Identification and Reconstitution of the First Two Enzymatic Steps for the Biosynthesis of Bioactive Meroterpenoids from Hericium erinaceus (Lion’s Mane Mushroom)
Abstract
:1. Introduction
2. Results and Discussion
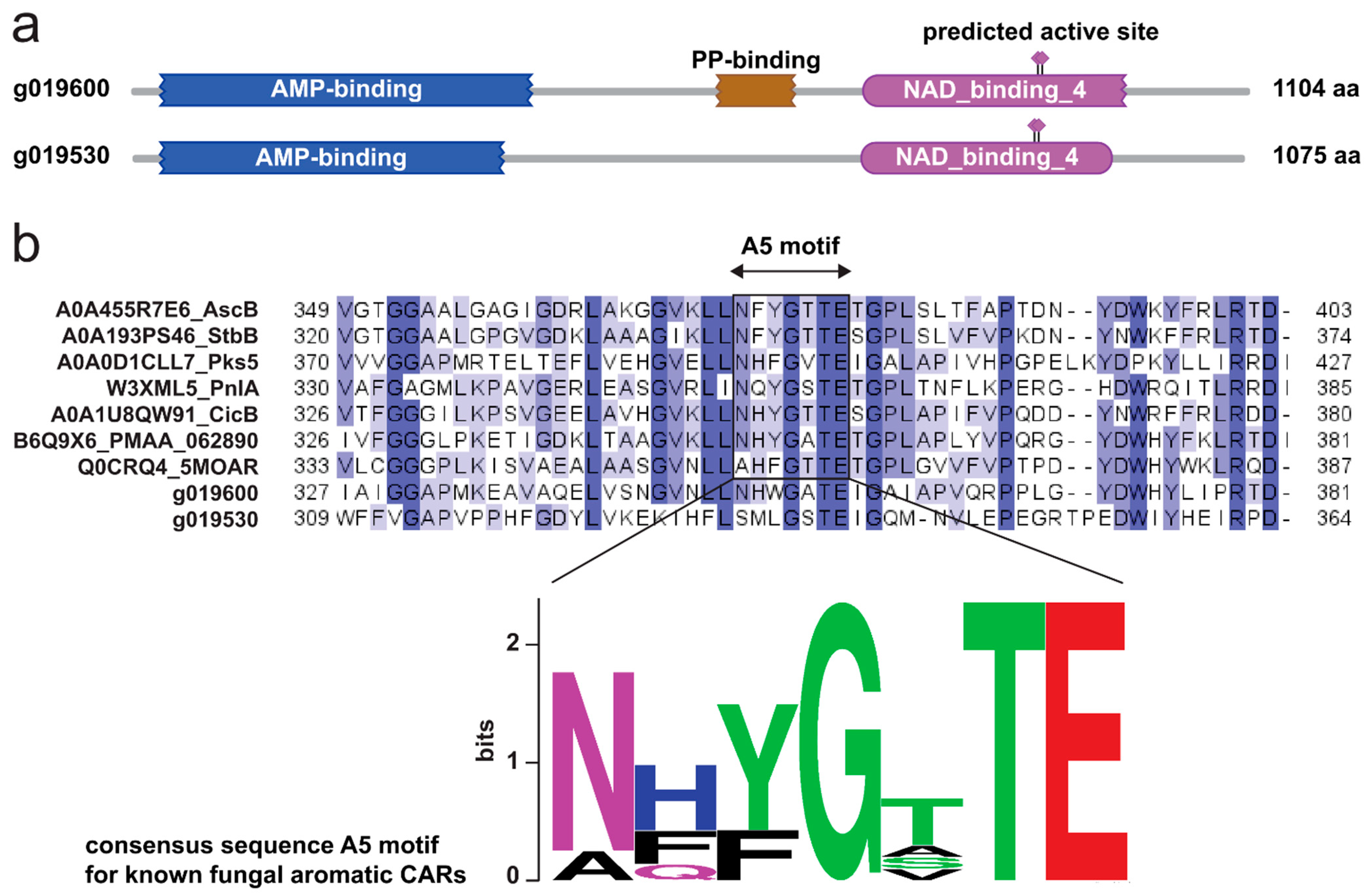
2.1. The Genome of Hericium erinaceus Encodes a Putative Hericenone BGC
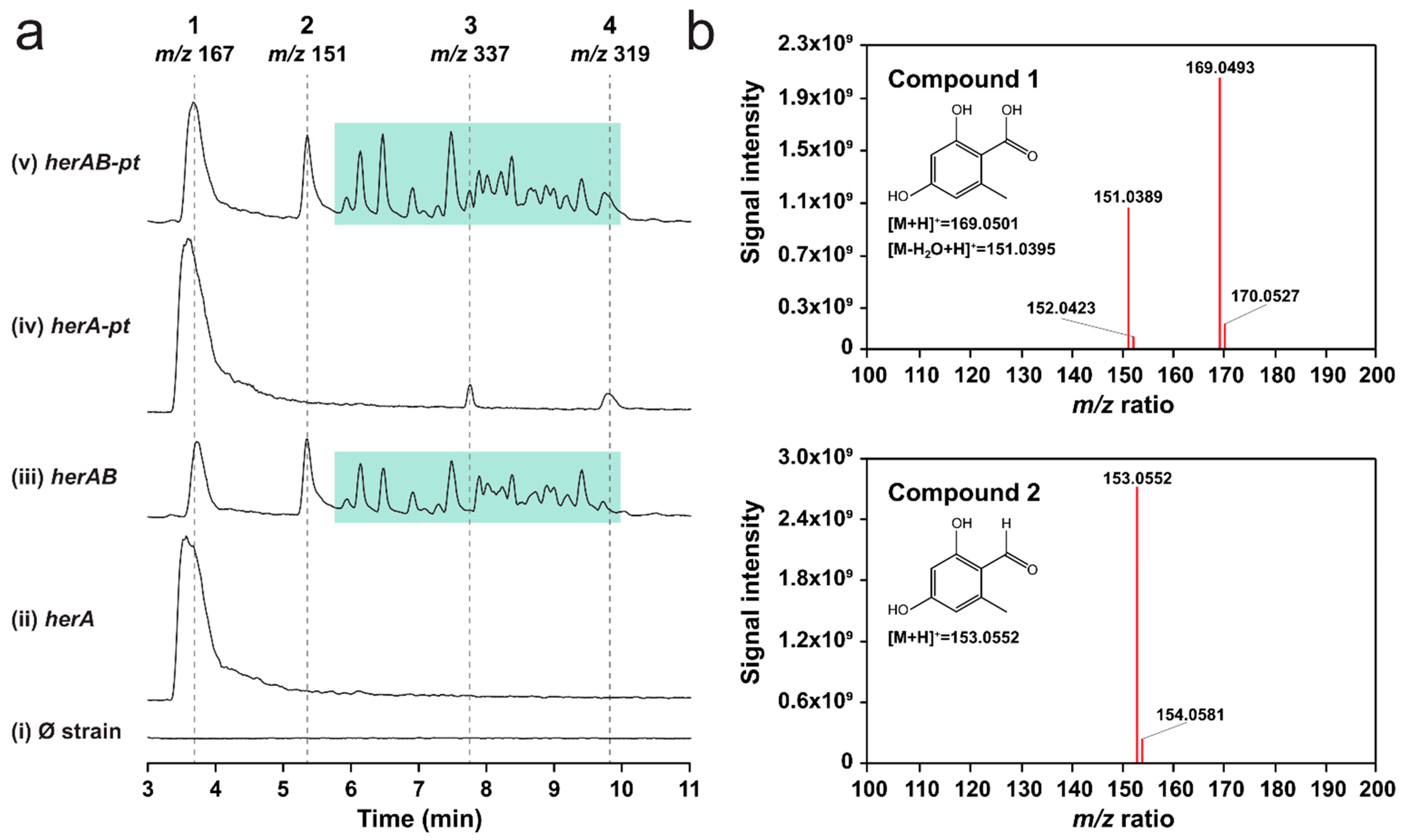
2.2. Overexpression of herA and herB in A. oryzae Results in the Production of Orsellinic Aldehyde
2.3. Overexpression of Gene g074890 Yields Two New Compounds That May Be Prenylated Variants of Orsellinic Acid
3. Conclusions
4. Materials and Methods
4.1. Bioinformatic Analyses
4.2. Fungal Strains
4.3. Amplification and Molecular Cloning of herA, herB, and g074890
4.4. Genetic Transformation of A. oryzae NSAR1
4.5. Cultivation and Extraction of Fungal Metabolites
4.6. HPLC-DAD-MS and HRMS-MS Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Shankar, A.; Sharma, K.K. Fungal secondary metabolites in food and pharmaceuticals in the era of multi-omics. Appl. Microbiol. Biotechnol. 2022, 106, 3465–3488. [Google Scholar] [CrossRef] [PubMed]
- Charria-Girón, E.; Surup, F.; Marin-Felix, Y. Diversity of biologically active secondary metabolites in the ascomycete order Sordariales. Mycol. Prog. 2022, 21, 43. [Google Scholar] [CrossRef]
- Helaly, S.E.; Thongbai, B.; Stadler, M. Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales. Nat. Prod. Rep. 2018, 35, 992–1014. [Google Scholar] [CrossRef] [PubMed]
- Shwab, E.K.; Keller, N.P. Regulation of secondary metabolite production in filamentous ascomycetes. Mycol. Res. 2008, 112, 225–230. [Google Scholar] [CrossRef]
- Sanchez, J.F.; Somoza, A.D.; Keller, N.P.; Wang, C.C.C. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat. Prod. Rep. 2012, 29, 351. [Google Scholar] [CrossRef]
- Assaf, C.E.H.; Zetina-Serrano, C.; Tahtah, N.; El Khoury, A.; Atoui, A.; Oswald, I.P.; Puel, O.; Lorber, S. Regulation of Secondary Metabolism in the Penicillium Genus. Int. J. Mol. Sci. 2020, 21, 9462. [Google Scholar] [CrossRef]
- Alberti, F.; Kaleem, S.; Weaver, J.A. Recent developments of tools for genome and metabolome studies in basidiomycete fungi and their application to natural product research. Biol. Open 2020, 9, bio056010. [Google Scholar] [CrossRef]
