0.4 | LPC 16:0 | 16:0/0:0 | 1.9 | 6.0 |
| 4.48 | 522.3582 | 522.3554 | −5.4 | LPC 18:1 | 18:1/0:0 | 0.0 | 0.6 |
| 4.94 | 524.3742 | 524.3711 | −5.9 | LPC 18:0 | 18:0/0:0 | 0.1 | 0.0 |
| 5.30 | 766.5019 | 766.5017 | −0.3 | PC 34:6; O | NI | 0.0 | 3.8 |
| 5.50 | 768.5175 | 768.5174 | −0.1 | PC 34:5;O | NI | 0.8 | 3.5 |
| 5.50 | 794.5332 | 794.5330 | −0.3 | PC 36:6; O | NI | 0.3 | 2.7 |
| 6.40 | 780.5535 | 780.5538 | 0.4 | PC 36:5 | NI | 9.3 | 8.4 |
| 5.50 | 754.5379 | 754.5381 | 0.3 | PC 34:4 | NI | 4.1 | 8.6 |
| 5.50 | 728.5227 | 728.5225 | −0.3 | PC 32:3 | NI | 7.0 | 9.9 |
| 6.60 | 782.5695 | 782.5694 | −0.1 | PC 36:4 | NI | 11.4 | 10.8 |
| 6.60 | 756.5539 | 756.5538 | −0.1 | PC 34:3 | NI | 12.0 | 8.3 |
| 6.95 | 784.5853 | 784.5851 | −0.3 | PC 36:3 | NI | 41.1 | 11.3 |
| 6.80 | 758.5696 | 758.5694 | −0.3 | PC 34:2 | NI | 4.1 | 2.7 |
| MGDG and DGDGs | |||||||
| 5.92 | 764.5437 | 764.5307 | 17.0 | MGDG 34:6 | 18:3/16:3 | 5.1 | 29.8 |
| 5.92 | 926.5840 | 926.5835 | −0.5 | DGDG 34:6 | 18:3/16:3 | 1.2 | 12.3 |
| 5.94 | 902.5841 | 902.5835 | −0.7 | DGDG 32:4 | 16:2/16:2; 16:3/16:1 # | 1.7 | 1.7 |
| 5.94 | 928.5988 | 928.5992 | 0.4 | DGDG 34:5 | 18:3/16:2; 18:2/16:3 # | 11.8 | 22.2 |
| 6.14 | 930.6156 | 930.6148 | −0.9 | DGDG 34:4 | 18:2/16:2 | 49.5 | 20.0 |
| 6.24 | 932.6296 | 932.6305 | 1.0 | DGDG 34:3 | 18:3/16:0; 18:2/16:1; 18:1/16:2 # | 8.0 | 3.8 |
| 6.25 | 958.6458 | 958.6461 | 0.3 | DGDG 36:4 | 18:2/18:2 | 4.4 | 2.6 |
| 6.38 | 934.6461 | 934.6461 | 0.0 | DGDG 34:2 | 18:2/16:0 | 12.9 | 6.1 |
| 6.50 | 936.6614 | 936.6618 | 0.4 | DGDG 34:1 | 18:1/16:0 | 5.4 | 1.6 |
| SQDG | |||||||
| 5.94 | 812.5552 | 812.5552 | 0.0 | SQDG 32:0 | 16:0/16:0 | ||
 high abundance.
high abundance.| Amino Acids (AA) | 20 L Photobioreactor | 25,000 L Photobioreactor |
|---|---|---|
| Aspartic acid (Asp) | 59.6 ± 2.1 | 66.4 ± 3.1 |
| Serine (Ser) | 25.0 ± 0.2 | 34.9 ± 1.7 |
| Glutamic acid (Glu) | 75.4 ± 1.9 | 84.8 ± 4.4 |
| Glycine (Gly) | 50.7 ± 1.3 | 59.1 ± 2.7 |
| Histidine (His) | 22.9 ± 0.6 | 26.2 ± 1.4 |
| Arginine (Arg) | 56.1 ± 2.6 | 66.9 ± 2.8 |
| Threonine (Thr) | 24.7 ± 0.5 | 33.8 ± 1.6 |
| Alanine (Ala) | 48.0 ± 0.5 | 53.9 ± 2.7 |
| Proline (Pro) | 37.2 ± 0.4 | 45.3 ± 2.2 |
| Cysteine (Cys) | - | - |
| Tyrosine (Tyr) | 48.0 ± 2.1 | 57.6 ± 2.3 |
| Valine (Val) | 52.1 ± 1.5 | 59.3 ± 3.5 |
| Methionine (Met) | 21.2 ± 2.3 | 22.5 ± 3.0 |
| Lysine (Lys) | 34.9 ± 2.2 | 41.6 ± 2.6 |
| Isoleucine (Ile) | 37.5 ± 1.2 | 39.7 ± 2.4 |
| Leucine (Leu) | 86.9 ± 1.3 | 89.1 ± 4.3 |
| Phenylalanine (Phe) | 68.7 ± 2.6 | 71 ± 3.8 |
| Total amino acids (mg/g) | 748.9 ± 6.4 | 852.2 ± 39.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banskota, A.H.; Stefanova, R.; Hui, J.P.M.; Bermarija, T.; Stemmler, K.; McGinn, P.J.; O’Leary, S.J.B. Comprehensive Analysis of Biomass from Chlorella sorokiniana Cultivated with Industrial Flue Gas as the Carbon Source. Molecules 2024, 29, 3368. https://doi.org/10.3390/molecules29143368
Banskota AH, Stefanova R, Hui JPM, Bermarija T, Stemmler K, McGinn PJ, O’Leary SJB. Comprehensive Analysis of Biomass from Chlorella sorokiniana Cultivated with Industrial Flue Gas as the Carbon Source. Molecules. 2024; 29(14):3368. https://doi.org/10.3390/molecules29143368
Chicago/Turabian StyleBanskota, Arjun H., Roumiana Stefanova, Joseph P. M. Hui, Tessa Bermarija, Kevin Stemmler, Patrick J. McGinn, and Stephen J. B. O’Leary. 2024. "Comprehensive Analysis of Biomass from Chlorella sorokiniana Cultivated with Industrial Flue Gas as the Carbon Source" Molecules 29, no. 14: 3368. https://doi.org/10.3390/molecules29143368
APA StyleBanskota, A. H., Stefanova, R., Hui, J. P. M., Bermarija, T., Stemmler, K., McGinn, P. J., & O’Leary, S. J. B. (2024). Comprehensive Analysis of Biomass from Chlorella sorokiniana Cultivated with Industrial Flue Gas as the Carbon Source. Molecules, 29(14), 3368. https://doi.org/10.3390/molecules29143368









