Metabolomic Analysis of Carotenoids Biosynthesis by Sphingopyxis sp. USTB-05
Abstract
:1. Introduction
2. Results
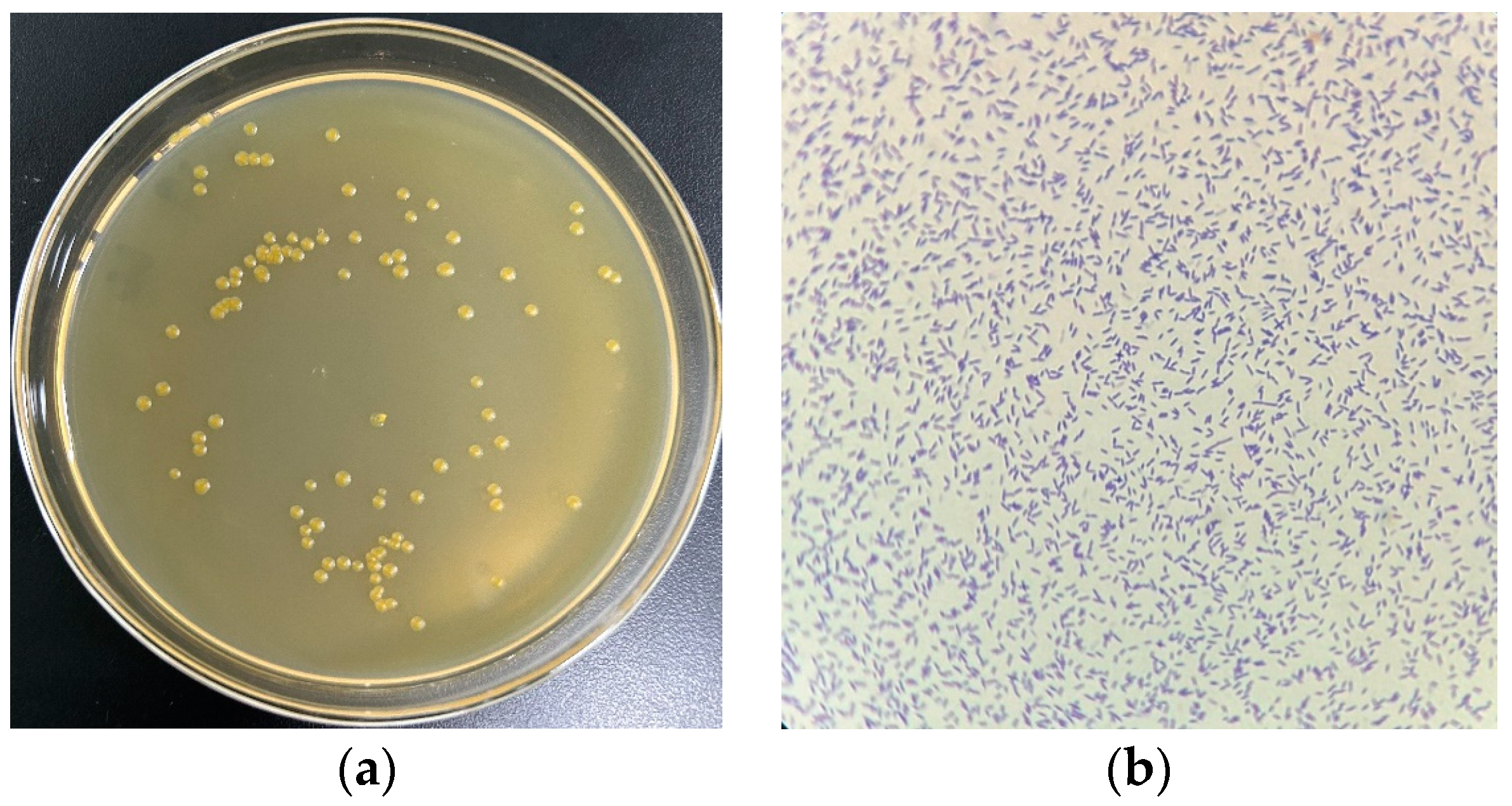
2.1. Culture of Strain USTB-05
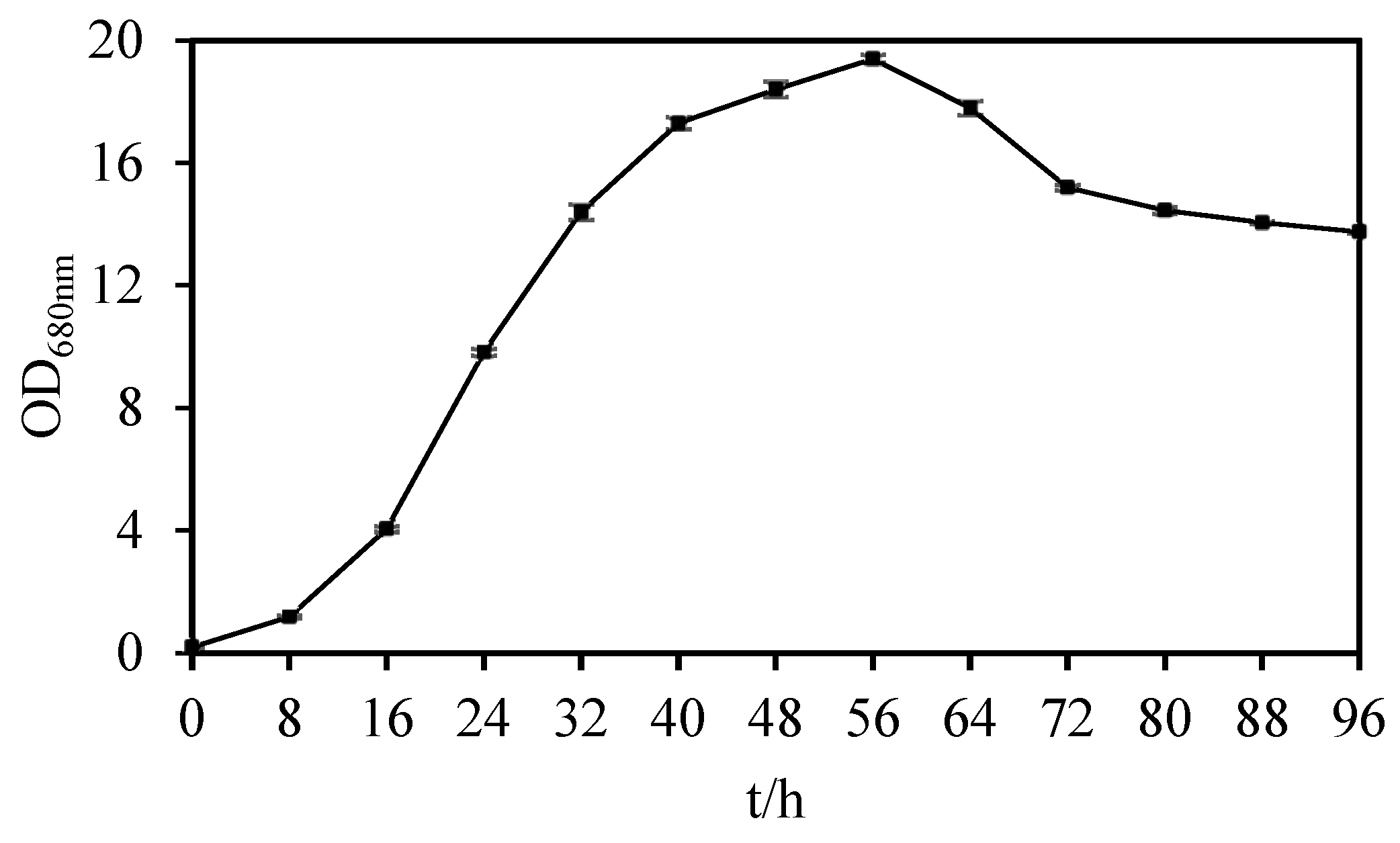
2.2. Determination of Growth Curve of Strain USTB-05
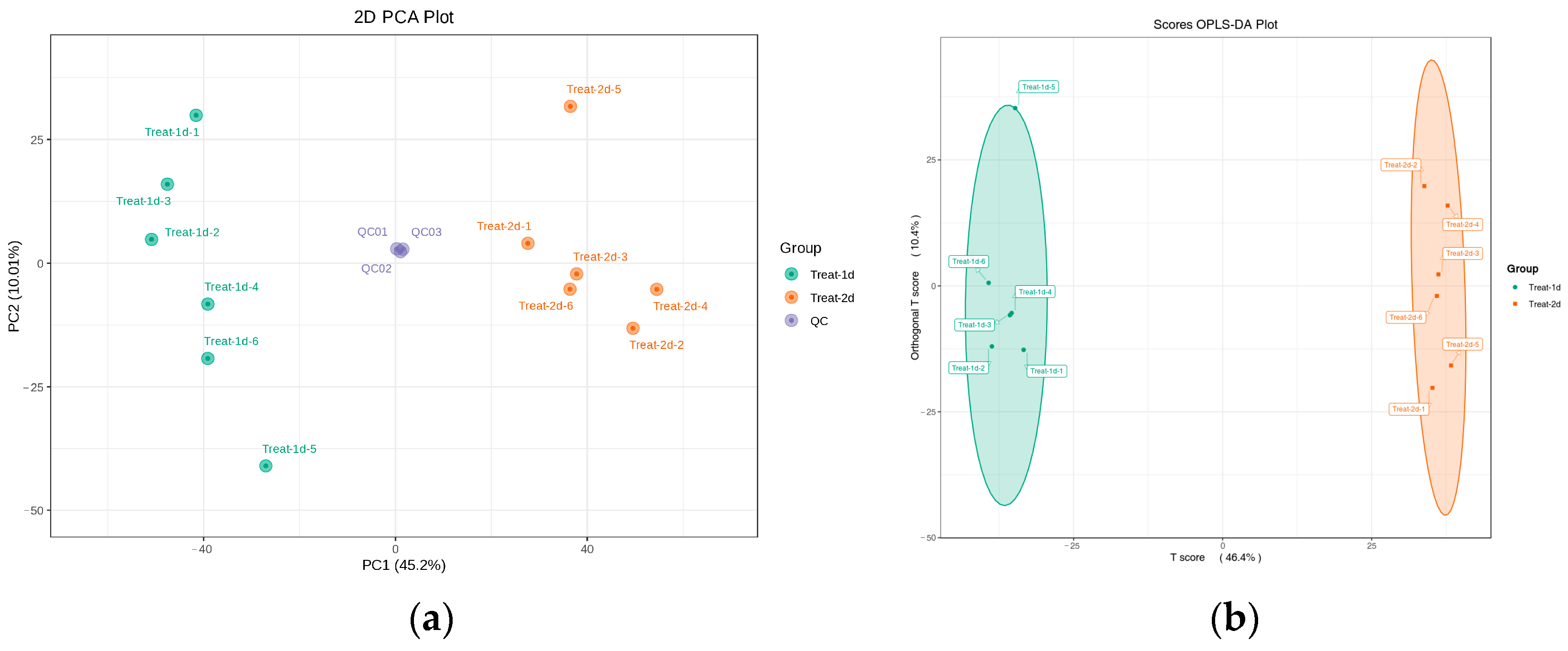
2.3. Analysis of Bacterial Composition Metabolites by UHPLC Q-TOF/MS
2.4. Identification and Content Alterations of Carotenoids during Cultivation
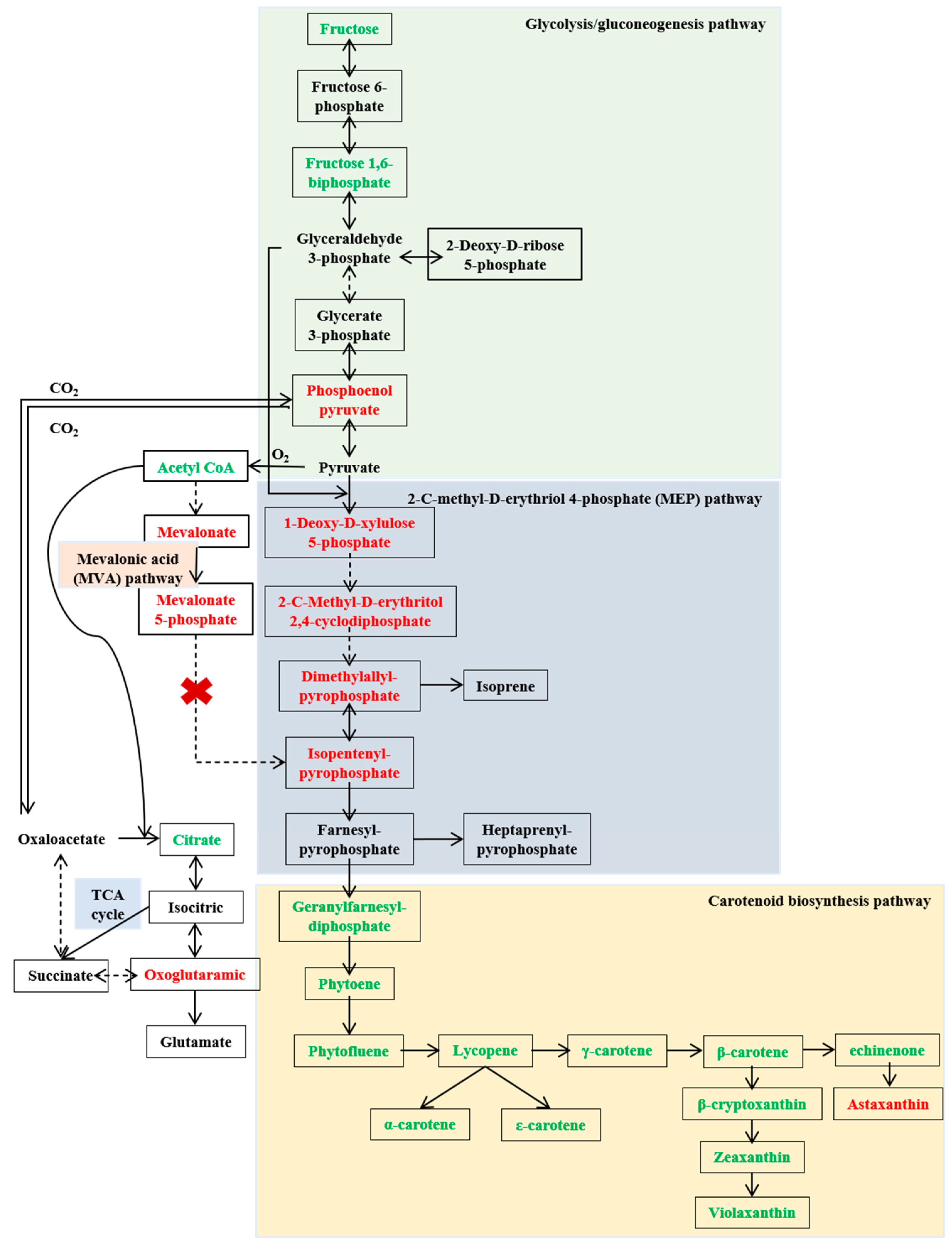
2.5. Elucidating the Biosynthetic Pathway of Carotenoids in Sphingopyxis sp. USTB-05
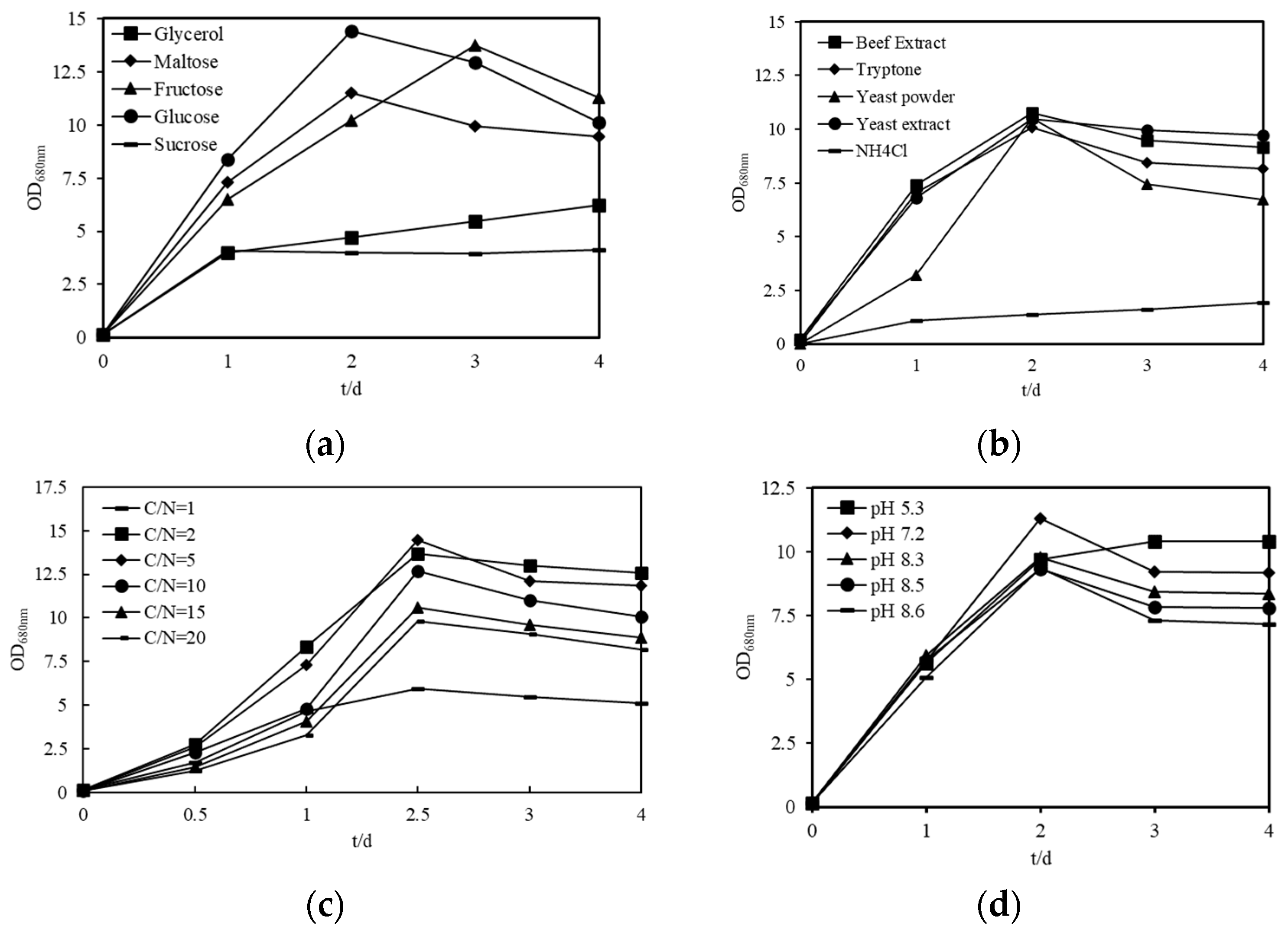
2.6. Optimization of Culture Conditions of Sphingopyxis sp. USTB-05
2.7. Determination of Carotenoid Content in Fermentation Broth of Strain USTB-05 at Different Growth Stages
3. Discussion
3.1. The Function of Sphingomonas
3.2. The Antioxidant Properties of Carotenoids
3.3. The Types and Contents of Carotenoids
3.4. The Biosynthetic Pathway of Carotenoids
4. Materials and Methods
4.1. Bacterial Strain and Culture Conditions
4.2. Bacterial Sample Treatment
4.3. Untargeted Metabolomics Analysis
4.4. Extraction and Measurement of CAROTENOIDS
4.4.1. Determination of Carotenoid in Bacterial Cells
4.4.2. Determination of Total Carotenoid Content in Bacterial Culture Medium
4.5. Data Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cezare-Gomes, E.A.; Mejia-da-Silva, L.d.C.; Pérez-Mora, L.S.; Matsudo, M.C.; Ferreira-Camargo, L.S.; Singh, A.K.; de Carvalho, J.C.M. Potential of microalgae carotenoids for industrial application. Appl. Biochem. Biotechnol. 2019, 188, 602–634. [Google Scholar] [CrossRef]
- Ye, J.; Liu, M.; He, M.; Ye, Y.; Huang, J. Illustrating and enhancing the biosynthesis of astaxanthin and docosahexaenoic acid in Aurantiochytrium sp. SK4. Mar. Drugs 2019, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.Y.; Niu, J.; Shang, F.; Yang, L.; Chang, T.Y.; Wang, J.J. Characterization of the geranylgeranyl diphosphate synthase gene in Acyrthosiphon pisum (Hemiptera: Aphididae) and its association with carotenoid biosynthesis. Front. Physiol. 2019, 10, 1398. [Google Scholar] [CrossRef]
- Khachik, F.; Bernstein, P.S.; Garland, D.L. Identification of lutein and zeaxanthin oxidation products in human and monkey retinas. Investig. Ophthalmol. Vis. Sci. 1997, 38, 1802–1811. [Google Scholar]