- Lin, H.-C.; Hewage, R.T.; Lu, Y.-C.; Chooi, Y.-H. Biosynthesis of bioactive natural products from Basidiomycota. Org. Biomol. Chem. 2019, 17, 1027–1036. [Google Scholar] [CrossRef]
- Gressler, M.; Löhr, N.A.; Schäfer, T.; Lawrinowitz, S.; Seibold, P.S.; Hoffmeister, D. Mind the mushroom: Natural product biosynthetic genes and enzymes of Basidiomycota. Nat. Prod. Rep. 2021, 38, 702–722. [Google Scholar] [CrossRef]
- Mau, J.-L.; Lin, H.-C.; Song, S.-F. Antioxidant properties of several specialty mushrooms. Food Res. Int. 2002, 35, 519–526. [Google Scholar] [CrossRef]
- Friedman, M. Chemistry, Nutrition, and Health-Promoting Properties of Hericium erinaceus (Lion’s Mane) Mushroom Fruiting Bodies and Mycelia and Their Bioactive Compounds. J. Agric. Food Chem. 2015, 63, 7108–7123. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M. Mushroom Polysaccharides: Chemistry and Antiobesity, Antidiabetes, Anticancer, and Antibiotic Properties in Cells, Rodents, and Humans. Foods 2016, 5, 80. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, X.; Fang, J.; Chang, Y.; Ning, N.; Guo, H.; Huang, L.; Huang, X.; Zhao, Z. Structures, biological activities, and industrial applications of the polysaccharides from Hericium erinaceus (Lion’s Mane) mushroom: A review. Int. J. Biol. Macromol. 2017, 97, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.-J.; Shen, J.-W.; Yu, H.-Y.; Ruan, Y.; Wu, T.-T.; Zhao, X. Hericenones and erinacines: Stimulators of nerve growth factor (NGF) biosynthesis in Hericium erinaceus. Mycology 2010, 1, 92–98. [Google Scholar] [CrossRef]
- Li, W.; Zhou, W.; Kim, E.J.; Shim, S.H.; Kang, H.K.; Kim, Y.H. Isolation and identification of aromatic compounds in Lion’s Mane Mushroom and their anticancer activities. Food Chem. 2015, 170, 336–342. [Google Scholar] [CrossRef]
- Ryu, S.H.; Hong, S.M.; Khan, Z.; Lee, S.K.; Vishwanath, M.; Turk, A.; Yeon, S.W.; Jo, Y.H.; Lee, D.H.; Lee, J.K.; et al. Neurotrophic isoindolinones from the fruiting bodies of Hericium erinaceus. Bioorg. Med. Chem. Lett. 2021, 31, 127714. [Google Scholar] [CrossRef]
- Kobayashi, S.; Tamura, T.; Koshishiba, M.; Yasumoto, T.; Shimizu, S.; Kintaka, T.; Nagai, K. Total Synthesis, Structure Revision, and Neuroprotective Effect of Hericenones C–H and Their Derivatives. J. Org. Chem. 2021, 86, 2602–2620. [Google Scholar] [CrossRef]
