Network Pharmacology Analysis of Liquid-Cultured Armillaria ostoyae Mycelial Metabolites and Their Molecular Mechanism of Action against Gastric Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Liquid Culture of A. ostoyae Mycelia
2.3. Scanning Electron Microscopy (SEM) Observations of Mycelial Morphology
2.4. Sample Preparation
2.5. UHPLC–QTOF/MS-Based Analysis of Secondary Metabolites in the A. ostoyae Mycelia
2.6. Network Pharmacology Analysis
2.7. Molecular Docking
2.8. Cytotoxicity Analysis
2.8.1. Cell Culture and Treatment
2.8.2. Cell Viability Assay
2.8.3. Cell Adhesion Analysis
2.8.4. Wound Healing Analysis
2.8.5. Transwell Migration Analysis
2.8.6. Immunoblotting
2.9. Data and Statistical Analyses
3. Results
3.1. Growth Pattern of Pure Parent Strain in Solid Culture
3.2. Mycelial Growth Pattern in the Artificial Liquid Culture of A. ostoyae
3.3. UHPLC–QTOF/MS-Based Secondary Metabolite Analysis
3.4. Network Pharmacology Analysis
3.4.1. Screening of Active Ingredients
3.4.2. Identification of Potent Substances and Disease Targets
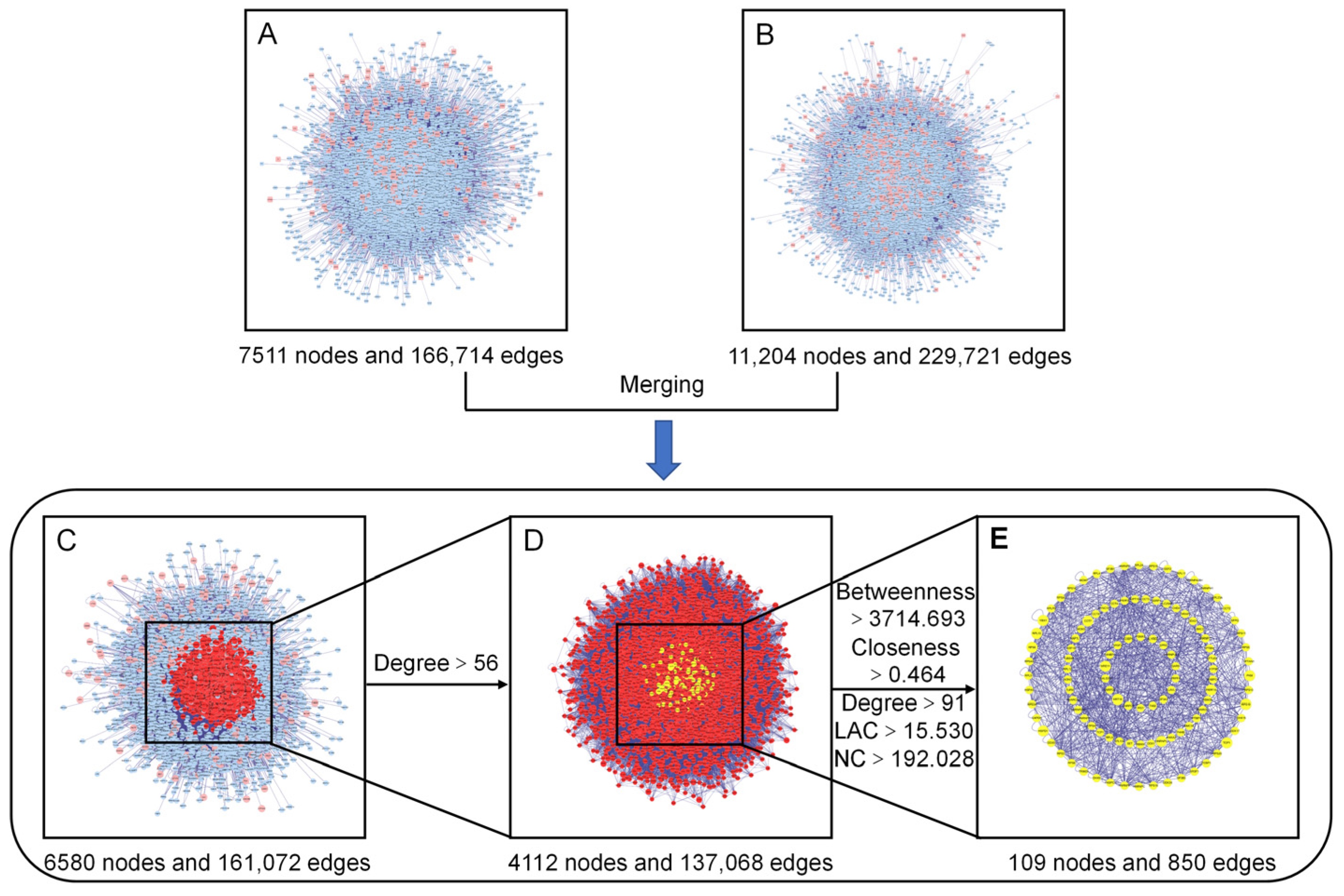
3.4.3. PPI Network Construction, Merging, and Analysis
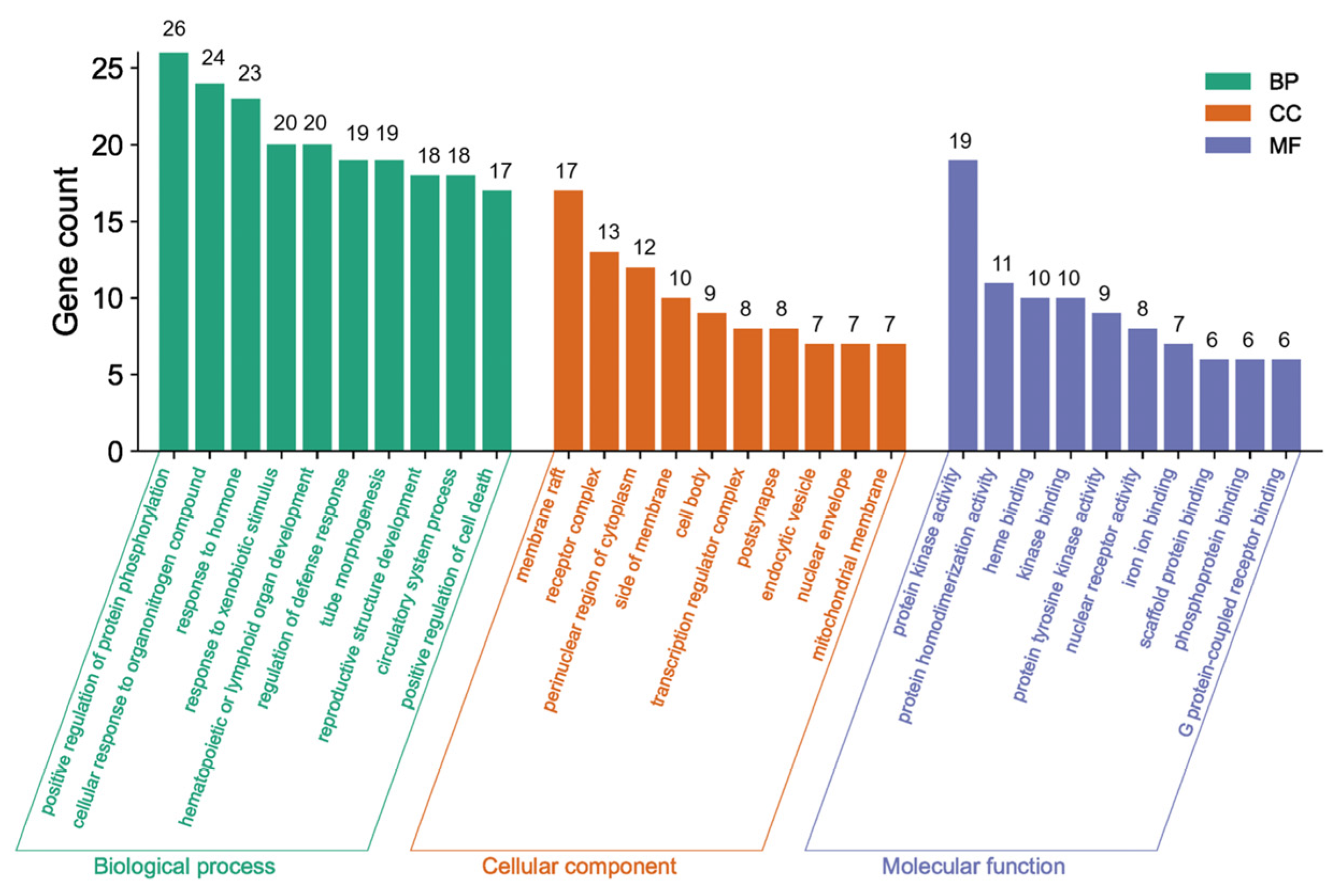
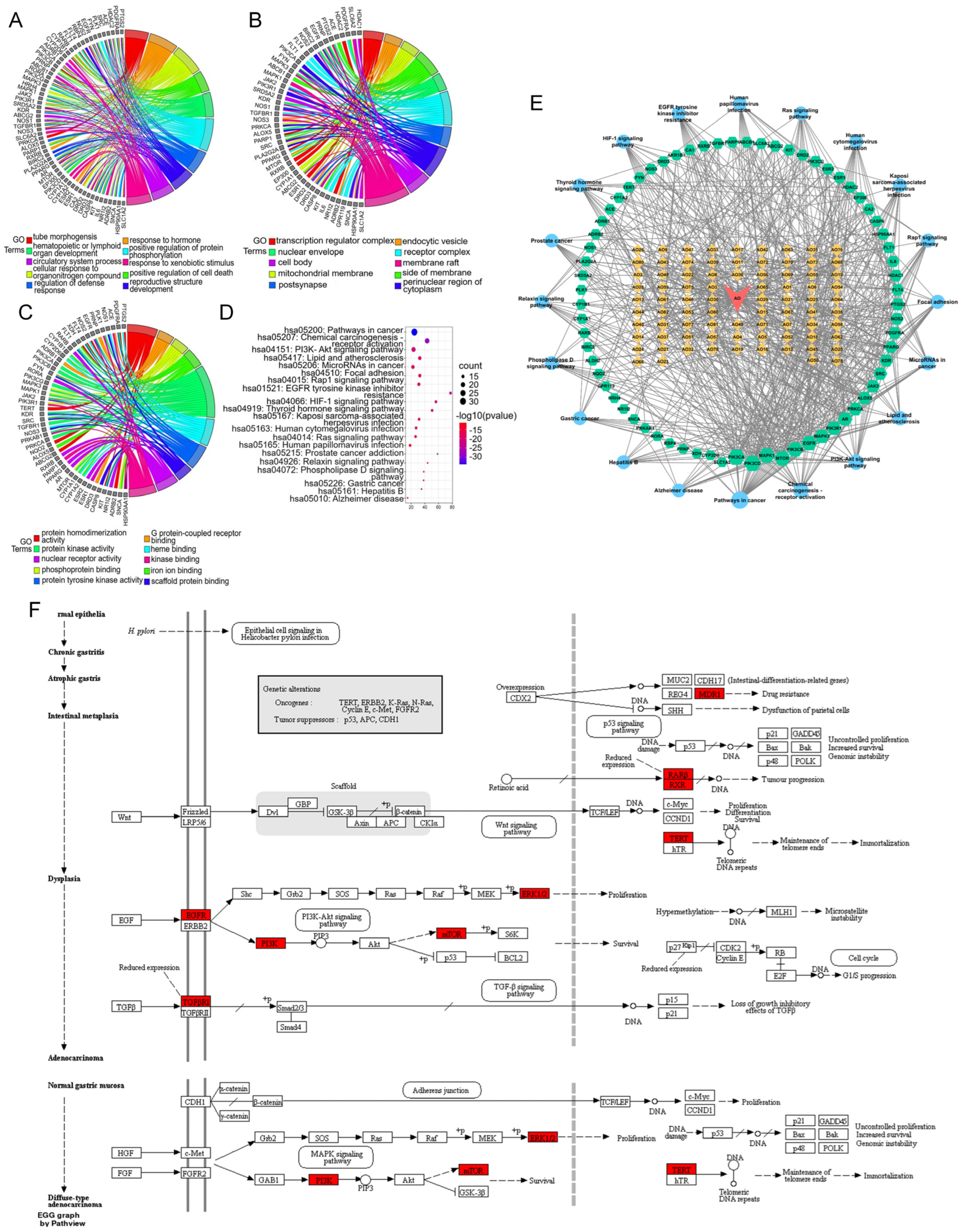
3.4.4. GO and KEGG Pathway Analyses of the Core Network