- Davies, N.P.; Morland, A.B. Macular pigments: Their characteristics and putative role. Prog. Retin. Eye Res. 2004, 23, 533–559. [Google Scholar] [CrossRef] [PubMed]
- Mrowicka, M.; Mrowicki, J.; Kucharska, E.; Majsterek, I. Lutein and zeaxanthin and their roles in age-related macular degeneration-neurodegenerative disease. Nutrients 2022, 14, 827. [Google Scholar] [CrossRef]
- Scripsema, N.K.; Hu, D.N.; Rosen, R.B. Lutein, zeaxanthin, and meso-zeaxanthin in the clinical management of eye disease. J. Ophthalmol. 2015, 2015, 865179. [Google Scholar] [CrossRef]
- Liu, M.; Sandmann, G.; Chen, F.; Huang, J. Enhanced coproduction of cell-bound zeaxanthin and secreted exopolysaccharides by Sphingobium sp. via metabolic engineering and optimized fermentation. J. Agric. Food Chem. 2019, 67, 12228–12236. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Sun, J.; Xue, C.; Mao, X. Biotechnological production of zeaxanthin by microorganisms. Trends Food Sci. Technol. 2018, 71, 225–234. [Google Scholar] [CrossRef]
- Shen, H.J.; Cheng, B.Y.; Zhang, Y.M.; Tang, L.; Li, Z.; Bu, Y.F.; Li, X.R.; Tian, G.Q.; Liu, J.Z. Dynamic control of the mevalonate pathway expression for improved zeaxanthin production in Escherichia coli and comparative proteome analysis. Metab. Eng. 2016, 38, 180–190. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Generalić Mekinić, I.; Šimat, V.; Rathod, N.B.; Hamed, I.; Čagalj, M. Algal carotenoids: Chemistry, sources, and application. Foods 2023, 12, 2768. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.; Yu, X.; Li, W.J.; Nie, Z.K.; Ji, X.J. Advances in heterologous biosynthesis of carotenoids in Yarrowia lipolytica. J. Nanjing Technol. Univ. 2022, 44, 546–555. [Google Scholar]
- Hamano, Y.; Dairi, T.; Yamamoto, M.; Kuzuyama, T.; Itoh, N.; Seto, H. Growth-phase dependent expression of the mevalonate pathway in a terpenoid antibiotic-producing Streptomyces strain. Biosci. Biotechnol. Biochem. 2002, 66, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Kikukawa, H.; Okaya, T.; Maoka, T.; Miyazaki, M.; Murofushi, K.; Kato, T.; Hirono-Hara, Y.; Katsumata, M.; Miyahara, S.; Hara, K.Y. Carotenoid nostoxanthin production by Sphingomonas sp. SG73 isolated from deep sea sediment. Mar. Drugs 2021, 19, 274. [Google Scholar] [CrossRef]