- Nazir, M.; Saleem, M.; Tousif, M.I.; Anwar, M.A.; Surup, F.; Ali, I.; Wang, D.; Mamadalieva, N.Z.; Alshammari, E.; Ashour, M.L.; et al. Meroterpenoids: A Comprehensive Update Insight on Structural Diversity and Biology. Biomolecules 2021, 11, 957. [Google Scholar] [CrossRef]
- Jiang, M.; Wu, Z.; Liu, L.; Chen, S. The chemistry and biology of fungal meroterpenoids (2009–2019). Org. Biomol. Chem. 2021, 19, 1644–1704. [Google Scholar] [CrossRef]
- Bentley, R. Mycophenolic Acid: A One Hundred Year Odyssey from Antibiotic to Immunosuppressant. Chem. Rev. 2000, 100, 3801–3826. [Google Scholar] [CrossRef] [PubMed]
- Regueira, T.B.; Kildegaard, K.R.; Hansen, B.G.; Mortensen, U.H.; Hertweck, C.; Nielsen, J. Molecular Basis for Mycophenolic Acid Biosynthesis in Penicillium brevicompactum. Appl. Environ. Microbiol. 2011, 77, 3035–3043. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.N.; Shahbazi, F.; Rondeau-Gagné, S.; Trant, J.F. The biosynthesis of the cannabinoids. J. Cannabis Res. 2021, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.A.; Whalley, B.J. The proposed mechanisms of action of CBD in epilepsy. Epileptic Disord. 2020, 22, S10–S15. [Google Scholar] [CrossRef] [PubMed]
- Iijima, M.; Munakata, R.; Takahashi, H.; Kenmoku, H.; Nakagawa, R.; Kodama, T.; Asakawa, Y.; Abe, I.; Yazaki, K.; Kurosaki, F.; et al. Identification and Characterization of Daurichromenic Acid Synthase Active in Anti-HIV Biosynthesis. Plant Physiol. 2017, 174, 2213–2230. [Google Scholar] [CrossRef]
- Okada, M.; Saito, K.; Wong, C.P.; Li, C.; Wang, D.; Iijima, M.; Taura, F.; Kurosaki, F.; Awakawa, T.; Abe, I. Combinatorial Biosynthesis of (+)-Daurichromenic Acid and Its Halogenated Analogue. Org. Lett. 2017, 19, 3183–3186. [Google Scholar] [CrossRef]
- Wong, K.-H.; Naidu, M.; David, P.; Abdulla, M.A.; Abdullah, N.; Kuppusamy, U.R.; Sabaratnam, V. Peripheral Nerve Regeneration Following Crush Injury to Rat Peroneal Nerve by Aqueous Extract of Medicinal Mushroom Hericium erinaceus (Bull.: Fr) Pers. (Aphyllophoromycetideae). Evid.-Based Complement. Altern. Med. 2011, 2011, 580752. [Google Scholar] [CrossRef]
- Wong, K.H.; Naidu, M.; David, P.; Bakar, R.; Sabaratnam, V. Neuroregenerative potential of lion’s mane mushroom, Hericium erinaceus (Bull.: Fr.) Pers. (Higher Basidiomycetes), in the treatment of peripheral nerve injury (review). Int. J. Med. Mushrooms 2012, 14, 427–446. [Google Scholar] [CrossRef]
