Research Progress on Detection of Pathogens in Medical Wastewater by Electrochemical Biosensors
Abstract
1. Introduction
2. Pathogens in Medical Wastewater
2.1. Bacteria
2.1.1. Escherichia coli
2.1.2. Salmonella
2.1.3. Staphylococcus aureus
2.1.4. Clostridium difficile
2.2. Viruses
2.2.1. Enterovirus
2.2.2. Respiratory Viruses
2.2.3. Hepatitis Virus
2.3. Parasites
2.4. Fungi
3. Electrochemical Biosensors for Detecting Pathogens in Medical Wastewater
3.1. Volt-Ampere Type Biosensor
3.2. Conductive Biosensor
3.3. Impedance Biosensor
3.4. Photoelectric Chemical Biosensor
3.5. Electrochemical Luminescent Biosensor
| Detection Method | Target Detector | LOD | References |
|---|---|---|---|
| Volt-ampere type biosensor | E. coli O157 | 2.5 × 10−2 CFU/L | [91] |
| Volt-ampere type biosensor | Schistosomiasis antigen | 1.13 × 101~2.3 × 103 ng/mL | [92] |
| Conductive biosensor | E. coli O157 | 1 × 10−2~1 × 10−1 cells/L | [97] |
| Conductive biosensor | Bacillus anthracis | 1 × 10−3 CFU/L | [98] |
| Impedance biosensor | Salmonella typhi | 7.3 × 10−2 CFU/L | [103] |
| Impedance biosensor | E. coli DH5a, Enterobacter cloacae, B. subtilis, Saccharomycetes | high sensitivity | [104] |
| Photoelectric chemical biosensor | E. coli | 4.6 × 101 CFU/L | [105] |
| Photoelectric chemical biosensor | Vibrio parahemolyticus | 2.5 × 101 CFU/L | [106] |
| Electrochemical luminescent biosensor | Shiga toxin E. coli | 0.3 pmol/L | [113] |
| Electrochemical luminescent biosensor | Mycotoxin | 0.034 pg/mL | [115] |
4. Discussion and Prospect
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mackuľak, T.; Cverenkárová, K.; Vojs Staňová, A.; Fehér, M.; Tamáš, M.; Škulcová, A.B.; Gál, M.; Naumowicz, M.; Špalková, V.; Bírošová, L. Hospital Wastewater—Source of Specific Micropollutants, Antibiotic-Resistant Microorganisms, Viruses, and Their Elimination. Antibiotics 2021, 10, 1070. [Google Scholar] [CrossRef]
- Silverman, A.I.; Boehm, A.B. Systematic Review and Meta-Analysis of the Persistence of Enveloped Viruses in Environmental Waters and Wastewater in the Absence of Disinfectants. Environ. Sci. Technol. 2021, 55, 14480–14493. [Google Scholar] [CrossRef]
- Prado, T.; Silva, D.M.; Guilayn, W.C.; Rose, T.L.; Gaspar, A.M.C.; Miagostovich, M.P. Quantification and molecular characterization of enteric viruses detected in effluents from two hospital wastewater treatment plants. Water Res. 2011, 45, 1287–1297. [Google Scholar] [CrossRef]
- Varela, A.R.; Manaia, C.M. Human health implications of clinically relevant bacteria in wastewater habitats. Environ. Sci. Pollut. Res. 2013, 20, 3550–3569. [Google Scholar] [CrossRef]
- Hocquet, D.; Muller, A.; Bertrand, X. What happens in hospitals does not stay in hospitals: Antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef]
- Arroyo, M.G.; Ferreira, A.M.; Frota, O.P.; Brizzotti-Mazuchi, N.S.; Peresi, J.T.M.; Rigotti, M.A.; Macedo, C.E.; Sousa, A.F.L.d.; Andrade, D.d.; Almeida, M.T.G.d. Broad Diversity of Fungi in Hospital Water. Sci. World J. 2020, 2020, 9358542. [Google Scholar] [CrossRef]
- Ogwugwa, V.H.; Oyetibo, G.O.; Amund, O.O. Taxonomic profiling of bacteria and fungi in freshwater sewer receiving hospital wastewater. Environ. Res. 2021, 192, 110319. [Google Scholar] [CrossRef]
- Freudenthal, J.; Ju, F.; Bürgmann, H.; Dumack, K. Microeukaryotic gut parasites in wastewater treatment plants: Diversity, activity, and removal. Microbiome 2022, 10, 27. [Google Scholar] [CrossRef]
- Pauwels, B.; Verstraete, W. The treatment of hospital wastewater: An appraisal. J. Water Health 2006, 4, 405–416. [Google Scholar] [CrossRef]
- Shen, Z.; Liu, H.; Zhang, Y. The Current Situation, Issues and Strategies of “Toilet Revolution” in China. Chin. J. Environ. Manag. 2018, 10, 45–48. [Google Scholar]
- Akin, B.S. Contaminant Properties of Hospital Clinical Laboratory Wastewater: A Physiochemical and Microbiological Assessment. J. Environ. Prot. 2016, 7, 635–642. [Google Scholar] [CrossRef]
- Cai, L.; Zhang, T. Detecting human bacterial pathogens in wastewater treatment plants by a high-throughput shotgun sequencing technique. Environ. Sci. Technol. 2013, 47, 5433–5441. [Google Scholar] [CrossRef]
- Olaolu, D.T.; Akpor, O.B.; Akor, C.O. Pollution indicators and pathogenic microorganisms in wastewater treatment: Implication on receiving water bodies. Int. J. Environ. Prot. Policy 2014, 2, 205–212. [Google Scholar] [CrossRef]
