Bioprospecting Endophytic Fungi of Forest Plants for Bioactive Metabolites with Antibacterial, Antifungal, and Antioxidant Potentials
Abstract
1. Introduction
2. Results and Discussion
2.1. Screening Fungal Extracts for Antibacterial and Antifungal Activities
2.2. Screening Fungal Extracts for Antioxidant Potential
2.3. Identification of the Selected Fungal Strains
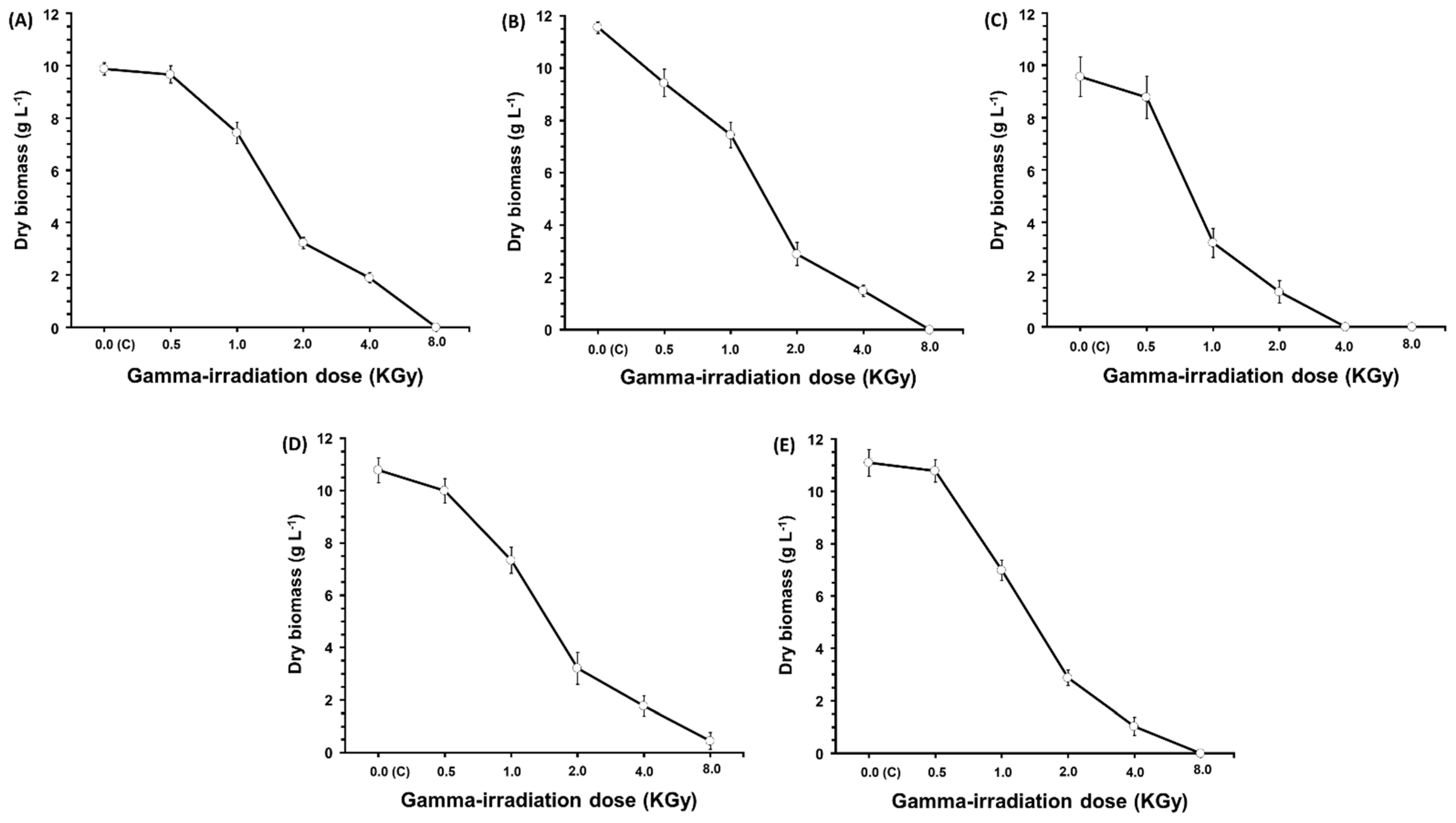
2.4. Gamma Irradiation Effect on Growth and the Antibacterial, Antifungal, and Antioxidant Activities
2.5. GC-MS Analysis
3. Materials and Methods
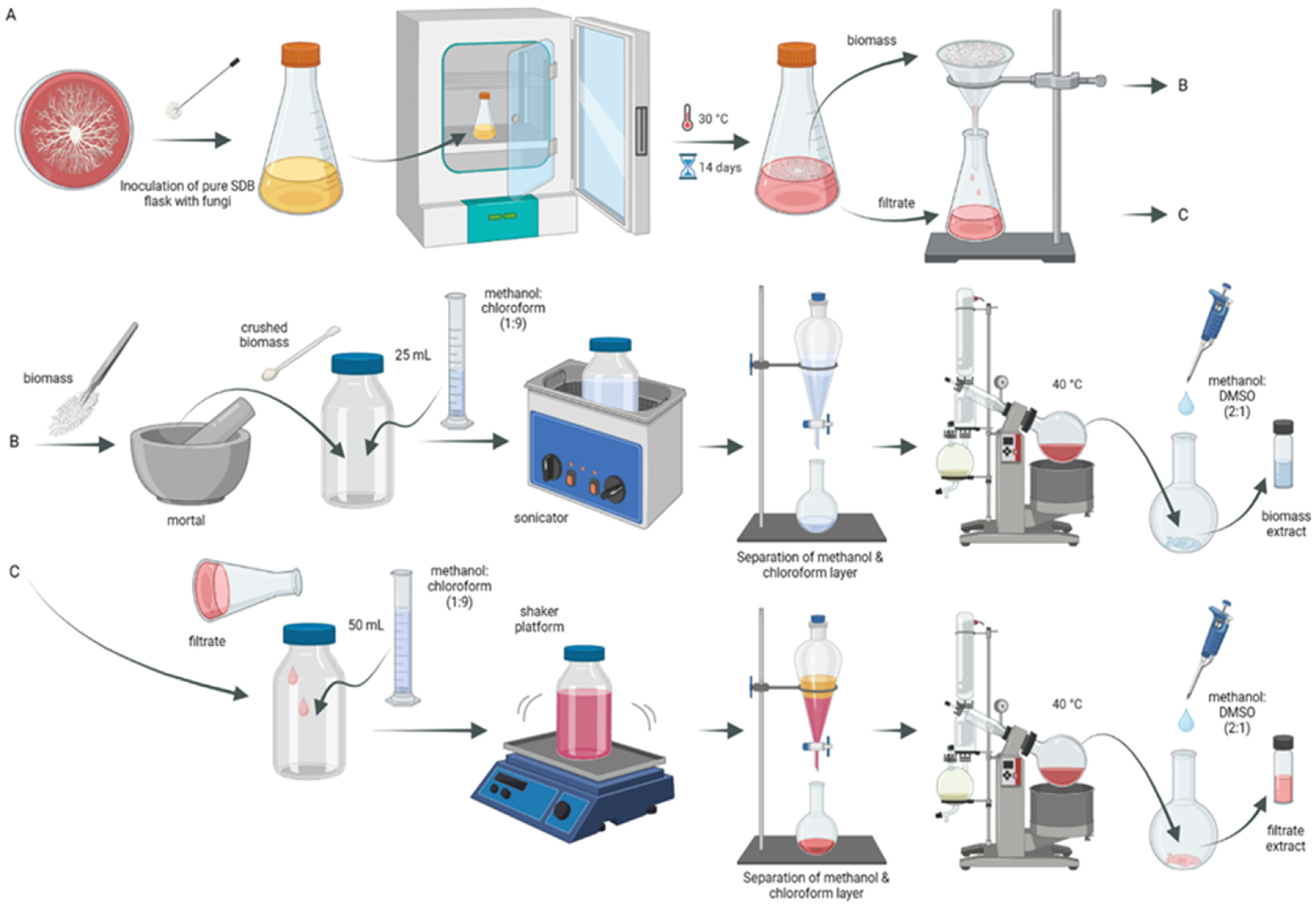
3.1. Cultivation Conditions and Preparation of Fungal Crude Extracts
3.2. Antimicrobial Activity Screening
3.3. DPPH Scavenging Activity Assay
3.4. Fungal Strains
3.5. Identification of the Selected Endophytic Fungi
3.6. Effect of 60Co Gamma Irradiation on Growth and the Tested Activities
3.7. Analytical Methods
3.7.1. Thin-Layer Chromatography (TLC) Fractionation
3.7.2. GC-MS Analysis
3.8. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Antibiotic Resistance. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 20 September 2023).
- Adeleke, B.S.; Babalola, O.O. Pharmacological potential of fungal endophytes associated with medicinal plants: A review. J. Fungi 2021, 7, 147. [Google Scholar] [CrossRef] [PubMed]
- Parveen, A.; Akash, M.S.; Rehman, K.; Kyunn, W.W. Recent Investigations for Discovery of Natural Antioxidants: A Comprehensive Review. Crit. Rev. Eukaryot. Gene Expr. 2016, 26, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.X.; Zou, W.X. Endophytes: A rich source of functional metabolites. Nat. Prod. Rep. 2001, 18, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhao, Q.; Zhang, X.M.; Li, W.J. Research progress on bioactive products from endophytes. J. Microbiol. 2018, 38, 103–113. [Google Scholar] [CrossRef]
- Tsipinana, S.; Husseiny, S.; Alayande, K.A.; Raslan, M.; Amoo, S.; Adeleke, R. Contribution of endophytes towards improving plant bioactive metabolites: A rescue option against red-taping of medicinal plants. Front. Plant Sci. 2023, 14, 1248319. [Google Scholar] [CrossRef]
- El-Sayed, E.R. Discovery of the anticancer drug vinblastine from the endophytic Alternaria alternata and yield improvement by gamma irradiation mutagenesis. J. Appl. Microbiol. 2021, 131, 2886–2898. [Google Scholar] [CrossRef]
- Bielecka, M.; Pencakowski, B.; Nicoletti, R. Using next-generation sequencing technology to explore genetic pathways in endophytic fungi in the syntheses of plant bioactive metabolites. Agriculture 2022, 12, 187. [Google Scholar] [CrossRef]
- Mengistu, A.A. Endophytes: Colonization, behaviour, and their role in defense mechanism. Int. J. Microbiol. 2020, 2020, 6927219. [Google Scholar] [CrossRef]
- Xingyuan, Z.; Linjun, M.; Fang, C. The medicinal potential of bioactive metabolites from endophytic fungi in plants. eFood 2022, 3, e28. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Molecul. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- El-Nagar, D.; Salem, S.H.; El-Zamik, F.I.; El-Basit, H.M.I.A.; Galal, Y.G.M.; Soliman, S.M.; Aziz, H.A.A.; Rizk, M.A.; El-Sayed, E.R. Bioprospecting endophytic fungi for bioactive metabolites with seed germination promoting potentials. BMC Microbiol. 2024, 24, 200. [Google Scholar] [CrossRef] [PubMed]