- Zhang, J.; An, S.; Hu, W.; Teng, M.; Wang, X.; Qu, Y.; Liu, Y.; Yuan, Y.; Wang, D. The Neuroprotective Properties of Hericium erinaceus in Glutamate-Damaged Differentiated PC12 Cells and an Alzheimer’s Disease Mouse Model. Int. J. Mol. Sci. 2016, 17, 1810. [Google Scholar] [CrossRef]
- Lee, S.-L.; Hsu, J.-Y.; Chen, T.-C.; Huang, C.-C.; Wu, T.-Y.; Chin, T.-Y. Erinacine A Prevents Lipopolysaccharide-Mediated Glial Cell Activation to Protect Dopaminergic Neurons against Inflammatory Factor-Induced Cell Death In Vitro and In Vivo. Int. J. Mol. Sci. 2022, 23, 810. [Google Scholar] [CrossRef]
- Kawagishi, H.; Ando, M.; Mizuno, T. Hericenone A and B as cytotoxic principles from the mushroom. Tetrahedron Lett. 1990, 31, 373–376. [Google Scholar] [CrossRef]
- Tao, H.; Abe, I. Enzymology and biosynthesis of the orsellinic acid derived medicinal meroterpenoids. Curr. Opin. Biotechnol. 2021, 69, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, P.-L.; Ran, H.; Fan, J.; Yin, W.-B. Characterization of a NRPS-like Protein from Pestalotiopsis fici for Aldehyde Generation. J. Fungi 2022, 8, 1001. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Matsuda, Y.; Gao, H.; Hu, D.; Yao, X.S.; Abe, I. Biosynthesis of LL-Z1272β: Discovery of a New Member of NRPS-like Enzymes for Aryl-Aldehyde Formation. ChemBioChem 2016, 17, 904–907. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Awakawa, T.; Matsuzaki, M.; Cho, R.; Matsuda, Y.; Hoshino, S.; Shinohara, Y.; Yamamoto, M.; Kido, Y.; Inaoka, D.K.; et al. Complete biosynthetic pathways of ascofuranone and ascochlorin in Acremonium egyptiacum. Proc. Natl. Acad. Sci. USA 2019, 116, 8269–8274. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; E Augustijn, H.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Chen, J.; Zeng, X.; Yang, Y.L.; Xing, Y.M.; Zhang, Q.; Li, J.M.; Ma, K.; Liu, H.W.; Guo, S.X. Genomic and transcriptomic analyses reveal differential regulation of diverse terpenoid and polyketides secondary metabolites in Hericium erinaceus. Sci. Rep. 2017, 7, 10151. [Google Scholar] [CrossRef]