- Fukuda, S.; Ohno, H. Gut microbiome and metabolic diseases. Semin. Immunopathol. 2014, 36, 103–114. [Google Scholar] [CrossRef]
- Harris, K.G.; Chang, E.B. The intestinal microbiota in the pathogenesis of inflammatory bowel diseases: New insights into complex disease. Clin. Sci. 2018, 132, 2013–2028. [Google Scholar] [CrossRef]
- Cipe, G.; Idiz, U.O.; Firat, D.; Bektasoglu, H. Relationship between intestinal microbiota and colorectal cancer. World J. Gastrointest. Oncol. 2015, 7, 233–240. [Google Scholar] [CrossRef]
- Ren, L.; Ye, J.; Zhao, B.; Sun, J.; Cao, P.; Yang, Y. The Role of Intestinal Microbiota in Colorectal Cancer. Front. Pharmacol. 2021, 12, 674807. [Google Scholar] [CrossRef]
- Ramírez-Coronel, A.A.; Mohammadi, M.J.; Majdi, H.S.; Zabibah, R.S.; Taherian, M.; Prasetio, D.B.; Gabr, G.A.; Asban, P.; Kiani, A.; Sarkohaki, S. Hospital wastewater treatment methods and its impact on human health and environments. Rev. Environ. Health 2023, ahead of print. [CrossRef]
- Carraro, E.; Bonetta, S.; Bertino, C.; Lorenzi, E.; Bonetta, S.; Gilli, G. Hospital effluents management: Chemical, physical, microbiological risks and legislation in different countries. J. Environ. Manag. 2016, 168, 185–199. [Google Scholar] [CrossRef]
- Khan, M.T.; Shah, I.A.; Ihsanullah, I.; Naushad, M.; Ali, S.; Shah, S.H.A.; Mohammad, A.W. Hospital wastewater as a source of environmental contamination: An overview of management practices, environmental risks, and treatment processes. J. Water Process Eng. 2021, 41, 101990. [Google Scholar] [CrossRef]
- Pramanik, S.; McEvoy, J.; Siripattanakul, S.; Khan, E. Effects of cell entrapment on nucleic acid content and microbial diversity of mixed cultures in biological wastewater treatment. Bioresour. Technol. 2011, 102, 3176–3183. [Google Scholar] [CrossRef]
- Doron, S.; Horowitz, G. Medical appropriateness and economics of nucleic acid amplification testing for infectious diseases. Clin. Biochem. 2023, 117, 48–52. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Nguyen, A.; McDonald, D.; Zong, Y.; Ronquillo, N.; Ren, J.; Zou, J.; Farmer, S.; Humphrey, G.; Henderson, D.; et al. Rapid, Large-Scale Wastewater Surveillance and Automated Reporting System Enable Early Detection of Nearly 85% of COVID-19 Cases on a University Campus. mSystems 2021, 6, e0079321. [Google Scholar] [CrossRef]
- Basu, P.; Choudhury, S.; Shridhar, V.; Huilgol, P.; Roychoudhury, S.; Nandi, I.; Chaudhuri, A.; Mitra, A. Surveillance of SARS-CoV-2 RNA in open-water sewage canals contaminated with untreated wastewater in resource-constrained regions. Access Microbiol. 2022, 4, 000318. [Google Scholar] [CrossRef]
- Hui, Q.; Pan, Y.; Yang, Z. Paper-based devices for rapid diagnostics and testing sewage for early warning of COVID-19 outbreak. Case Stud. Chem. Environ. Eng. 2020, 2, 100064. [Google Scholar] [CrossRef]
- Zhang, C.; Yu, Z.; Wang, X. A review of electrochemical oxidation technology for advanced treatment of medical wastewater. Front. Chem. 2022, 10, 1002038. [Google Scholar] [CrossRef]
- El Ekiaby, M.; Lelie, N.; Allain, J.P. Nucleic acid testing (NAT) in high prevalence-low resource settings. Biol. J. Int. Assoc. Biol. Stand. 2010, 38, 59–64. [Google Scholar] [CrossRef]
- Jain, R.; Aggarwal, P.; Gupta, G.N. Need for nucleic Acid testing in countries with high prevalence of transfusion-transmitted infections. ISRN Hematol. 2012, 2012, 718671. [Google Scholar] [CrossRef]
- Abe, M. Pitfall of Laboratory Testing -Immunological Test -. Rinsho Byori Jpn. J. Clin. Pathol. 2016, 64, 573–580. [Google Scholar]
- Campaña, A.L.; Florez, S.L.; Noguera, M.J.; Fuentes, O.P.; Ruiz Puentes, P.; Cruz, J.C.; Osma, J.F. Enzyme-Based Electrochemical Biosensors for Microfluidic Platforms to Detect Pharmaceutical Residues in Wastewater. Biosensors 2019, 9, 41. [Google Scholar] [CrossRef]
- Yáñez-Sedeño, P.; Agüí, L.; Campuzano, S.; Pingarrón, J.M. What Electrochemical Biosensors Can Do for Forensic Science? Unique Features and Applications. Biosensors 2019, 9, 127. [Google Scholar] [CrossRef]
- Crini, G.; Lichtfouse, E. Advantages and disadvantages of techniques used for wastewater treatment. Environ. Chem. Lett. 2019, 17, 145–155. [Google Scholar] [CrossRef]
- Sikosana, M.L.; Sikhwivhilu, K.; Moutloali, R.; Madyira, D.M. Municipal wastewater treatment technologies: A review. Procedia Manuf. 2019, 35, 1018–1024. [Google Scholar] [CrossRef]