- Jiang, W.; Sun, J.; Gao, H.; Tang, Y.; Wang, C.; Jiang, Y.; Zhang, W.; Xin, F.; Jiang, M. Carotenoids production and genome analysis of a novel carotenoid producing Rhodococcus aetherivorans N1. Enzym. Microb. Technol. 2023, 164, 110190. [Google Scholar] [CrossRef]
- Takaichi, S.; Mochimaru, M.; Uchida, H.; Murakami, A.; Hirose, E.; Maoka, T.; Tsuchiya, T.; Mimuro, M. Opposite chilarity of α-carotene in unusual cyanobacteria with unique chlorophylls, Acaryochloris and Prochlorococcus. Plant Cell Physiol. 2012, 53, 1881–1888. [Google Scholar] [CrossRef]
- Serrano, S.; Mendo, S.; Caetano, T. Haloarchaea have a high genomic diversity for the biosynthesis of carotenoids of biotechnological interest. Res. Microbiol. 2022, 173, 103919. [Google Scholar] [CrossRef]
- Asker, D.; Awad, T.; Ohta, Y. Lipids of Haloferax alexandrinus strain TM(T): An extremely halophilic canthaxanthin-producing archaeon. J. Biosci. Bioeng. 2002, 93, 37–43. [Google Scholar] [CrossRef]
- Fang, C.; Ku, K.; Lee, M.; Su, N. Influence of nutritive factors on C50 carotenoids production by Haloferax mediterranei ATCC 33500 with two-stage cultivation. Bioresour. Technol. 2010, 101, 6487–6493. [Google Scholar] [CrossRef]
- Luo, Y.; Tian, Y.; Huang, X.; Kwon, K.; Yang, S.; Seo, H.; Kim, S.; Zheng, T. Sphingomonas polyaromaticivorans sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium from an oil port water sample. Int. J. Syst. Evol. Microbiol. 2012, 62, 1223–1227. [Google Scholar] [CrossRef] [PubMed]
- Perruchon, C.; Patsioura, V.; Vasileiadis, S.; Karpouzas, D.G. Isolation and characterisation of a Sphingomonas strain able to degrade the fungicide ortho-phenylphenol. Pest Manag. Sci. 2016, 72, 113–124. [Google Scholar] [CrossRef]
- Zhao, Q.; Yue, S.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. Comparative genomic analysis of 26 Sphingomonas and Sphingobium strains: Dissemination of bioremediation capabilities, biodegradation potential and horizontal gene transfer. Sci. Total Environ. 2017, 609, 1238–1247. [Google Scholar] [CrossRef]
- Mulla, S.; Wang, H.; Sun, Q.; Hu, A.; Yu, C. Characterization of triclosan metabolism in Sphingomonas sp. strain YL-JM2C. Sci. Rep. 2016, 6, 21965. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Sheng, L.; Tong, Q. A new strategy to enhance gellan production by two-stage culture in Sphingomonas paucimobilis. Carbohydr. Polym. 2013, 98, 829–834. [Google Scholar] [CrossRef]