- Humann, J.L.; Lee, T.; Ficklin, S.; Main, D. Structural and Functional Annotation of Eukaryotic Genomes with GenSAS. In Gene Prediction Methods in Molecular Biology; Kollmar, M., Ed.; Humana: New York, NY, USA, 2019; Volume 1962, pp. 29–51. [Google Scholar] [CrossRef]
- Lazarus, C.M.; Williams, K.; Bailey, A.M. Reconstructing fungal natural product biosynthetic pathways. Nat. Prod. Rep. 2014, 31, 1339–1347. [Google Scholar] [CrossRef]
- Jin, F.J.; Maruyama, J.I.; Juvvadi, P.R.; Arioka, M.; Kitamoto, K. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol. Lett. 2004, 239, 79–85. [Google Scholar] [CrossRef]
- Han, H.; Yu, C.; Qi, J.; Wang, P.; Zhao, P.; Gong, W.; Xie, C.; Xia, X.; Liu, C. High-efficient production of mushroom polyketide compounds in a platform host Aspergillus oryzae. Microb. Cell Fact. 2023, 22, 60. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. Available online: https://www.repeatmasker.org (accessed on 11 October 2022).
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef]
- Borodovsky, M.; Lomsadze, A. Eukaryotic Gene Prediction Using GeneMark.hmm-E and GeneMark-ES. Curr. Protoc. Bioinform. 2011, 35, 4.6.1–4.6.10. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome. Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Iacovelli, R.; He, T.; Allen, J.L.; Hackl, T.; Haslinger, K. Genome sequencing and molecular networking analysis of the wild fungus Anthostomella pinea reveal its ability to produce a diverse range of secondary metabolites. Fungal Biol. Biotechnol. 2024, 11, 1. [Google Scholar] [CrossRef]
- Hutner, S.H.; Provasoli, L.; Schatz, A.; Haskins, C.P. Some approaches to the study of the role of metals in the metabolism of microorganisms. Proc. Am. Philos. Soc. 1950, 94, 152–170. [Google Scholar]
- Iacovelli, R.; He, S.; Sokolova, N.; Fodran, P.; Haslinger, K. Discovery and heterologous expression of functional 4-O-dimethylallyl-L-tyrosine synthases from lichen-forming fungi. J. Nat. Prod. 2024, 87, 2243–2254. [Google Scholar] [CrossRef]
- Kanemori, Y.; Gomi, K.; Kitamoto, K.; Kumagai, C.; Tamura, G. Insertion Analysis of Putative Functional Elements in the Promoter Region of the Aspergillus oryzae Taka-amylase A Gene (amyB) Using a Heterologous Aspergillus nidulans amdS-lacZ Fusion Gene System. Biosci. Biotechnol. Biochem. 1999, 63, 180–183. [Google Scholar] [CrossRef]