- Venugopalan, A.; Srivastava, S. Endophytes as in vitro production platforms of high value plant secondary metabolites. Biotechnol. Adv. 2015, 33, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Manganyi, M.C.; Ateba, C.N. Untapped Potentials of Endophytic Fungi: A Review of Novel Bioactive Compounds with Biological Applications. Microorganisms 2020, 8, 1934. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Bae, H. Endophytic Fungi: Key Insights, Emerging Prospects, and Challenges in Natural Product Drug Discovery. Microorganisms 2022, 10, 360. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Saravanakumar, K.; Jin, T.; Wang, M.H. Mycosynthesis, characterization, anticancer and antibacterial activity of silver nanoparticles from endophytic fungus Talaromyces purpureogenus. Int. J. Nanomed. 2019, 14, 3427–3438. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Sharma, D.; Jadon, N.; Agrawal, P.K. Antimicrobial and phytochemical screening of endophytic fungi isolated from spikes of Pinus roxburghii. Arch. Clin. Microbiol. 2015, 6, 1–9. [Google Scholar]
- Tănase, C.; Coşarcă, S.; Toma, F.; Mare, A.; Man, A.; Miklos, A.; Imre, S.; Boz, I. Antibacterial activities of beech bark (Fagus sylvatica L.) polyphenolic extract. Environ. Eng. Manag. J. (EEMJ) 2018, 17. [Google Scholar] [CrossRef]
- Tanase, C.; Mocan, A.; Coșarcă, S.; Gavan, A.; Nicolescu, A.; Gheldiu, A.M.; Crișan, O. Biological and chemical insights of beech (Fagus sylvatica L.) bark: A source of bioactive compounds with functional properties. Antioxidants 2019, 8, 417. [Google Scholar] [CrossRef]
- Schulz, B.; Sucker, J.; Aust, H.J.; Krohn, K.; Ludewig, K.; Jones, P.G.; Döring, D.J.M.R. Biologically active secondary metabolites of endophytic Pezicula species. Mycol. Res. 1995, 99, 1007–1015. [Google Scholar] [CrossRef]
- Blondeau, D.; St-Pierre, A.; Bourdeau, N.; Bley, J.; Lajeunesse, A.; Desgagné-Penix, I. Antimicrobial activity and chemical composition of white birch (Betula papyrifera Marshall) bark extracts. Microbiol. Open 2020, 9, e00944. [Google Scholar] [CrossRef]
- Cioch, M.; Satora, P.; Skotniczny, M.; Semik-Szczurak, D.; Tarko, T. Characterisation of antimicrobial properties of extracts of selected medicinal plants. Polish J. Microbiol. 2017, 66, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Emrich, S.; Schuster, A.; Schnabel, T.; Oostingh, G.J. Antimicrobial activity and wound-healing capacity of birch, beech and larch bark extracts. Molecules 2022, 27, 2817. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, S.Y.; Ye, Y.B.; Zhao, W.H.; Sun, X.G.; Wang, Z.Q.; Zhang, Y.X. The antibacterial efficacy of an aceraceous plant [Shantung maple (Acer truncatum Bunge)] may be related to inhibition of bacterial β-oxoacyl-acyl carrier protein reductase (FabG). Biotechnol. Appl. Biochem. 2008, 51, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Cozma, L.S. The role of antioxidant therapy in cardiovascular disease. Curr. Opin. Lipidol. 2004, 15, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.J.; Zhang, W.W.; Li, X.M.; Wang, B.G. Evaluation of antioxidant property of extract and fractions obtained from a red alga, Polysiphonia urceolata. Food Chem. 2006, 95, 37–43. [Google Scholar] [CrossRef]