- Pang, Y.; Chen, M.; Lu, W.; Chen, M.; Yan, Y.; Lin, M.; Zhang, W.; Zhou, Z. Comparative genome characterization of Echinicola marina sp. nov., isolated from deep-sea sediment provide insight into carotenoid biosynthetic gene cluster evolution. Sci. Rep. 2021, 11, 24188. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhao, Z.; Xu, Q.; Zhang, H.; Liu, X.; Yin, C.; Yan, H.; Liu, Y. Comparative Genomic Analysis of Sphingomonas morindae sp. NBD5 and Sphingopyxis sp. USTB-05 for Producing Macular Pigment. Microorganisms 2023, 11, 266. [Google Scholar] [CrossRef] [PubMed]
- Yabuuchi, E.; Kosako, Y.; Fujiwara, N.; Naka, T.; Matsunaga, I.; Ogura, H.; Kobayashi, K. Emendation of the genus Sphingomonas yabuuchi et al. 1990 and junior objective synonymy of the species of three genera, Sphingobium, Novosphingobium and Sphingopyxis, in conjunction with Blastomonas ursincola. Int. J. Syst. Evol. Microbiol. 2002, 52, 1485–1496. [Google Scholar] [CrossRef]
- White, D.; Sutton, S.; Ringelberg, D. The genus Sphingomonas: Physiology and ecology. Curr. Opin. Biotechnol. 1996, 7, 301–306. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, S.; Lee, Y.; Cao, Y.; Zhai, L.; Zhang, X.; Su, J.; Ge, Y.; Kim, S.; Cheng, C. Sphingomonas morindae sp. nov., isolated from Noni (Morinda citrifolia L.) branch. Int. J. Syst. Evol. Microbiol. 2015, 65, 2817–2823. [Google Scholar] [CrossRef]
- Trujillo-Cayado, L.A.; Alfaro, M.C.; Raymundo, A.; Sousa, I.; Muñoz, J. Rheological behavior of aqueous dispersions containing blends of rhamsan and welan polysaccharides with an eco-friendly surfactant. Colloids Surf. B Biointerfaces 2016, 145, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.L.; Ullah, I.; Hussain, J.; Kang, S.M.; Al-Harrasi, A.; Al-Rawahi, A.; Lee, I.J. Regulations of essential amino acids and proteomics of bacterial endophytes Sphingomonas sp. Lk11 during cadmium uptake. Environ. Toxicol. 2016, 31, 887–896. [Google Scholar] [CrossRef]
- Wang, J.; Wu, P.; Chen, J.; Yan, H. Biodegradation of microcystin-RR by a new isolated Sphingopyxis sp. USTB-05. Chin. J. Chem. Eng. 2010, 18, 108–112. [Google Scholar] [CrossRef]
- Wang, H.S.; Yan, H.; Liu, X.L.; Yin, C.H.; Lv, L.; Xu, Q.Q. First step in the biodegradation of microcystins by Sphingopyxis sp. USTB-05. Adv. Mater. Res. 2013, 647, 338–343. [Google Scholar] [CrossRef]