3.4.5. Drug–Compound–Target–Pathway Network Analysis
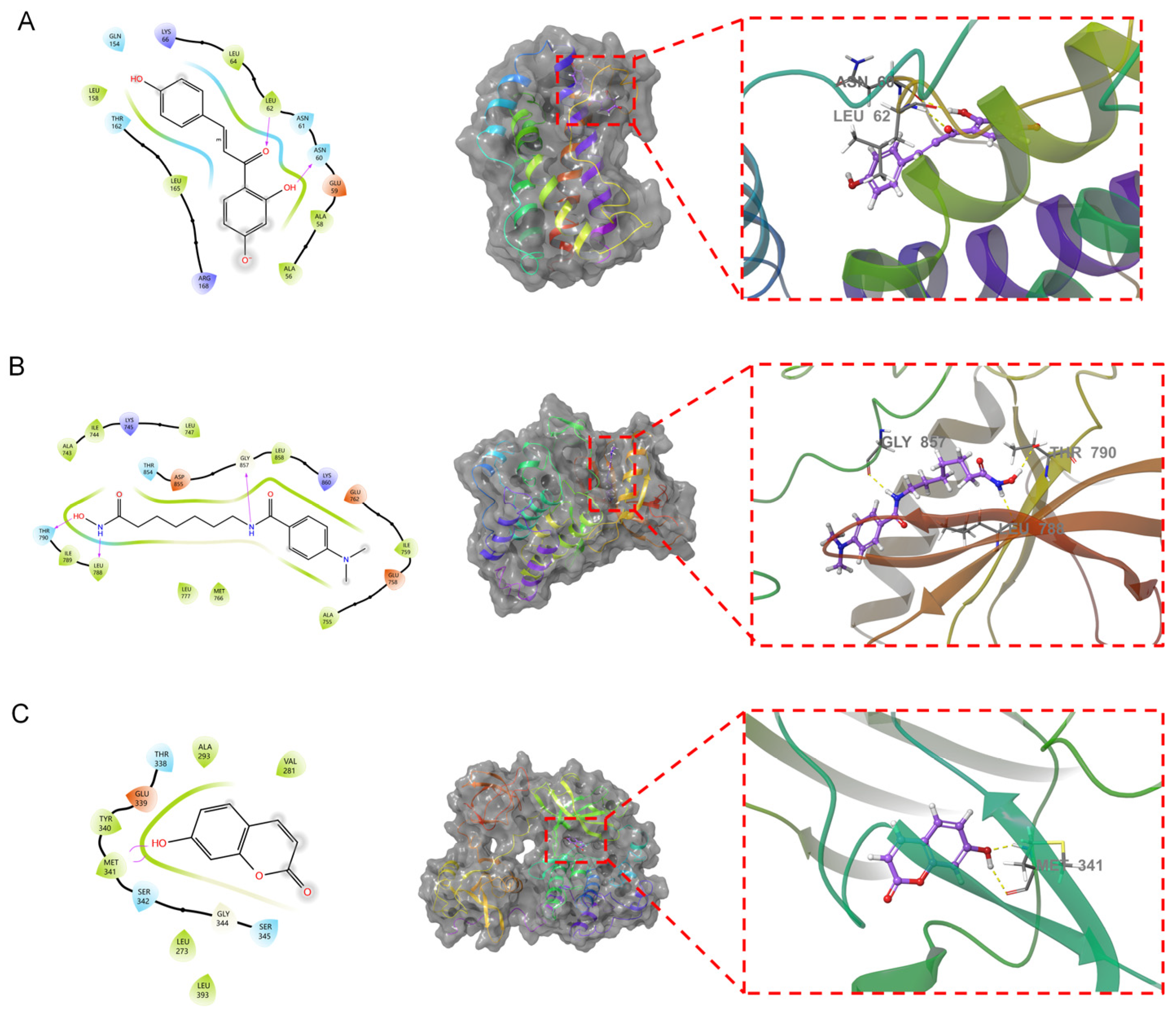
3.5. Molecular Docking
3.6. Cytotoxicity Analysis
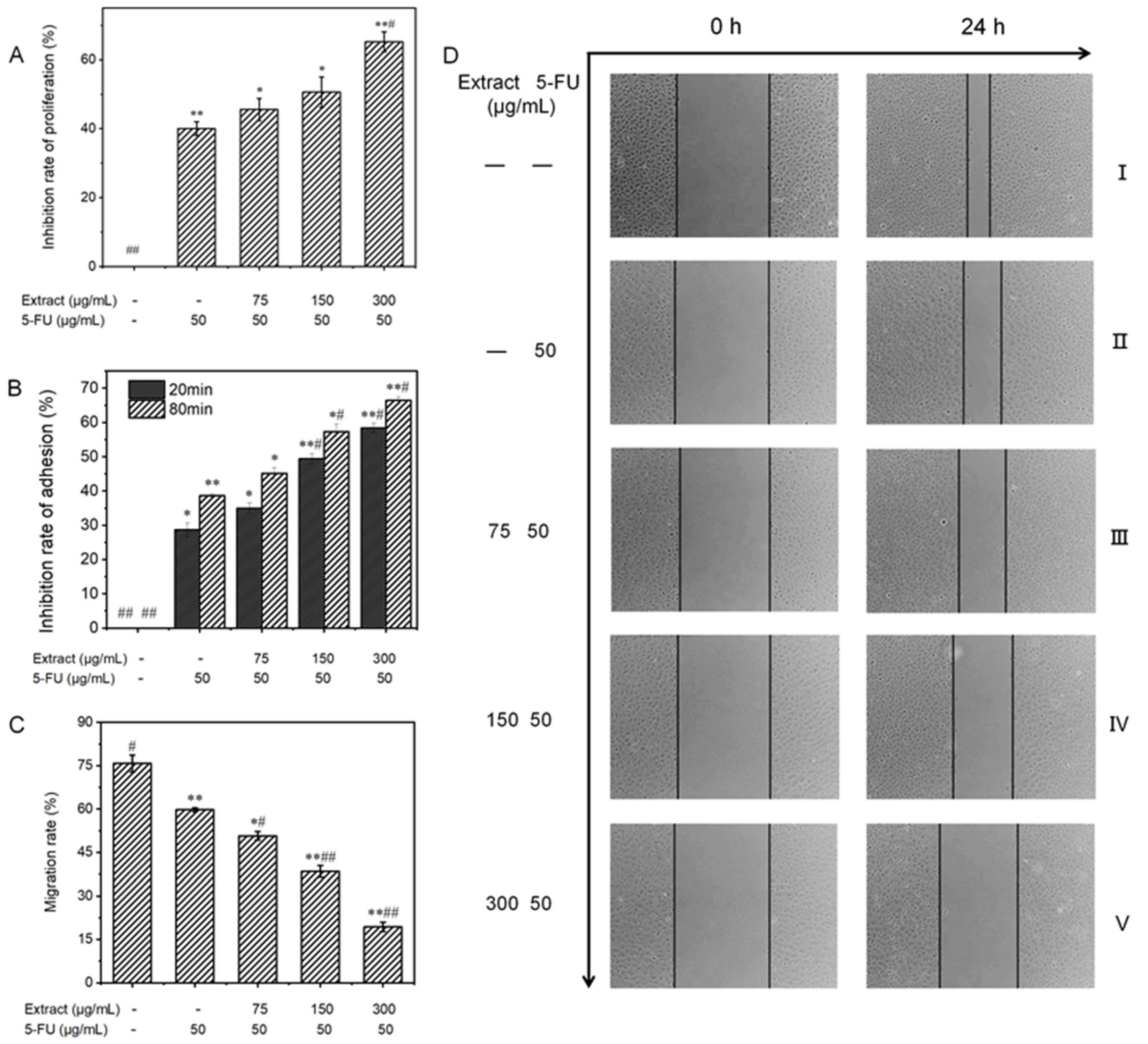
3.6.1. Effect on the Proliferative Capacity of MGC-803 Cells
3.6.2. Influence on the Adhesion Ability of MGC-803 Cells