- Pirvu, L.; Grigore, A.; Bubueanu, C.; Draghici, E. Comparative analytical and antioxidant activity studies on a series of Fagus sylvatica L. leaves extracts. JPC–J. Planar Chromatogr.–Modern TLC 2013, 26, 237–242. [Google Scholar] [CrossRef]
- Vek, V.; Oven, P.; Poljanšek, I. Review on lipophilic and hydrophilic extractives in tissues of common beech. Drv. Ind. 2016, 67, 85–96. [Google Scholar] [CrossRef]
- Formato, M.; Piccolella, S.; Zidorn, C.; Pacifico, S. UHPLC-HRMS analysis of Fagus sylvatica (Fagaceae) leaves: A renewable source of antioxidant polyphenols. Antioxidants 2021, 10, 1140. [Google Scholar] [CrossRef]
- Latif, A.L.; Hussain, S.H.; Mumtaz Ali, M.A.; Mohammad Arfan, M.A.; Manzoor Ahmed, M.A.; Cox, R.J.; Ghias Uddin, G.U. A new antioxidant triterpenoid from the stem wood of Sorbus lanata. Rec. Nat. Prod. 2014, 8, 119–124. [Google Scholar]
- Adepoju, F.O.; Duru, K.C.; Li, E.; Kovaleva, E.G.; Tsurkan, M.V. Pharmacological potential of betulin as a multitarget compound. Biomolecules 2023, 13, 1105. [Google Scholar] [CrossRef]
- Kumar, V.; Prasher, I.B. Phytochemical analysis and antioxidant activity of endophytic fungi isolated from Dillenia indica Linn. Appl. Biochem. Biotechnol. 2024, 196, 332–349. [Google Scholar] [CrossRef] [PubMed]
- Parekh, S.; Vinci, V.A.; Strobel, R.J. Improvement of microbial strains and fermentation processes. Appl. Microbiol. Biotechnol. 2000, 54, 287–301. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, E.R.; Mousa, S.A.; Strzała, T.; Boratyński, F. Enhancing bioprocessing of red pigment from immobilized culture of gamma rays mutant of the endophytic fungus Monascus ruber SRZ112. J. Biol. Eng. 2024, 18, 44. [Google Scholar] [CrossRef]
- El-Sayed, E.R.; Ahmed, A.S.; Al-Hagar, O.E.A. Agro-industrial wastes for production of paclitaxel by irradiated Aspergillus fumigatus under solid-state fermentation. J. Appl. Microbiol. 2020, 128, 1427–1439. [Google Scholar] [CrossRef]
- Anwar, M.M.; Aly, S.S.H.; Nasr, E.H.; El-Sayed, E.-S.R. Improving carboxymethyl cellulose edible coating using ZnO nanoparticles from irradiated Alternaria tenuissima. AMB Expr. 2022, 12, 116. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Parekh, S.R. Development of improved strains and optimization of fermentation processes. In Microbial Processes and Products. Methods in Biotechnology; Barredo, J.L., Ed.; Humana Press: Totowa, NJ, NSA, 2005; Volume 18. [Google Scholar]
- Konar, A.; Datta, S. Strain improvement of microbes. In Industrial Microbiology and Biotechnology; Verma, P., Ed.; Springer: Singapore, 2022. [Google Scholar] [CrossRef]
- El-Sayed, E.R.; El-Sayyad, G.S.; Abdel-Fatah, S.S.; El-Batal, A.I.; Boratyński, F. Novel nanoconjugates of metal oxides and natural red pigment from the endophyte Monascus ruber using solid-state fermentation. Microb. Cell Fact. 2024, 23, 259. [Google Scholar] [CrossRef]
- Paster, N.; Barkai-Golan, R.; Padova, R. Effect of gamma radiation on ochratoxin production by the fungus Aspergillus ochraceus. J. Sci. Food. Agric. 1985, 36, 445–449. [Google Scholar] [CrossRef]
- Ismaiel, A.A.; Ahmed, A.S.; El-Sayed, E.R. Immobilization technique for enhanced production of the immunosuppressant mycophenolic acid by ultraviolet and gamma-irradiated Penicillium roqueforti. J. Appl. Microbiol. 2015, 119, 112–126. [Google Scholar] [CrossRef]