- Donnenberg, M.S.; Finlay, B.B. Combating enteropathogenic Escherichia coli (EPEC) infections: The way forward. Trends Microbiol. 2013, 21, 317–319. [Google Scholar] [CrossRef]
- Lefoll, C.; Caubet, C.; Tasca, C.; Milon, A.; Boullier, S. Simultaneous inactivation of espB and tir abrogates the strong, but non-protective, inflammatory response induced by EPEC. Vet. Immunol. Immunopathol. 2010, 138, 34–44. [Google Scholar] [CrossRef]
- Dutta, S.; Pazhani, G.P.; Nataro, J.P.; Ramamurthy, T. Heterogenic virulence in a diarrheagenic Escherichia coli: Evidence for an EPEC expressing heat-labile toxin of ETEC. Int. J. Med. Microbiol. 2015, 305, 47–54. [Google Scholar] [CrossRef]
- Devane, M.L.; Moriarty, E.; Weaver, L.; Cookson, A.; Gilpin, B. Fecal indicator bacteria from environmental sources; strategies for identification to improve water quality monitoring. Water Res. 2020, 185, 116204. [Google Scholar] [CrossRef]
- Olapade, O.A.; Weage, E.A. Comparison of fecal indicator bacterial populations in surface waters of the Kalamazoo River, USA. Microbes Environ. 2010, 25, 41–44. [Google Scholar] [CrossRef]
- Wang, Z.; Xiao, G.; Zhou, N.; Qi, W.; Han, L.; Ruan, Y.; Guo, D.; Zhou, H. Comparison of two methods for detection of fecal indicator bacteria used in water quality monitoring of the Three Gorges Reservoir. J. Environ. Sci. 2015, 38, 42–51. [Google Scholar] [CrossRef]
- Sırıken, B. Salmonella pathogenicity islands. Mikrobiyoloji Bul. 2013, 47, 181–188. [Google Scholar] [CrossRef]
- Espigares, E.; Bueno, A.; Espigares, M.; Gálvez, R. Isolation of Salmonella serotypes in wastewater and effluent: Effect of treatment and potential risk. Int. J. Hyg. Environ. Health 2006, 209, 103–107. [Google Scholar] [CrossRef]
- Yan, T.; O’Brien, P.; Shelton, J.M.; Whelen, A.C.; Pagaling, E. Municipal Wastewater as a Microbial Surveillance Platform for Enteric Diseases: A Case Study for Salmonella and Salmonellosis. Environ. Sci. Technol. 2018, 52, 4869–4877. [Google Scholar] [CrossRef]
- Masarikova, M.; Manga, I.; Cizek, A.; Dolejska, M.; Oravcova, V.; Myskova, P.; Karpiskova, R.; Literak, I. Salmonella enterica resistant to antimicrobials in wastewater effluents and black-headed gulls in the Czech Republic, 2012. Sci. Total Environ. 2016, 542, 102–107. [Google Scholar] [CrossRef]
- Lu, S.; Li, F.; Liu, B.; Yang, K.; Tian, F.; Cheng, Z.; Ding, S.; Hou, K. Monodisperse Fluorescent Polystyrene Microspheres for Staphylococcus aureus Aerosol Simulation. Polymers 2023, 15, 3614. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Varela, A.R.; Pereira, T.V.; Fochat, R.C.; Manaia, C.M. Multidrug Resistance in Quinolone-Resistant Gram-Negative Bacteria Isolated from Hospital Effluent and the Municipal Wastewater Treatment Plant. Microb. Drug Resist. 2015, 22, 155–163. [Google Scholar] [CrossRef]
- Ben Said, M.; Abbassi, M.S.; Gómez, P.; Ruiz-Ripa, L.; Sghaier, S.; Ibrahim, C.; Torres, C.; Hassen, A. Staphylococcus aureus isolated from wastewater treatment plants in Tunisia: Occurrence of human and animal associated lineages. J. Water Health 2017, 15, 638–643. [Google Scholar] [CrossRef]
- Gómez, P.; Lozano, C.; Benito, D.; Estepa, V.; Tenorio, C.; Zarazaga, M.; Torres, C. Characterization of staphylococci in urban wastewater treatment plants in Spain, with detection of methicillin resistant Staphylococcus aureus ST398. Environ. Pollut. 2016, 212, 71–76. [Google Scholar] [CrossRef]
- Amirsoleimani, A.; Brion, G.M.; Diene, S.M.; François, P.; Richard, E.M. Prevalence and characterization of Staphylococcus aureus in wastewater treatment plants by whole genomic sequencing. Water Res 2019, 158, 193–202. [Google Scholar] [CrossRef]
- Schäffler, H.; Breitrück, A. Clostridium difficile—From Colonization to Infection. Front. Microbiol. 2018, 9, 646. [Google Scholar] [CrossRef]
- Vaishnavi, C. Clinical spectrum & pathogenesis of Clostridium difficile associated diseases. Indian J. Med. Res. 2010, 131, 487–499. [Google Scholar]
- Romano, V.; Pasquale, V.; Krovacek, K.; Mauri, F.; Demarta, A.; Dumontet, S. Toxigenic Clostridium difficile PCR Ribotypes from Wastewater Treatment Plants in Southern Switzerland. Appl. Environ. Microbiol. 2012, 78, 6643–6646. [Google Scholar] [CrossRef]