- Yan, H.; Wang, J.; Chen, J.; Wei, W.; Wang, H.; Wang, H. Characterization of the first step involved in enzymatic pathway for microcystin-RR biodegraded by Sphingopyxis sp. USTB-05. Chemosphere 2012, 87, 12–18. [Google Scholar] [CrossRef]
- Yan, H.; Wang, H.; Wang, J.; Yin, C.; Ma, S.; Liu, X.; Yin, X. Cloning and expression of the first gene for biodegrading microcystin LR by Sphingopyxis sp. USTB-05. J. Environ. Sci. 2012, 24, 1816–1822. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, H.; Xu, Q.; Lv, L.; Yin, C.; Liu, X.; Du, H.; Yan, H. Pathway for biodegrading microcystin-YR by Sphingopyxis sp. USTB-05. PLoS ONE 2015, 10, e0124425. [Google Scholar] [CrossRef]
- Wang, H.; Yan, H.; Ma, S.; Liu, X.; Yin, C.; Wang, H.; Xu, Q.; Lv, L. Characterization of the second and third steps in the enzymatic pathway for microcystin-RR biodegradation by Sphingopyxis sp. USTB-05. Ann. Microbiol. 2015, 65, 495–502. [Google Scholar] [CrossRef]
- Xu, Q.; Fan, J.; Yan, H.; Ahmad, S.; Zhao, Z.; Yin, C.; Liu, X.; Liu, Y.; Zhang, H. Structural basis of microcystinase activity for biodegrading microcystin-LR. Chemosphere 2019, 236, 124281. [Google Scholar] [CrossRef]
- Xu, Q.; Ma, H.; Fan, J.; Yan, H.; Zhang, H.; Yin, C.; Liu, X.; Liu, Y.; Wang, H. Cloning and expression of genes for biodegrading nodularin by Sphingopyxis sp. USTB-05. Toxins 2019, 11, 549. [Google Scholar] [CrossRef]
- Liu, C.; Xu, Q.; Zhao, Z.; Zhang, H.; Liu, X.; Yin, C.; Liu, Y.; Yan, H. Genomic analysis of Sphingopyxis sp. USTB-05 for biodegrading cyanobacterial hepatotoxins. Toxins 2022, 14, 333. [Google Scholar] [CrossRef]
- Liu, X.; Gai, Z.; Tao, F.; Tang, H.; Xu, P. Carotenoids play a positive role in the degradation of heterocycles by Sphingobium yanoikuyae. PLoS ONE 2012, 7, e39522. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lin, Y.; He, M.; Gong, Y.; Huang, J. Induced high-yield production of zeaxanthin, lutein, and β-carotene by a mutant of Chlorella zofingiensis. J. Agric. Food Chem. 2018, 66, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Liu, N.; Greisen, P.; Li, J.; Qiao, K.; Huang, S.; Stephanopoulos, G. Removal of lycopene substrate inhibition enables high carotenoid productivity in Yarrowia lipolytica. Nat. Commun. 2022, 13, 572. [Google Scholar] [CrossRef]