- Chopra, V.L. Mutagenesis: Investigating the process and processing the outcome for crop improvement. Curr. Sci. 2005, 89, 353–359. [Google Scholar]
- Rizi, E.S.; Jahadi, M.; Zia, M. Evaluation of gamma irradiation effect on morphological changes, macroscopic, microscopic characteristics and pigment production of Monascus purpureus. J. Food Process. Preserv. 2021, 46, e16129. [Google Scholar] [CrossRef]
- Abdel-Fatah, S.S.; El-Batal, A.I.; El-Sherbiny, G.M.; Khalaf, M.A.; El-Sayed, A.S. Production, bioprocess optimization and γ-irradiation of Penicillium polonicum, as a new Taxol producing endophyte from Ginko biloba. Biotechnol. Rep. 2021, 30, e00623. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, E.S.R.; Ahmed, A.S.; Hassan, I.A.; Ismaiel, A.A.; Karam El-Din, A.Z.A. Semi-continuous production of the anticancer drug taxol by Aspergillus fumigatus and Alternaria tenuissima immobilized in calcium alginate beads. Bioproc. Biosys. Eng. 2020, 43, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, E.S.R.; Hazaa, M.A.; Shebl, M.M.; Amer, M.M.; Mahmoud, S.R.; Khattab, A.A. Bioprospecting endophytic fungi for bioactive metabolites and use of irradiation to improve their bioactivities. AMB Expr. 2022, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, E.R.; Gach, J.; Olejniczak, T.; Boratyński, F. A new endophyte Monascus ruber SRZ112 as an efficient production platform of natural pigments using agro-industrial wastes. Sci. Rep. 2022, 12, 12611. [Google Scholar] [CrossRef]
- El-Sayed, E.S.R.; Mousa, S.A.; Abdou, D.A.; El-Seoud, M.A.A.; Elmehlawy, A.A.; Mohamed, S.S. Exploiting the exceptional biosynthetic potency of the endophytic Aspergillus terreus in enhancing production of Co3O4, CuO, Fe3O4, NiO, and ZnO nanoparticles using bioprocess optimization and gamma irradiation. Saudi J. Biol. Sci. 2022, 29, 2463–2474. [Google Scholar] [CrossRef]
- El-Batal, A.I.; El-Sayyad, G.S.; El-Ghamery, A.; Gobara, M. Response surface methodology optimization of melanin production by Streptomyces cyaneus and synthesis of copper oxide nanoparticles using gamma radiation. J. Clust. Sci. 2017, 28, 1083–1112. [Google Scholar] [CrossRef]
- Chen, C.; Ye, Y.; Wang, R.; Zhang, Y.; Wu, C.; Debnath, S.C.; Wu, M. Streptomyces nigra sp. nov. is a novel actinobacterium isolated from mangrove soil and exerts a potent antitumor activity in vitro. Front. Microbiol. 2018, 9, 1587. [Google Scholar] [CrossRef]
- Ser, H.L.; Palanisamy, U.D.; Yin, W.F.; Abd Malek, S.N.; Chan, K.G.; Goh, B.H.; Lee, L.H. Presence of antioxidative agent, Pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro-in newly isolated Streptomyces mangrovisoli sp. nov. Front. Microbiol. 2015, 6, 854. [Google Scholar] [CrossRef]
- Hoda, S.; Gupta, L.; Agarwal, H.; Raj, G.; Vermani, M.; Vijayaraghavan, P. Inhibition of Aspergillus fumigatus biofilm and cytotoxicity study of natural compound Cis-9-Hexadecenal. J. Pure Appl. Microbiol. 2019, 13, 1207–1216. [Google Scholar] [CrossRef]
- Wang, Z.J.; Liang, C.L.; Li, G.M.; Yu, C.Y.; Yin, M. Stearic acid protects primary cultured cortical neurons against oxidative stress. Acta Pharmacol. Sin. 2007, 28, 315–326. [Google Scholar] [CrossRef]
- Cheng, M.C.; Ker, Y.B.; Yu, T.H.; Lin, L.Y.; Peng, R.Y.; Peng, C.H. Chemical synthesis of 9 (Z)-octadecenamide and its hypolipidemic effect: A bioactive agent found in the essential oil of mountain celery seeds. J. Agric. Food Chem. 2010, 58, 1502–1508. [Google Scholar] [CrossRef]