- Freier, L.; Zacharias, N.; Gemein, S.; Gebel, J.; Engelhart, S.; Exner, M.; Mutters Nico, T. Environmental Contamination and Persistence of Clostridioides difficile in Hospital Wastewater Systems. Appl. Environ. Microbiol. 2023, 89, e00014–e00023. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, D.; Huber, A.; Weese, S.J.; Warriner, K. Persistence of Clostridium difficile in wastewater treatment-derived biosolids during land application or windrow composting. J. Appl. Microbiol. 2016, 120, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Ignatius, R.; Neuber, R.; Kietzmann, H.; Berg, C.; Wenzel, T.; Fuhrmann, J.; Müller, M. Clostridioides difficile in Outpatients: Application of a Diagnostic Algorithm Recommended by the European Society of Clinical Microbiology and Infectious Diseases. Eur. J. Microbiol. Immunol. 2019, 9, 88–90. [Google Scholar] [CrossRef] [PubMed]
- Cançado, G.G.L.; Silva, R.O.S.; Nader, A.P.; Lobato, F.C.F.; Vilela, E.G. Impact of simultaneous glutamate dehydrogenase and toxin A/B rapid immunoassay on Clostridium difficile diagnosis and treatment in hospitalized patients with antibiotic-associated diarrhea in a university hospital of Brazil. J. Gastroenterol. Hepatol. 2018, 33, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.S.; McDermott, B.P.; Parchuri, S.; Cunha, B.A. Lack of value of repeat stool testing for Clostridium difficile toxin. Am. J. Med. 2006, 119, e357–e358. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, R.E.; Tabor-Godwin, J.M.; Tsueng, G.; Feuer, R. Enterovirus infections of the central nervous system. Virology 2011, 411, 288–305. [Google Scholar] [CrossRef] [PubMed]
- Pennino, F.; Nardone, A.; Montuori, P.; Aurino, S.; Torre, I.; Battistone, A.; Delogu, R.; Buttinelli, G.; Fiore, S.; Amato, C.; et al. Large-Scale Survey of Human Enteroviruses in Wastewater Treatment Plants of a Metropolitan Area of Southern Italy. Food Environ. Virol. 2018, 10, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Pellegrinelli, L.; Galli, C.; Seiti, A.; Primache, V.; Hirvonen, A.; Schiarea, S.; Salmoiraghi, G.; Castiglioni, S.; Ammoni, E.; Cereda, D.; et al. Wastewater-based epidemiology revealed in advance the increase of enterovirus circulation during the COVID-19 pandemic. Sci. Total Environ. 2023, 902, 166539. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xue, L.; Li, Y.; Cai, W.; Miao, S.; Meng, L.; Ren, S.; Zhang, J.; Wang, J.; Wu, S.; et al. Rapid and sensitive lateral flow biosensor for the detection of GII human norovirus based on immunofluorescent nanomagnetic microspheres. J. Med. Virol. 2024, 96, e29487. [Google Scholar] [CrossRef]
- Ali, W.; Zhang, H.; Wang, Z.; Chang, C.; Javed, A.; Ali, K.; Du, W.; Niazi, N.K.; Mao, K.; Yang, Z. Occurrence of various viruses and recent evidence of SARS-CoV-2 in wastewater systems. J. Hazard. Mater. 2021, 414, 125439. [Google Scholar] [CrossRef]
- Ouardani, I.; Turki, S.; Aouni, M.; Romalde, J.L. Detection and Molecular Characterization of Hepatitis A Virus from Tunisian Wastewater Treatment Plants with Different Secondary Treatments. Appl. Env. Microbiol. 2016, 82, 3834–3845. [Google Scholar] [CrossRef] [PubMed]
- Rachida, S.; Taylor, M.B. Potentially Infectious Novel Hepatitis A Virus Strains Detected in Selected Treated Wastewater Discharge Sources, South Africa. Viruses 2020, 12, 1468. [Google Scholar] [CrossRef]
- Wu, R.; Meng, B.; Corredig, M.; Griffiths, M.W. Rapid Detection of Hepatitis A Virus in Foods Using a Bioluminescent Assay in Real-Time (BART) and Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) Technology. Food Environ. Virol. 2023, 15, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Bukkitgar, S.D.; Shetti, N.P.; Aminabhavi, T.M. Electrochemical investigations for COVID-19 detection-A comparison with other viral detection methods. Chem. Eng. J. 2021, 420, 127575. [Google Scholar] [CrossRef] [PubMed]
- Eyayu, T.; Kiros, T.; Workineh, L.; Sema, M.; Damtie, S.; Hailemichael, W.; Dejen, E.; Tiruneh, T. Prevalence of intestinal parasitic infections and associated factors among patients attending at Sanja Primary Hospital, Northwest Ethiopia: An institutional-based cross-sectional study. PLoS ONE 2021, 16, e0247075. [Google Scholar] [CrossRef] [PubMed]
- Mahvi, A.H.; Kia, E.B. Helminth eggs in raw and treated wastewater in the Islamic Republic of Iran. EMHJ-East. Mediterr. Health J. 2006, 12, 137–143. [Google Scholar]