- Ukibe, K.; Katsuragi, T.; Tani, Y.; Takagi, H. Efficient screening for astaxanthin-overproducing mutants of the yeast Xanthophyllomyces dendrorhous by flow cytometry. FEMS Microbiol. Lett. 2008, 286, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Suwaleerat, T.; Thanapimmetha, A.; Srisaiyoot, M.; Chisti, Y.; Srinophakun, P. Enhanced production of carotenoids and lipids by Rhodococcus opacus PD630. J. Chem. Technol. Biotechnol. 2018, 93, 2160–2169. [Google Scholar] [CrossRef]
- Imran, M.; Ghorat, F.; Ul-Haq, I.; Ur-Rehman, H.; Aslam, F.; Heydari, M.; Shariati, M.A.; Okuskhanova, E.; Yessimbekov, Z.; Thiruvengadam, M.; et al. Lycopene as a natural antioxidant used to prevent human health disorders. Antioxidants 2020, 9, 706. [Google Scholar] [CrossRef]
- Khan, U.M.; Sevindik, M.; Zarrabi, A.; Nami, M.; Ozdemir, B.; Kaplan, D.N.; Selamoglu, Z.; Hasan, M.; Kumar, M.; Alshehri, M.M.; et al. Lycopene: Food Sources, Biological Activities, and Human Health Benefits. Oxidative Med. Cell. Longev. 2021, 2021, 2713511. [Google Scholar] [CrossRef]
- Leh, H.E.; Lee, L.K. Lycopene: A potent antioxidant for the amelioration of type II diabetes mellitus. Molecules 2022, 27, 2335. [Google Scholar] [CrossRef]
- Aziz, E.; Batool, R.; Akhtar, W.; Rehman, S.; Shahzad, T.; Malik, A.; Shariati, M.A.; Laishevtcev, A.; Plygun, S.; Heydari, M.; et al. Xanthophyll: Health benefits and therapeutic insights. Life Sci. 2020, 240, 117104. [Google Scholar] [CrossRef]
- Zhao, D.; Li, C.; Zhang, N.; Li, B. Integrative analysis of genomic and metabolomic data reveals key metabolic pathways involved in lipid and carotenoid biosynthesis in oleaginous red yeast Rhodosporidiobolus odoratus XQR. Microbiol. Res. 2023, 270, 127339. [Google Scholar] [CrossRef]
- Lange, B.M.; Rujan, T.; Martin, W.; Croteau, R. Isoprenoid biosynthesis: The evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13172–13177. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, N.; Tripathi, A.K. Biosynthesis of carotenoids in Azospirillum brasilense Cd is mediated via squalene (C30) route. Biochem. Biophys. Res. Commun. 2024, 722, 150154. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.L.; Wu, Y.H.; Cheng, H.; Zhang, X.Q.; Wang, C.S.; Xu, X.W. Complete genome sequence of astaxanthin-producing bacterium Altererythrobacter ishigakiensis NBRC 107699. Mar. Genom. 