- Ivanov, I.; Petrov, K.; Lozanov, V.; Hristov, I.; Wu, Z.; Liu, Z.; Petrova, P. Bioactive compounds produced by the accompanying microflora in Bulgarian yoghurt. Processes 2021, 9, 114. [Google Scholar] [CrossRef]
- Fabbrini, E.; Serafini, M.; Colic Baric, I.; Hazen, S.L.; Klein, S. Effect of plasma uric acid on antioxidant capacity, oxidative stress, and insulin sensitivity in obese subjects. Diabetes 2014, 63, 976–981. [Google Scholar] [CrossRef]
- Baskaran, R.; Mohan, P.M.; Sivakumar, K.; Kumar, A. Antimicrobial activity and phylogenetic analysis of Streptomyces parvulus DOSMB-D105 isolated from the mangrove sediments of Andaman Islands. Acta Microbiol. Immunol. Hung. 2016, 63, 27–46. [Google Scholar] [CrossRef]
- Kim, S.K.; Karadeniz, F. Biological importance and applications of squalene and squalane. Adv. Food Nutr. Res. 2012, 65, 223–233. [Google Scholar]
- Rajeswari, G.; Murugan, M.; Mohan, V.R. GC-MS analysis of bioactive components of Hugonia mystax L. (Linaceae). J. Pharm. Biomed. Sci. 2013, 29, 818–824. [Google Scholar]
- Alabi, K.A.; Lajide, L.; Owolabi, B.J. Biological activity of oleic acid and its primary amide: Experimental and Computational studies. J. Chem. Soc. Nigeria 2018, 43, 38–49. [Google Scholar]
- Xie, Y.; Peng, Q.; Ji, Y.; Xie, A.; Yang, L.; Mu, S.; Zhang, Q. Isolation and identification of antibacterial bioactive compounds from Bacillus megaterium L2. Front. Microbiol. 2021, 12, 645484. [Google Scholar] [CrossRef]
- Magaldi, S.; Mata-Essayag, S.; De Capriles, C.H.; Pérez, C.; Colella, M.T.; Olaizola, C.; Ontiveros, Y. Well diffusion for antifungal susceptibility testing. Int. J. Infect. Dis. 2004, 8, 39–45. [Google Scholar] [CrossRef]
- Ebrahimzadeh, M.A.; Nabavi, S.M.; Nabavi, S.F.; Bahramian, F.; Bekhradnia, A.R. Antioxidant and free radical scavenging activity of H. officinalis L. var. angustifolius, V. odorata, B. hyrcana and C. speciosum. Pak. J. Pharm. Sci. 2010, 23, 29–34. [Google Scholar]
- De Coster, W.; Rademakers, R. NanoPack2: Population-scale evaluation of long-read sequencing data. Bioinformatics 2023, 39, btad311. [Google Scholar] [CrossRef]
- Silva, G.G.; Dutilh, B.E.; Matthews, T.D.; Elkins, K.; Schmieder, R.; Dinsdale, E.A.; Edwards, R.A. Combining de novo and reference-guided assembly with scaffold_builder. Source Code Biol. Med. 2013, 8, 23. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and high-performance computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]

| Fungi Code No. | Diameter of Inhibition Zone (mm) | ||||
|---|---|---|---|---|---|
| E. coli | S. aureus | A. brasiliensis | F. oxysporum | C. albicans | |
| 3 | 18.67 ± 0.58 | 18.00 ± 1.00 | 15.67 ± 1.53 | 14.67 ± 0.58 | 16.67 ± 0.58 |
| 4 | 19.00 ± 1.00 | 16.00 ± 1.00 | 13.00 ± 0.58 | 12.00 ± 1.53 | 15.33 ± 0.58 |
| 7 | 19.33 ± 1.53 | 17.67 ± 0.58 | Nil | Nil | Nil |
| 10 | Nil | Nil | 12.67 ± 1.53 | 11.33 ± 0.58 | 10.00 ± 1.00 |
| 12 | Nil | Nil | Nil | Nil | 9.67 ± 1.53 |
| 13 | Nil | Nil | 18.67 ± 0.58 | 10.67 ± 0.58 | 11.00 ± 1.00 |
| 15 | 13.00 ± 1.00 | 12.67.00 ± 0.58 | Nil | Nil | Nil |
| 17 | Nil | Nil | Nil | Nil | 10.00 ± 1.00 |
| 20 | 10.67 ± 1.58 | 11.00 ± 1.00 | Nil | Nil | Nil |