- Bourouache, M.; Mimouni, R.; Ait Alla, A.; Hamadi, F.; El Boulani, A.; Bihadassen, B.; Laktib, A.; Moustaoui, F.; Aghrouch, M. Occurrence and removal of intestinal parasites in two wastewater treatment plants in the south of Morocco. J. Environ. Health Sci. Eng. 2021, 19, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Chowdhari, S.; Rana, S.; Rana, S.; Morrison, C.M.; Abney, S.E.; Singh, R.; Gurian, P.L.; Kumar, A.; Kumar, A.; Betancourt, W.Q.; et al. Quantitative Assessment of Microbial Pathogens and Indicators of Wastewater Treatment Performance for Safe and Sustainable Water Reuse in India. Microbiol. Spectr. 2022, 10, e0172022. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Nobile, C.J.; Johnson, A.D. Candida albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef]
- Ferreira, A.V.; Prado, C.G.; Carvalho, R.R.; Dias, K.S.; Dias, A.L. Candida albicans and non-C. albicans Candida species: Comparison of biofilm production and metabolic activity in biofilms, and putative virulence properties of isolates from hospital environments and infections. Mycopathologia 2013, 175, 265–272. [Google Scholar] [CrossRef]
- Yuan, T.; Pian, Y. Hospital wastewater as hotspots for pathogenic microorganisms spread into aquatic environment: A review. Front. Environ. Sci. 2023, 10, 1091734. [Google Scholar] [CrossRef]
- Pan, L.; Gao, X.; Wang, Y.; Zhang, F.; Gao, S.; Wang, J. Medical institutions sewage fecal coliform rapid detection method research. Chin. J. Health Lab. Technol. 2010, 20, 2091–2092. [Google Scholar]
- Yang, R.; Tong, Y.; Zhao, Y.; Tang, C.; Liu, J.; Zhu, C.; Zeng, Y.; Yu, J.; Tang, J. Establishment of Insulated Isothermal PCR Detection Method for Salmonella in Food. Mod. Food Sci. Technol. 2021, 37, 285–293. [Google Scholar] [CrossRef]
- Pandey, S.K.; Vinayaka, A.C.; Rishi, D.B.; Rishi, P.; Suri, C.R. Immuno-fluorescence based Vi capsular polysaccharide detection for specific recognition of Salmonella enterica serovar Typhi in clinical samples. Anal. Chim. Acta 2014, 841, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Planche, T.D.; Davies, K.A.; Coen, P.G.; Finney, J.M.; Monahan, I.M.; Morris, K.A.; O’Connor, L.; Oakley, S.J.; Pope, C.F.; Wren, M.W.; et al. Differences in outcome according to Clostridium difficile testing method: A prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect. Dis. 2013, 13, 936–945. [Google Scholar] [CrossRef]
- Heim, A.; Ebnet, C.; Harste, G.; Pring-Akerblom, P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 2003, 70, 228–239. [Google Scholar] [CrossRef]
- Hennechart-Collette, C.; Fourniol, L.; Fraisse, A.; Martin-Latil, S.; Perelle, S. Evaluation of a Proteinase K-Based Extraction Method to Detect Hepatitis A Virus, Hepatitis E Virus and Norovirus in Artificially Contaminated Dairy Products. Foods 2023, 12, 1489. [Google Scholar] [CrossRef]
- Daigle, J.; Racher, K.; Hazenberg, J.; Yeoman, A.; Hannah, H.; Duong, D.; Mohammed, U.; Spreitzer, D.; Gregorchuk Branden, S.J.; Head Breanne, M.; et al. A Sensitive and Rapid Wastewater Test for SARS-COV-2 and Its Use for the Early Detection of a Cluster of Cases in a Remote Community. Appl. Environ. Microbiol. 2022, 88, e01740-21. [Google Scholar] [CrossRef]
- Ganguli, A.; Lim, J.; Mostafa, A.; Saavedra, C.; Rayabharam, A.; Aluru, N.R.; Wester, M.; White, K.C.; Kumar, J.; McGuffin, R.; et al. A culture-free biphasic approach for sensitive and rapid detection of pathogens in dried whole-blood matrix. Proc. Natl. Acad. Sci. USA 2022, 119, e2209607119. [Google Scholar] [CrossRef]
- Roy, R.; Singh, G.; Dahiya, U.R.; Pandey, M.; Xess, I.; Kalyanasundaram, D. Rapid detection of Mucorales in human blood and urine samples by functionalized Heusler magnetic nanoparticle assisted customized loop-mediated isothermal amplification. Med. Mycol. 2024, 62, myae007. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-B.; Park, C.; Cho, S.-Y.; Chun, H.-S.; Lee, D.-G. Development of multiplex real-time PCR for rapid identification and quantitative analysis of Aspergillus species. PLoS ONE 2020, 15, e0229561. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-X.; Lou, Y.-R.; Duan, L.-J.; Zhou, Q.-J.; Xu, Z.-J.; Chen, F.-J.; Chen, H.-X.; Xu, G.-Z.; Du, A.-F.; Chen, J. Parallel detection of multiple zoonotic parasites using a real-time fluorogenic loop-mediated isothermal amplification-based quadruple-sample microfluidic chip. Front. Microbiol. 2023, 14, 1238376. [Google Scholar] [CrossRef] [PubMed]