2016, 30, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Zhou, Y.; Li, X.; Zhu, F.; Cheng, Y.; Liu, Y.; Deng, Z.; Liu, T. Genome mining of astaxanthin biosynthetic genes from Sphingomonas sp. ATCC 55669 for heterologous overproduction in Escherichia coli. Biotechnol. J. 2016, 11, 228–237. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, X.; Li, K.; Wang, H.; Zhang, C.; Shi, Y.; Yao, M.; Wang, Y.; Xiao, W. Systems metabolic engineering for efficient violaxanthin production in yeast. J. Agric. Food Chem. 2024, 72, 10459–10468. [Google Scholar] [CrossRef]
- Goss, R.; Latowski, D. Lipid dependence of xanthophyll cycling in higher plants and algae. Front. Plant Sci. 2020, 11, 455. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, R.; Diksha-Kumari, A.; Panwar, A. Astaxanthin: A super antioxidant from microalgae and its therapeutic potential. J. Basic Microbiol. 2022, 62, 1064–1082. [Google Scholar] [CrossRef]
- Davinelli, S.; Nielsen, M.E.; Scapagnini, G. Astaxanthin in skin health, repair, and disease: A comprehensive review. Nutrients 2018, 10, 522. [Google Scholar] [CrossRef]

| Number | Category | Negative Ion Mode | Positive Ion Mode | Total |
|---|---|---|---|---|
| 1 | Amino acid and its metabolites | 360 | 564 | 924 |
| 2 | Benzene and substituted derivatives | 609 | 464 | 1073 |
| 3 | Heterocyclic compounds | 469 | 449 | 918 |
| 4 | Organic acid and its derivatives | 360 | 224 | 584 |
| 5 | Aldehyde, ketones, esters | 237 | 253 | 490 |
| 6 | Alcohol and amines | 142 | 156 | 298 |
| 7 | Glycerophospholipids | 71 | 121 | 192 |
| 8 | Fatty acids | 87 | 104 | 191 |
| 9 | Nucleotide and its metabolites | 100 | 66 | 166 |
| 10 | Hormones and hormone related compounds | 44 | 52 | 96 |
| 11 | Flavonoids | 50 | 32 | 82 |
| 12 | Terpenoids | 36 | 49 | 85 |
| 13 | Alkaloids | 35 | 45 | 80 |
| 14 | Carbohydrates and its metabolites | 60 | 13 | 73 |
| 15 | Glycerolipids | 7 | 32 | 39 |
| 16 | CoEnzyme and vitamins | 10 | 23 | 33 |
| 17 | Lignans and coumarins | 13 | 14 | 27 |
| 18 | Sphingolipids | 6 | 17 | 23 |
| 19 | Steroids | 7 | 13 | 20 |
| 20 | Tryptamines, cholines, pigments | 9 | 10 | 19 |
| 21 | Bile acids | 7 | 9 | 16 |
| 22 | Phenolic acids | 7 | 0 | 7 |
| 23 | Quinones | 2 | 1 | 3 |
| 24 | Tannins | 0 | 1 | 1 |
| 25 | Others | 224 | 217 | 441 |
| Type | Category | R.T | Mother Ion Molecular Weight (Da) | Characteristic Fragment Ion Molecular Weight (Da) | Molecular Weight (Da) |