| 22 | Nil | Nil | Nil | Nil | 12.33 ± 0.55 |
| 24 | 10.33 ± 0.58 | 12.00 ± 1.00 | Nil | Nil | Nil |
| 27 | Nil | Nil | 10.00 ± 1.00 | 10.67 ± 1.58 | 15.33 ± 0.58 |
| 28 | 10.33 ± 0.58 | 12.00 ± 1.00 | Nil | Nil | Nil |
| 30 | 10.33 ± 0.58 | 12.00 ± 1.00 | Nil | Nil | Nil |
| 32 | Nil | Nil | Nil | Nil | 11.00 ± 1.00 |
| 34 | 14.67 ± 1.58 | 12.00 ± 1.00 | Nil | Nil | Nil |
| 35 | Nil | Nil | Nil | Nil | Nil |
| 37 | 12.33 ± 0.55 | 10.00 ± 1.00 | Nil | Nil | Nil |
| 39 | Nil | Nil | Nil | Nil | 14.33 ± 0.58 |
| 40 | 13.67 ± 1.58 | 14.33 ± 0.58 | Nil | Nil | Nil |
| 42 | Nil | Nil | 14.00 ± 1.00 | 11.67 ± 0.58 | 12.67 ± 0.58 |
| 43 | 15.00 ± 1.00 | 14.67 ± 0.58 | 11.33 ± 1.00 | 12.00 ± 1.00 | 15.00 ± 1.00 |
| Control | 9.00 | 9.00 | 11.00 | 10.00 | 12.00 |
| LSD | 0.875 | 0.619 | 0.557 | 0.398 | 0.661 |
| Fungi Code No. | Diameter of Inhibition Zone (mm) | ||||
|---|---|---|---|---|---|
| E. coli | S. aureus | A. brasiliensis | F. oxysporum | C. albicans | |
| 3 | 10.33 ± 0.58 | 10.00 ± 1.00 | Nil | Nil | Nil |
| 4 | 12.00 ± 1.00 | 13.67 ± 1.58 | Nil | Nil | Nil |
| 5 | Nil | Nil | Nil | Nil | 10.00 ± 1.00 |
| 7 | 19.00 ± 1.00 | 18.33 ± 0.58 | 12.00 ± 1.00 | 10.33 ± 0.58 | 15.67 ± 1.53 |
| 10 | Nil | Nil | 10.00 ± 1.00 | 9.67 ± 1.58 | 12.33 ± 0.55 |
| 13 | 10.00 ± 1.00 | 11.33 ± 1.55 | Nil | Nil | 11.00 ± 1.00 |
| 15 | 14.33 ± 0.58 | 13.00 ± 1.00 | Nil | Nil | Nil |
| 17 | Nil | Nil | Nil | Nil | 12.00 ± 1.00 |
| 20 | Nil | Nil | Nil | Nil | 14.00 ± 1.00 |
| 22 | 13.00 ± 1.00 | 12.67 ± 1.58 | Nil | Nil | Nil |
| 23 | Nil | Nil | Nil | Nil | 9.33 ± 0.55 |
| 24 | 14.33 ± 0.58 | 16.00 ± 1.00 | Nil | Nil | Nil |
| 26 | 12.33 ± 0.55 | 11.67 ± 1.58 | Nil | Nil | Nil |
| 27 | Nil | Nil | Nil | Nil | 15.33 ± 0.58 |
| 28 | 14.00 ± 1.00 | 14.33 ± 0.55 | Nil | Nil | Nil |
| 30 | 11.67 ± 1.58 | 10.33 ± 0.55 | Nil | Nil | Nil |
| 31 | Nil | Nil | 15.00 ± 1.00 | 13.33 ± 0.58 | Nil |
| 32 | Nil | Nil | Nil | Nil | 12.00 ± 1.00 |
| 34 | 10.00 ± 1.00 | 11.33 ± 0.55 | Nil | Nil | Nil |
| 37 | 14.00 ± 1.00 | 12.67 ± 1.58 | Nil | Nil | Nil |
| 39 | Nil | Nil | Nil | Nil | 10.33 ± 0.58 |
| 40 | 10.33 ± 0.55 | 10.67 ± 1.58 | Nil | Nil | Nil |
| 42 | 20.00 ± 1.00 | 18.33 ± 0.55 | 10.00 ± 1.00 | 12.67 ± 1.58 | 17.00 ± 1.00 |
| 43 | Nil | Nil | 9.00 ± 1.00 | 10.33 ± 0.55 | 13.67 ± 1.58 |
| Control | 9.00 | 9.00 | 11.00 | 10.00 | 12.00 |
| LSD | 0.739 | 0.821 | 0.872 | 0.298 | 0.871 |
| Fungi Code No. | Scavenging Activity (%) | |
|---|---|---|
| Biomass Extract | Cell-Free Filtrate Extract | |
| 1 | Nil | 21.76 ± 1.47 |
| 3 | 54.84 ± 10.73 | Nil |
| 4 | 44.96 ± 8.45 | Nil |
| 6 | Nil | 13.21 ± 2.56 |
| 7 | 67.41 ± 10.7 | 25.76 ± 5.87 |
| 8 | 12.91 ± 0.05 | 10.44 ± 1.36 |
| 9 | 21.93 ± 0.15 | 9.48 ± 1.05 |
| 10 | 12.09 ± 0.33 | 14.36 ± 1.58 |
| 15 | Nil | 62.47 ± 10.13 |
| 18 | Nil | 10.46 ± 0.34 |
| 20 | 34.55 ± 6.21 | 54.37 ± 1.48 |
| 22 | 4.87 ± 5.21 | 38.21 ± 11.46 |
| 23 | 20.77 ± 11.2 | Nil |
| 24 | 44.87 ± 9.21 | 25.57 ± 1.58 |
| 29 | Nil | 14.56 ± 1.48 |
| 30 | 7.90 ± 0.79 | Nil |
| 31 | 11.57 ± 1.45 | 27.58 ± 0.88 |
| 32 | 18.43 ± 1.21 | 12.00 ± 1.00 |
| 34 | 12.76 ± 1.05 | 43.65 ± 7.78 |
| 35 | 24.76 ± 1.89 | 26.01 ± 5.38 |
| 39 | 7.43 ± 0.17 | 13.57 ± 1.21 |
| 42 | 31.55 ± 2.32 | 46.46 ± 11.91 |
| 43 | 45.65 ± 3.67 | Nil |
| Control | 85.32 | 85.32 |