- Schmid, R.; Heuckeroth, S.; Korf, A.; Smirnov, A.; Myers, O.; Dyrlund, T.S.; Bushuiev, R.; Murray, K.J.; Hoffmann, N.; Lu, M.; et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol. 2023, 41, 447–449. [Google Scholar] [CrossRef]

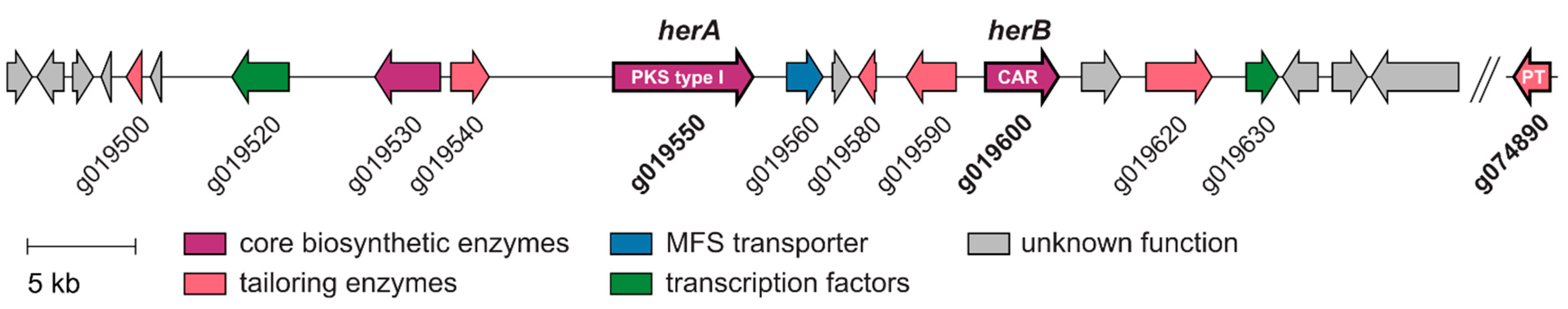
| Gene | Predicted Function | stb BGC Homolog (aa % Identity; % Similarity) | asc BGC Homolog (aa % Identity; % Similarity) |
|---|---|---|---|
| g019500 | Flavoprotein | ||
| g019520 | Fungal-specific transcription factor | ||
| g019530 | NRPS-like reductase | ||
| g019540 | Aldehyde dehydrogenase | ||
| g019550_herA | Type I PKS | BAV19379.1_StbA (24.89; 41.98) | BBF25315.1_AscC (25.02; 41.92) |
| g019560 | MFS 1 family transporter | ||
| g019580 | short-chain dehydrogenase/reductase SDR | ||
| g019590 | Monooxygenase FAD-binding | ||
| g019600_herB | Carboxylic acid reductase | BAV19380.1_StbB (31.71; 50.75) | BBF25314.1_AscB(32.10; 50.13) |
| g019620 | Phenylalanine-specific permease | ||
| g019630 | Serine/threonine protein kinase | ||
| g074890 | Prenyltransferase | BAV19381.1_StbC (25.79; 44.99) | BBF25313.1_AscA (26.52; 41.44) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacovelli, R.; Poon, F.; Haslinger, K. Identification and Reconstitution of the First Two Enzymatic Steps for the Biosynthesis of Bioactive Meroterpenoids from Hericium erinaceus (Lion’s Mane Mushroom). Molecules 2024, 29, 5576. https://doi.org/10.3390/molecules29235576
Iacovelli R, Poon F, Haslinger K. Identification and Reconstitution of the First Two Enzymatic Steps for the Biosynthesis of Bioactive Meroterpenoids from Hericium erinaceus (Lion’s Mane Mushroom). Molecules. 2024; 29(23):5576. https://doi.org/10.3390/molecules29235576
Chicago/Turabian StyleIacovelli, Riccardo, Fons Poon, and Kristina Haslinger. 2024. "Identification and Reconstitution of the First Two Enzymatic Steps for the Biosynthesis of Bioactive Meroterpenoids from Hericium erinaceus (Lion’s Mane Mushroom)" Molecules 29, no. 23: 5576. https://doi.org/10.3390/molecules29235576
APA StyleIacovelli, R., Poon, F., & Haslinger, K. (2024). Identification and Reconstitution of the First Two Enzymatic Steps for the Biosynthesis of Bioactive Meroterpenoids from Hericium erinaceus (Lion’s Mane Mushroom). Molecules, 29(23), 5576. https://doi.org/10.3390/molecules29235576