|---|---|---|---|---|---|
| α-carotene | carotenes | 5.93 | 537.5 | 123.2 | 536.4382 |
| β-carotene | carotenes | 6.29 | 537.6 | 177.1 | 536.4382 |
| phytoene | carotenes | 4.98 | 545.3 | 81.0 | 544.5008 |
| ε-carotene | carotenes | 5.53 | 537.6 | 123.2 | 536.4382 |
| phytofluene | carotenes | 1.91 | 543.5 | 81.2 | 542.4852 |
| γ-carotene | carotenes | 7.40 | 537.4 | 177.3 | 536.4382 |
| lycopene | carotenes | 8.35 | 537.4 | 81.0 | 536.4382 |
| echinenone | xanthophylls | 5.55 | 551.6 | 203.1 | 550.9000 |
| zeaxanthin | xanthophylls | 4.64 | 569.4 | 477.5 | 568.4280 |
| violaxanthin | xanthophylls | 1.59 | 601.4 | 221.0 | 600.4179 |
| β-cryptoxanthin | xanthophylls | 5.53 | 553.5 | 177.4 | 552.4331 |
| β-citraurin | xanthophylls | 2.79 | 433.3 | 341.1 | 432.6000 |
| astaxanthin | xanthophylls | 3.42 | 597.3 | 147.1 | 596.8400 |
| Type | Category | Molecular Formula | Cultivation 24 h (μg/g) | Cultivation 48 h (μg/g) | Change Amount (%) |
|---|---|---|---|---|---|
| α-carotene | carotenes | C40H56 | 0.3 ± 0.03 a | 0.23 ± 0.01 b | −23.21 |
| β-carotene | carotenes | C40H56 | 0.14 ± 0.02 a | 0.02 ± 0.01 b | −84.46 |
| phytoene | carotenes | C40H64 | 2.32 ± 0.24 a | 0.35 ± 0.18 b | −84.85 |
| phytofluene | carotenes | C40H62 | 0.5 ± 0.04 a | 0.42 ± 0.03 b | −17.34 |
| zeaxanthin | xanthophylls | C40H56O2 | 37.06 ± 0.81 a | 35.25 ± 0.79 b | −4.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Xu, Q.; Liu, Y.; Song, M.; Cao, X.; Du, X.; Yan, H. Metabolomic Analysis of Carotenoids Biosynthesis by Sphingopyxis sp. USTB-05. Molecules 2024, 29, 4235. https://doi.org/10.3390/molecules29174235
Liu C, Xu Q, Liu Y, Song M, Cao X, Du X, Yan H. Metabolomic Analysis of Carotenoids Biosynthesis by Sphingopyxis sp. USTB-05. Molecules. 2024; 29(17):4235. https://doi.org/10.3390/molecules29174235
Chicago/Turabian StyleLiu, Chao, Qianqian Xu, Yang Liu, Meijie Song, Xiaoyu Cao, Xinyue Du, and Hai Yan. 2024. "Metabolomic Analysis of Carotenoids Biosynthesis by Sphingopyxis sp. USTB-05" Molecules 29, no. 17: 4235. https://doi.org/10.3390/molecules29174235
APA StyleLiu, C., Xu, Q., Liu, Y., Song, M., Cao, X., Du, X., & Yan, H. (2024). Metabolomic Analysis of Carotenoids Biosynthesis by Sphingopyxis sp. USTB-05. Molecules, 29(17), 4235. https://doi.org/10.3390/molecules29174235