| LSD | 5.985 | 6.921 |
| Fungal Strain | Dose (kGy) | Diameter of Inhibition Zone (mm) | Scavenging Activity (%) | ||||
|---|---|---|---|---|---|---|---|
| E. coli | S. aureus | A. brasiliensis | F. oxysporum | C. albicans | |||
| T. harzianum BUK-T | 0.0 (C) | 18.00 ± 1.00 | 17.67 ± 1.58 | 14.33 ± 0.55 | 12.00 ± 1.00 | 16.33 ± 0.55 | 55.02 ± 9.89 |
| 0.5 | 20.33 ± 0.55 | 19.00 ± 1.00 | 14.00 ± 1.00 | 14.33 ± 0.55 | 18.00 ± 1.00 | 58.43 ± 7.31 | |
| 1 | 22.67 ± 1.67 | 24.00 ± 1.00 | 16.67 ± 1.58 | 14.67 ± 1.58 | 21.00 ± 1.0 | 75.32 ± 10.03 | |
| 2 | 10.00 ± 1.00 | 9.33 ± 0.55 | 10.00 ± 1.00 | 9.33 ± 0.55 | 12.67 ± 1.58 | 43.76 ± 7.55 | |
| 4 | Nil | Nil | Nil | Nil | Nil | 10.32 ± 1.76 | |
| 8 | Nil | Nil | Nil | Nil | Nil | Nil | |
| A. ochraceus ROB-L1 | 0.0 (C) | 19.33 ± 0.55 | 15.67 ± 1.58 | 14.00 ± 1.00 | 12.33 ± 0.55 | 14.67 ± 1.58 | 45.06 ± 10.21 |
| 0.5 | 19.67 ± 1.58 | 16.00 ± 1.00 | 14.67 ± 1.58 | 14.00 ± 1.00 | 15.33 ± 0.55 | 47.91 ± 11.45 | |
| 1 | 20.00 ± 1.00 | 18.33 ± 0.55 | 16.00 ± 1.00 | 15.67 ± 1.58 | 18.00 ± 1.00 | 56.97 ± 5.81 | |
| 2 | 23.00 ± 1.00 | 25.00 ± 1.00 | 18.67 ± 1.58 | 17.00 ± 1.00 | 23.33 ± 0.55 | 67.37 ± 12.41 | |
| 4 | 10.67 ± 1.58 | 11.33 ± 0.55 | 12.33 ± 0.55 | 11.67 ± 1.58 | 10.00 ± 1.00 | 32.90 ± 7.38 | |
| 8 | Nil | Nil | Nil | Nil | Nil | Nil | |
| C. cochliodes KLON-L1 | 0.0 (C) | 18.67 ± 1.58 | 17.67 ± 1.58 | 12.33 ± 0.55 | 11.00 ± 1.00 | 14.33 ± 0.55 | 26.41 ± 8.91 |
| 0.5 | 21.00 ± 1.00 | 19.33 ± 0.55 | 14.00 ± 1.00 | 14.33 ± 0.55 | 23.00 ± 1.00 | 31.44 ± 5.89 | |
| 1 | 19.33 ± 0.55 | 16.00 ± 1.00 | 10.67 ± 1.58 | 12.00 ± 1.00 | 16.33 ± 0.55 | 22.19 ± 10.34 | |
| 2 | 10.66 ± 1.58 | 9.00 ± 1.00 | 9.33 ± 0.55 | 10.67 ± 1.58 | 9.00 ± 1.00 | 6.37 ± 1.28 | |
| 4 | Nil | Nil | Nil | Nil | Nil | Nil | |
| 8 | Nil | Nil | Nil | Nil | Nil | Nil | |
| F. tricinctum KLON-L2 | 0.0 (C) | 19.67 ± 1.58 | 18.00 ± 1.00 | 10.33 ± 0.55 | 11.33 ± 0.55 | 16.67 ± 1.58 | 45.81 ± 10.52 |
| 0.5 | 19.33 ± 0.55 | 19.00 ± 1.00 | 12.00 ± 1.00 | 13.00 ± 1.00 | 18.33 ± 0.55 | 61.32 ± 12.31 | |
| 1 | 26.00 ± 1.00 | 25.67 ± 1.58 | 16.00 ± 1.00 | 15.67 ± 1.58 | 24.00 ± 1.00 | 87.51 ± 14.02 | |
| 2 | 20.33 ± 0.55 | 21.00 ± 1.00 | 14.33 ± 0.55 | 14.00 ± 1.00 | 20.33 ± 0.55 | 77.37 ± 12.43 | |
| 4 | 9.00 ± 1.00 | 10.33 ± 0.58 | 11.67 ± 1.58 | 9.00 ± 1.00 | 10.00 ± 1.00 | Nil | |
| 8 | Nil | Nil | Nil | Nil | Nil | Nil | |
| P. chrysogenum SOS-B2 | 0.0 (C) | 14.00 ± 1.00 | 14.33 ± 0.55 | 10.67 ± 1.58 | 11.67 ± 1.58 | 14.67 ± 1.58 | 44.98 ± 7.31 |
| 0.5 | 14.00 ± 1.00 | 15.00 ± 1.00 | 12.00 ± 1.00 | 12.33 ± 0.55 | 15.00 ± 1.00 | 67.32 ± 12.41 | |
| 1 | 16.67 ± 1.58 | 16.67 ± 1.58 | 13.00 ± 1.00 | 14.00 ± 1.00 | 15.33 ± 0.55 | 71.22 ± 14.57 | |
| 2 | 20.00 ± 1.0 | 19.00 ± 1.00 | 18.67 ± 1.58 | 14.67 ± 1.58 | 18.00 ± 1.00 | 73.49 ± 10.21 | |
| 4 | Nil | Nil | Nil | Nil | Nil | 3.67 ± 1.22 | |
| 8 | Nil | Nil | Nil | Nil | Nil | Nil | |
| Control | 9.00 | 9.00 | 11.00 | 10.00 | 12.00 | 85.32 | |
| LSD | 0.840 | 0.872 | 0.698 | 0.871 | 0.569 | 6.921 | |
| Fungus | S.N. | RT (min) | Detected Compounds | Bioactivity | Reference |
|---|---|---|---|---|---|
| T. harzianum BUK-T | 1 | 41.19 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- | Anticancer, antioxidant | [50] |
| 2 | 47.12 | Palmitic acid | Anti-inflammatory | [51] | |