- Zendejas-Heredia, P.A.; Colella, V.; Hii, S.F.; Traub, R.J. Comparison of the egg recovery rates and limit of detection for soil-transmitted helminths using the Kato-Katz thick smear, faecal flotation and quantitative real-time PCR in human stool. PLoS Negl. Trop. Dis. 2021, 15, e0009395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, J.; Du, X. Electrochemical Biosensors for Detection of Foodborne Pathogens. Micromachines 2019, 10, 222. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.; Mathew, M.R.; Sam, S.; Keerthi, K.; Kumar, K.G. Recent advances and challenges in electrochemical biosensors for emerging and re-emerging infectious diseases. J. Electroanal. Chem. 2020, 878, 114596. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Lopez-Tellez, J.; Ramirez-Montes, S.; Ferreira, T.A.; Santos, E.; Rodriguez, J. Application of Voltammetric Sensors for Pathogen Bacteria Detection: A Review. Chemosensors 2022, 10, 424. [Google Scholar] [CrossRef]
- Xu, M.; Wang, R.; Li, Y. Electrochemical biosensors for rapid detection of Escherichia coli O157:H7. Talanta 2017, 162, 511–522. [Google Scholar] [CrossRef]
- Odundo, J.; Noah, N.; Andala, D.; Kiragu, J.; Masika, E. Development of an electrochemical nano-biosensor for rapid and sensitive diagnosis of bilharzia in Kenya. S. Afr. J. Chem. 2018, 71, 127–134. [Google Scholar] [CrossRef]
- Wei, J.; Bu, S.; Zhou, H.; Sun, H.; Hao, Z.; Qu, G.; Wan, J. Hybrid nanoflower-based electrochemical lateral flow immunoassay for Escherichia coli O157 detection. Microchim. Acta 2024, 191, 453. [Google Scholar] [CrossRef]
- Nambiar, S.; Yeow, J.T. Conductive polymer-based sensors for biomedical applications. Biosens. Bioelectron. 2011, 26, 1825–1832. [Google Scholar] [CrossRef]
- Ghorbani Zamani, F.; Moulahoum, H.; Ak, M.; Odaci Demirkol, D.; Timur, S. Current trends in the development of conducting polymers-based biosensors. TrAC Trends Anal. Chem. 2019, 118, 264–276. [Google Scholar] [CrossRef]
- Ramanavicius, S.; Samukaite-Bubniene, U.; Ratautaite, V.; Bechelany, M.; Ramanavicius, A. Electrochemical molecularly imprinted polymer based sensors for pharmaceutical and biomedical applications (review). J. Pharm. Biomed. Anal. 2022, 215, 114739. [Google Scholar] [CrossRef]
- Pandey, A.; Gurbuz, Y.; Ozguz, V.; Niazi, J.H.; Qureshi, A. Graphene-interfaced electrical biosensor for label-free and sensitive detection of foodborne pathogenic E. coli O157:H7. Biosens. Bioelectron. 2017, 91, 225–231. [Google Scholar] [CrossRef]
- Xu, G.; Adeloju, S.B.; Wu, Y.; Zhang, X. Modification of polypyrrole nanowires array with platinum nanoparticles and glucose oxidase for fabrication of a novel glucose biosensor. Anal. Chim. Acta 2012, 755, 100–107. [Google Scholar] [CrossRef]
- Bredar, A.; Chown, A.; Burton, A.; Farnum, B. Electrochemical Impedance Spectroscopy of Metal Oxide Electrodes for Energy Applications. ACS Appl. Energy Mater. 2020, 3, 66–98. [Google Scholar] [CrossRef]
- Huang, R.; Carr, C.G.; Gopal, C.B.; Haile, S.M. Broad Applicability of Electrochemical Impedance Spectroscopy to the Measurement of Oxygen Nonstoichiometry in Mixed Ion and Electron Conductors. ACS Appl. Mater. Interfaces 2022, 14, 19629–19643. [Google Scholar] [CrossRef] [PubMed]
- Randviir, E.P.; Banks, C.E. A review of electrochemical impedance spectroscopy for bioanalytical sensors. Anal. Methods Adv. Methods Appl. 2022, 14, 4602–4624. [Google Scholar] [CrossRef]
- Isaac, J.A.; Mangani, L.R.; Devaux, D.; Bouchet, R. Electrochemical Impedance Spectroscopy of PEO-LATP Model Multilayers: Ionic Charge Transport and Transfer. ACS Appl. Mater. Interfaces 2022, 14, 13158–13168. [Google Scholar] [CrossRef]
- Zhang, L.; Du, X.; Chen, Z.; Chen, C.; Gong, N.; Song, Y.; Song, Y.; Han, Q.; Xia, X.; Luo, H.; et al. Instrument-Free and Visual Detection of Salmonella Based on Magnetic Nanoparticles and an Antibody Probe Immunosensor. Int. J. Mol. Sci. 2019, 20, 4645. [Google Scholar] [CrossRef] [PubMed]
- Xi, F.; Gao, J.; Wang, J.; Wang, Z. Discrimination and detection of bacteria with a label-free impedimetric biosensor based on self-assembled lectin monolayer. J. Electroanal. Chem. 2011, 656, 252–257. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, Y.; Zhang, D. An integrated multifunctional photoelectrochemical platform for simultaneous capture, detection, and inactivation of pathogenic bacteria. Sens. Actuators B Chem. 2018, 274, 228–234. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, P.; Lv, F.; Liu, L.; Wang, S. Photoelectrochemical Strategy for Discrimination of Microbial Pathogens Using Conjugated Polymers. Chem. Asian J. 2018, 13, 3469–3473. [Google Scholar] [CrossRef]