| 3 | 53.7 | cis-9-Hexadecenal | Antifungal, antimelanogenic | [52] | |
| 4 | 54.4 | Stearic acid | Neuroprotection | [53] | |
| 5 | 59.97 | 9-Octadecenamide, (Z)- | Lipid metabolism | [54] | |
| A. ochraceus ROB-L1 | 1 | 43.351 | Cyclo(leucyloprolyl) derivative | ||
| 2 | 46.523 | Cyclo(leucyloprolyl) | Antimicrobial | [55] | |
| 3 | 47.078 | Cyclo(leucyloprolyl) derivative | |||
| 4 | 60.33 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl)- | Anticancer | [50] | |
| 5 | 66.7 | unknown | |||
| C. cochliodes KLON-L1 | 1 | 39.305 | 7-Oxabicyclo[4.1.0]heptane, 3-oxiranyl- | ||
| 2 | 39.614 | Uric acid | Antioxidant | [56] | |
| 3 | 43.351 | Cyclo(leucyloprolyl) derivative | |||
| 4 | 44.36 | 1-(3-Methylbutyryl)pyrrolidine | |||
| 5 | 46.523 | Cyclo(leucyloprolyl) | Antimicrobial | [55] | |
| 6 | 47.078 | Cyclo(leucyloprolyl) derivative | |||
| 7 | 47.313 | Proline | |||
| 8 | 47.489 | Cyclo(leucyloprolyl) derivative | |||
| 9 | 48.226 | Palmitic acid | Anti-inflammatory | [51] | |
| 10 | 56.264 | Cyclo(leucyloprolyl) derivative | |||
| 11 | 58.995 | Dehydroergotamine | Antimicrobial | [57] | |
| 12 | 59.966 | 9-Octadecenamide, (Z)- | Lipid metabolism | [54] | |
| 13 | 60.33 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl)- | Anticancer | [50] | |
| F. tricinctum KLON-L2 | 1 | 59.29 | Squalene | Antioxidant, anticancer | [58] |
| 2 | 64.9 | Octacosane, 2-methyl- | |||
| P. chrysogenum SOS-B2 | 1 | 41 | Pentadecanoic acid | ||
| 2 | 48.7 | Linolelaidic acid, methyl ester | |||
| 3 | 50.51 | 9,12-Octadecadien-1-ol, (Z,Z) | Anti-inflammatory, hypocholesterolemic, hepatoprotective | [59] | |
| 4 | 50.7 | Oleic Acid | Antifungal, antitermite | [60] | |
| 5 | 51.86 | Behenic acid | Antibacterial | [61] | |
| 6 | 58.84 | Oleic acid amide | Antifungal, antitermite | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sayed, E.-S.R.; Baskaran, A.; Pomarańska, O.; Mykhailova, D.; Dunal, A.; Dudek, A.; Satam, S.; Strzała, T.; Łyczko, J.; Olejniczak, T.; et al. Bioprospecting Endophytic Fungi of Forest Plants for Bioactive Metabolites with Antibacterial, Antifungal, and Antioxidant Potentials. Molecules 2024, 29, 4746. https://doi.org/10.3390/molecules29194746
El-Sayed E-SR, Baskaran A, Pomarańska O, Mykhailova D, Dunal A, Dudek A, Satam S, Strzała T, Łyczko J, Olejniczak T, et al. Bioprospecting Endophytic Fungi of Forest Plants for Bioactive Metabolites with Antibacterial, Antifungal, and Antioxidant Potentials. Molecules. 2024; 29(19):4746. https://doi.org/10.3390/molecules29194746
Chicago/Turabian StyleEl-Sayed, El-Sayed R., Abirami Baskaran, Oliwia Pomarańska, Daria Mykhailova, Anna Dunal, Anita Dudek, Sahil Satam, Tomasz Strzała, Jacek Łyczko, Teresa Olejniczak, and et al. 2024. "Bioprospecting Endophytic Fungi of Forest Plants for Bioactive Metabolites with Antibacterial, Antifungal, and Antioxidant Potentials" Molecules 29, no. 19: 4746. https://doi.org/10.3390/molecules29194746
APA StyleEl-Sayed, E.-S. R., Baskaran, A., Pomarańska, O., Mykhailova, D., Dunal, A., Dudek, A., Satam, S., Strzała, T., Łyczko, J., Olejniczak, T., & Boratyński, F. (2024). Bioprospecting Endophytic Fungi of Forest Plants for Bioactive Metabolites with Antibacterial, Antifungal, and Antioxidant Potentials. Molecules, 29(19), 4746. https://doi.org/10.3390/molecules29194746