- Hao, H.; Hao, S.; Hou, H.; Zhang, G.; Hou, Y.; Zhang, Z.; Bi, J.; Yan, S. A novel label-free photoelectrochemical immunosensor based on CdSe quantum dots sensitized Ho3+/Yb3+-TiO2 for the detection of Vibrio parahaemolyticus. Methods 2019, 168, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhou, T.; Huang, R. Recent Advances in Electrochemiluminescence Sensors for Pathogenic Bacteria Detection. Micromachines 2019, 10, 532. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.P.; Zhang, Y.W.; Mao, C.J. A silver nanoparticle-assisted signal amplification electrochemiluminescence biosensor for highly sensitive detection of mucin 1. J. Mater. Chem. B 2020, 8, 2471–2475. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Inverse Polymerase Chain Reaction (PCR). Cold Spring Harb. Protoc. 2019, 2019, pdb-prot095166. [Google Scholar] [CrossRef]
- Jaria, G.; Calisto, V.; Otero, M.; Esteves, V.I. Monitoring pharmaceuticals in the aquatic environment using enzyme-linked immunosorbent assay (ELISA)-a practical overview. Anal. Bioanal. Chem. 2020, 412, 3983–4008. [Google Scholar] [CrossRef]
- das Neves Almeida, R.; Braz-de-Melo, H.A.; Corrêa, R.; Magalhães, K.G. Enzyme-Linked Immunosorbent Assay and Quantitative Reverse Transcription PCR as a Technique to Analyze Inflammation. Methods Mol. Biol. 2020, 2142, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Wang, M.; Ma, Q. A visual electrochemiluminescence resonance energy transfer/surface plasmon coupled electrochemiluminescence nanosensor for Shiga toxin-producing Escherichia coli detection. Green Chem. 2018, 20, 5520–5527. [Google Scholar] [CrossRef]
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen detection with electrochemical biosensors: Advantages, challenges and future perspectives. J. Electroanal. Chem. 2021, 882, 114989. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Qian, X.; Jin, Y.; Miao, X.; Deng, A.; Li, J. Ultrasensitive detection of zearalenone based on electrochemiluminescent immunoassay with Zr-MOF nanoplates and Au@MoS2 nanoflowers. Anal. Chim. Acta 2024, 1299, 342451. [Google Scholar] [CrossRef] [PubMed]
- Starke, J.C.; Bell, N.S.; Martinez, C.M.; Friberg, I.K.; Lawley, C.; Sriskantharajah, V.; Hirschberg, D.L. Measuring SARS-CoV-2 RNA concentrations in neighborhood wastewater. Sci. Total Environ. 2024, 926, 172021. [Google Scholar] [CrossRef] [PubMed]
- Toribio-Avedillo, D.; Gómez-Gómez, C.; Sala-Comorera, L.; Galofré, B.; Muniesa, M. Adapted methods for monitoring influenza virus and respiratory syncytial virus in sludge and wastewater. Sci. Total Environ. 2024, 918, 170636. [Google Scholar] [CrossRef] [PubMed]
- Mutesa, L.; Ndishimye, P.; Butera, Y.; Souopgui, J.; Uwineza, A.; Rutayisire, R.; Ndoricimpaye, E.L.; Musoni, E.; Rujeni, N.; Nyatanyi, T.; et al. A pooled testing strategy for identifying SARS-CoV-2 at low prevalence. Nature 2021, 589, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Zhao, H.; Dunham, K.E.; Cao, Q.; Lavrik, N.V.; Venton, B.J. 3D-Printed Carbon Nanoneedle Electrodes for Dopamine Detection in Drosophila. Angew. Chem. Int. Ed. 2024, 63, e202405634. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.H.; Kim, J.; Yesuraj, J.; Kim, N.K.; An, T.; Kim, K. Electrostatic spray catalytic particle coating on carbon electrode for enhancing electrochemical reaction. Int. J. Hydrog. Energy 2023, 48, 15796–15808. [Google Scholar] [CrossRef]
- Baba, H.; Kuroda, M.; Sekizuka, T.; Kanamori, H. Highly sensitive detection of antimicrobial resistance genes in hospital wastewater using the multiplex hybrid capture target enrichment. mSphere 2023, 8, e00100-23. [Google Scholar] [CrossRef]
- Jin, Y.; Yao, X.; Liu, Q.; Li, J. Hairpin DNA probe based electrochemical biosensor using methylene blue as hybridization indicator. Biosens. Bioelectron. 2007, 22, 1126–1130. [Google Scholar] [CrossRef]
- Choong, C.-L.; Bendall, J.S.; Milne, W.I. Carbon nanotube array: A new MIP platform. Biosens. Bioelectron. 2009, 25, 652–656. [Google Scholar] [CrossRef]
- Li, X.; Tian, J.; Shen, W. Quantitative biomarker assay with microfluidic paper-based analytical devices. Anal. Bioanal. Chem. 2010, 396, 495–501. [Google Scholar] [CrossRef]
- Naguib, M.; Mochalin, V.N.; Barsoum, M.W.; Gogotsi, Y. 25th Anniversary Article: MXenes: A New Family of Two-Dimensional Materials. Adv. Mater. 2014, 26, 992–1005. [Google Scholar] [CrossRef]
- Shi, M.; Zhao, S.; Huang, Y.; Zhao, L.; Liu, Y.-M. Signal amplification in capillary electrophoresis based chemiluminescent immunoassays by using an antibody–gold nanoparticle–DNAzyme assembly. Talanta 2014, 124, 14–20. [Google Scholar] [CrossRef]
- Wang, L.-J.; Chang, Y.-C.; Sun, R.; Li, L. A multichannel smartphone optical biosensor for high-throughput point-of-care diagnostics. Biosens. Bioelectron. 2017, 87, 686–692. [Google Scholar] [CrossRef]
- Cui, F.; Yue, Y.; Zhang, Y.; Zhang, Z.; Zhou, H.S. Advancing Biosensors with Machine Learning. ACS Sens. 2020, 5, 3346–3364. [Google Scholar] [CrossRef]
- Yang, Z.; Guo, J.; Wang, L.; Zhang, J.; Ding, L.; Liu, H.; Yu, X. Nanozyme-Enhanced Electrochemical Biosensors: Mechanisms and Applications. Small 2024, 20, 2307815. [Google Scholar] [CrossRef]
- Wang, Z.-G.; Hu, Y.; Liu, H.-Y.; Wen, H.-Y.; Qi, B.-P.; Liu, S.-L. Electrochemiluminescence-Based Single-Particle Tracking of the Biomolecules Moving along Intercellular Membrane Nanotubes between Live Cells. Anal. Chem. 2024, 96, 7231–7239. [Google Scholar] [CrossRef]
- Jia, Y.; Zhu, M.; Zhang, X.; Jia, D.; Tian, T.; Shi, B.; Ru, Z.; Ma, H.; Wan, Y.; Wei, Q. Nanobody-Based Microfluidic Immunosensor Chip Using Tetraphenylethylene-Derived Covalent Organic Frameworks as Aggregation-Induced Electrochemiluminescence Emitters for the Detection of Thymic Stromal Lymphopoietin. Anal. Chem. 2024, 96, 10116–10120. [Google Scholar] [CrossRef]
- Tumrani, S.H.; Soomro, R.A.; Thabet, H.K.; Karakuş, S.; El-Bahy, Z.M.; Küçükdeniz, T.; Khoso, S. Au-decorated Ti3C2Tx/porous carbon immunoplatform for ECM1 breast cancer biomarker detection with machine learning computation for predictive accuracy. Talanta 2024, 278, 126507. [Google Scholar] [CrossRef]
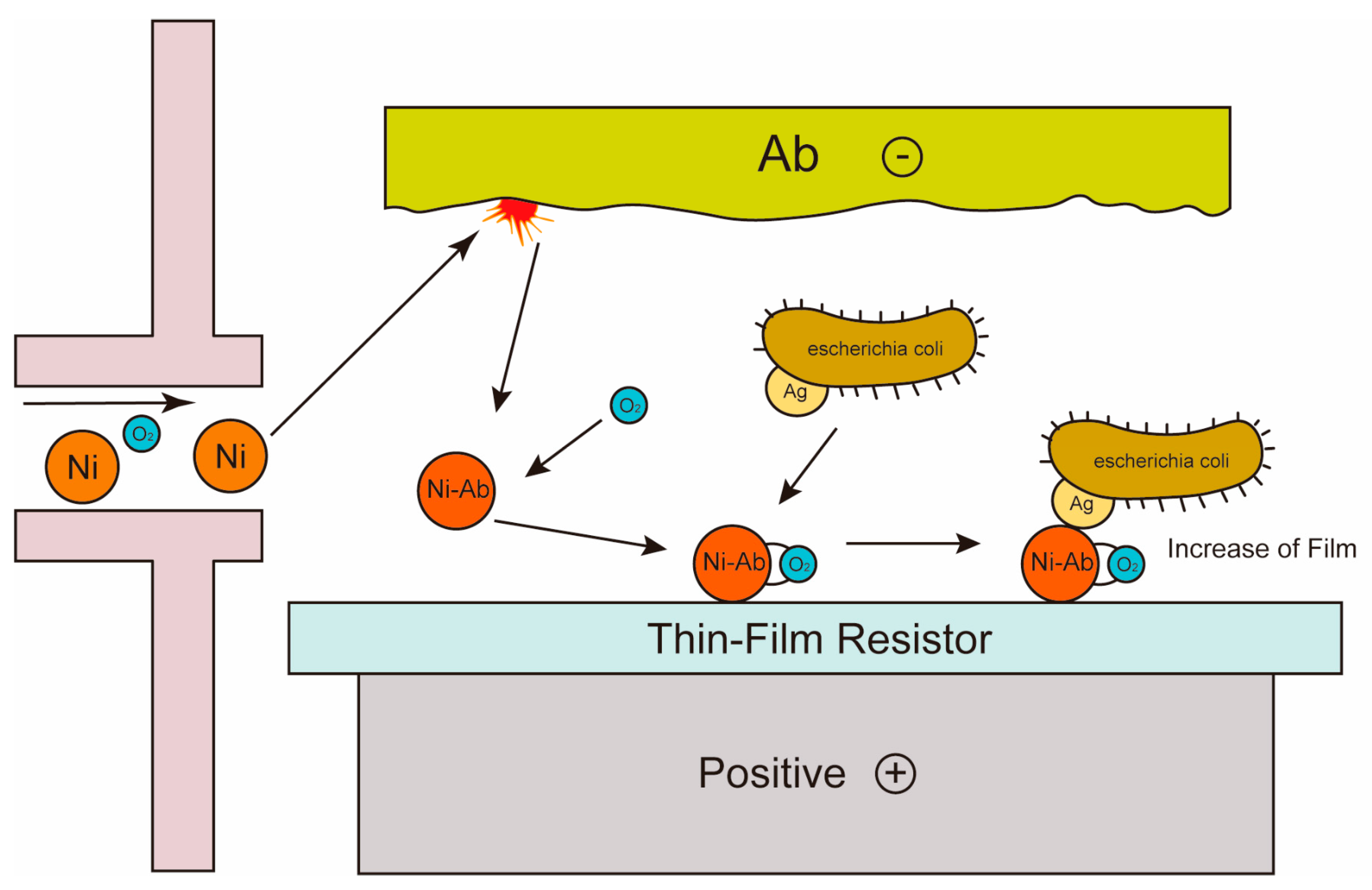
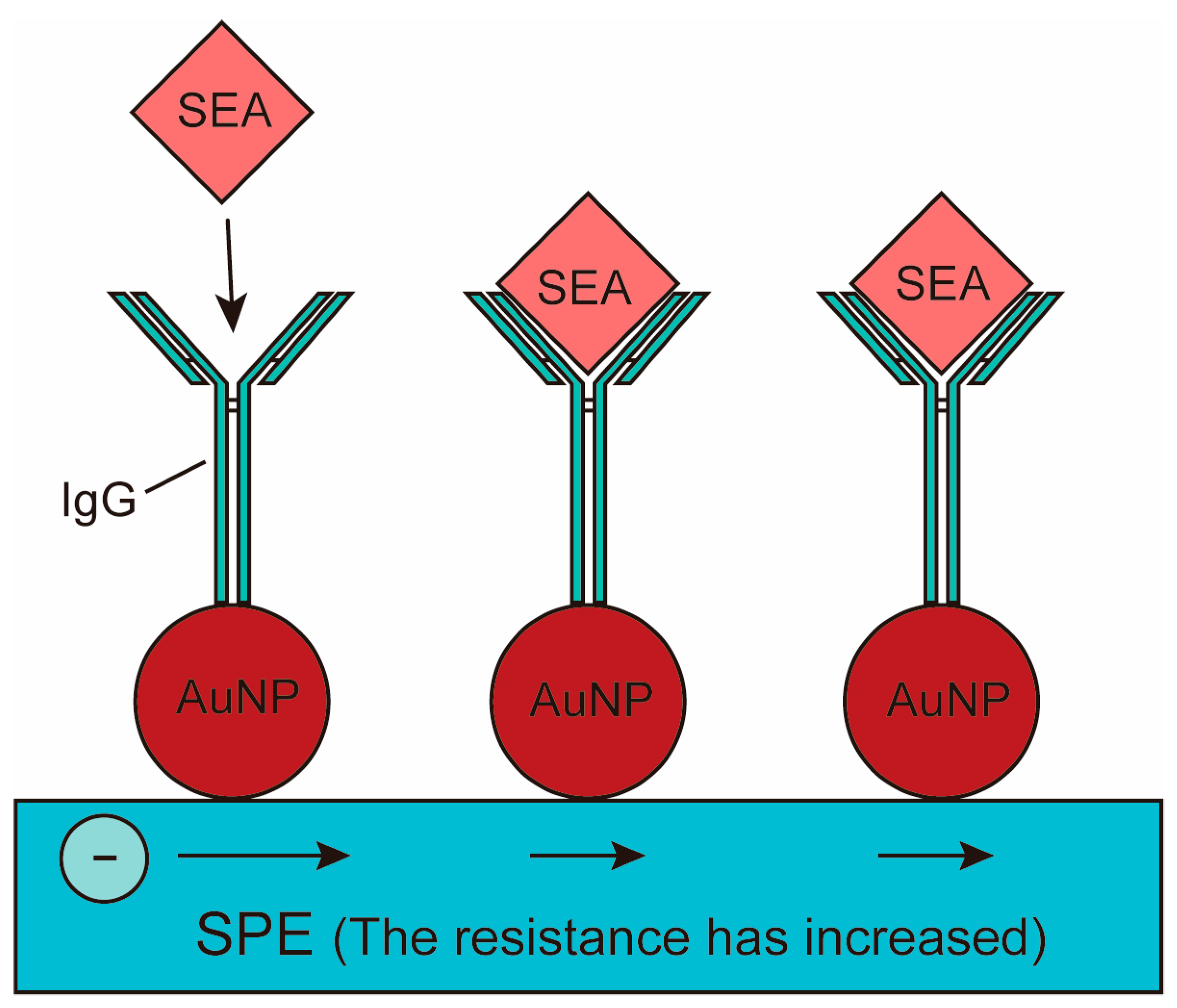

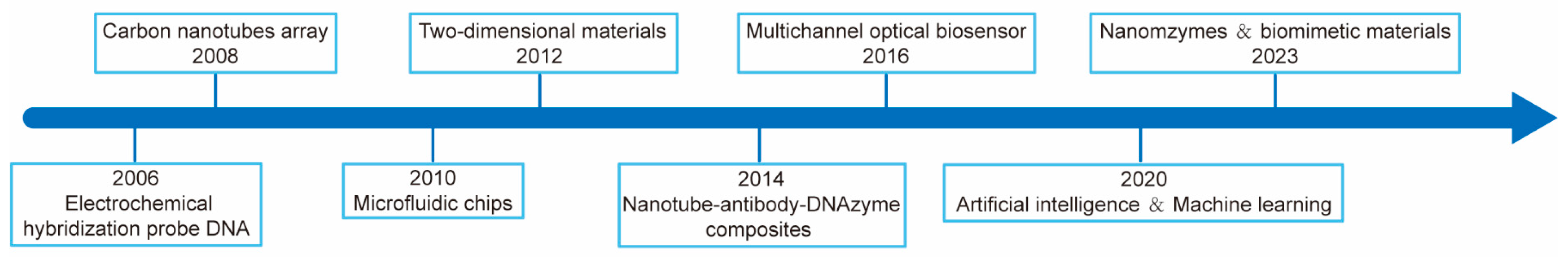
| Pathogen | Source | Disease | Detection Type | References |
|---|---|---|---|---|
| Bacteria | ||||
| E. coli | Faeces of patients | Gastroenteritis | MTF LOQ: 1 × 103 CFU/L | [74] |
| Salmonella | Faeces of patients | Salmonellosis, bacteremia | RT-PCR LOD: 7.5 × 10−2 CFU/L | [75] |
| S. aureus | Tissue fragments | Gastroenteritis, bromatoxism | Immunofluorescence technique LOD: 1 × 10−2 CFU/L | [76] |
| C. difficile | Faeces of patients | Pseudomembranous colitis, antibiotic-associated diarrhoea | EIASA Sensitivity: 69%~99% | [77] |
| Virus | ||||
| Adenovirus | Aerosol | Respiratory tract infection | RT-PCR LOQ: 1.5 × 101 copies/mL | [78] |
| HAV | Faeces of patients | Infectious hepatitis | RT-PCR LOD95: 8.02 × 102~2.80 × 103 copies/mL | [79] |
| Norovirus | Faeces of patients | Gastroenteritis | RT-PCR LOD95: 8.02 × 102~2.80 × 103 copies/mL | [79] |
| SARS-CoV-2 | Aerosol | Respiratory infections | RT-PCR LOD: 5 × 101 copies/mL | [80] |
| Fungi | ||||
| C. albicans | Wash water of the ward | Candidemia | Microfluidics LOD: 1.2 × 10−3 CFU/L | [81] |
| Rhizopus | Wash water of the ward | Mucormycosis | LAMP LOD: 1 × 10−1 pg/μL | [82] |
| Aspergillus fumigatus | Aerosol | Aspergillosis | RT-PCR LOD: 40 fg; LOQ: 400 fg | [83] |
| Parasite | ||||
| Cryptosporidium | Faeces of patients | Severe chronic diarrhoea | LAMP LOD: 1 × 10−2~1 × 10−3 pg/μL | [84] |
| Tapeworm | Faeces of patients | Cestodiasis | LAMP LOD: 1 × 10−2~1 × 10−3 pg/μL | [84] |
| Ascaris spp. | Faeces of patients | Ascariasis | Microscopy LOD: 50 seeded eggs/gr faeces | [85] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, B.; He, J.; Tian, K.; Qu, J.; Hong, L.; Lin, Q.; Yang, K.; Ma, L.; Xu, X. Research Progress on Detection of Pathogens in Medical Wastewater by Electrochemical Biosensors. Molecules 2024, 29, 3534. https://doi.org/10.3390/molecules29153534
Chen B, He J, Tian K, Qu J, Hong L, Lin Q, Yang K, Ma L, Xu X. Research Progress on Detection of Pathogens in Medical Wastewater by Electrochemical Biosensors. Molecules. 2024; 29(15):3534. https://doi.org/10.3390/molecules29153534
Chicago/Turabian StyleChen, Bangyao, Jiahuan He, Kewei Tian, Jie Qu, Lihui Hong, Qin Lin, Keda Yang, Lei Ma, and Xiaoling Xu. 2024. "Research Progress on Detection of Pathogens in Medical Wastewater by Electrochemical Biosensors" Molecules 29, no. 15: 3534. https://doi.org/10.3390/molecules29153534
APA StyleChen, B., He, J., Tian, K., Qu, J., Hong, L., Lin, Q., Yang, K., Ma, L., & Xu, X. (2024). Research Progress on Detection of Pathogens in Medical Wastewater by Electrochemical Biosensors. Molecules, 29(15), 3534. https://doi.org/10.3390/molecules29153534