3.6.3. Influence on the Migratory Ability of MGC-803 Cells
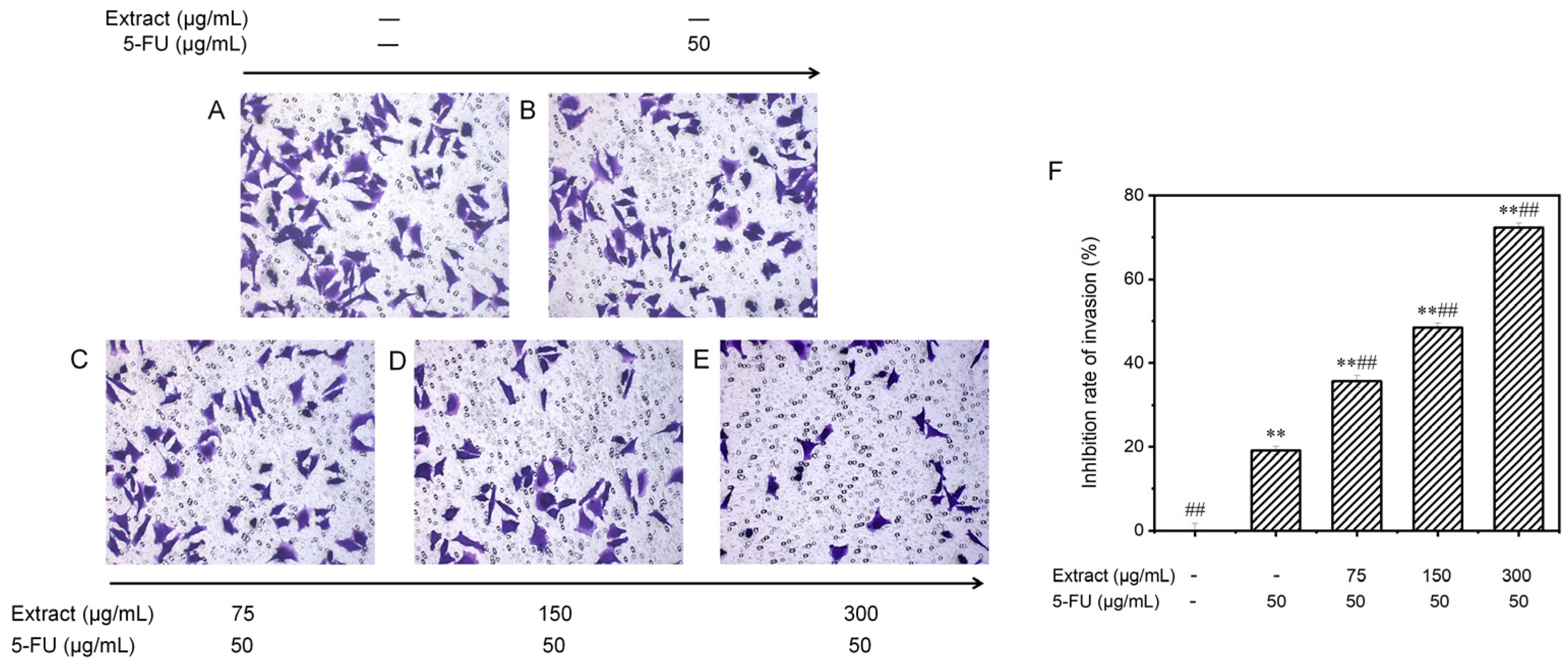
3.6.4. Effect on the Invasion Potential of MGC-803 Cells
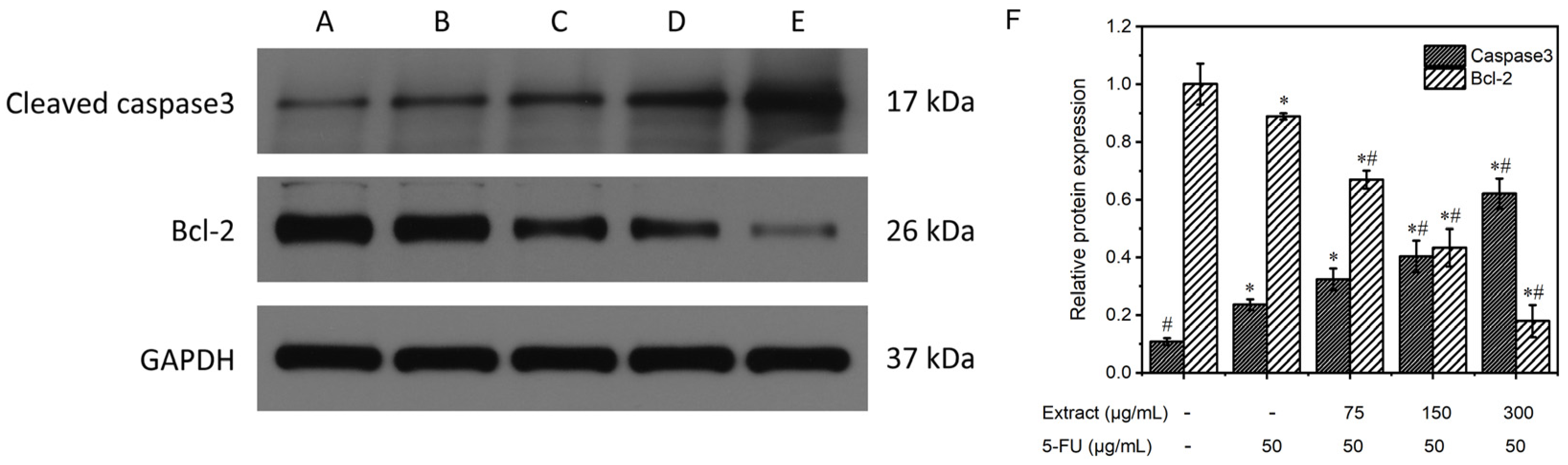
3.6.5. Study of the Mechanism of the Extract on MGC-803 Cell Apoptosis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
Appendix A. PCR Amplification Sequences
References
- Hou, C.L.; Liu, L.Y.; Ren, J.Y.; Huang, M.; Yuan, E.D. Structural characterization of two Hericium erinaceus polysaccharides and their protective effects on the alcohol-induced gastric mucosal injury. Food Chem. 2022, 375, 131896. [Google Scholar] [CrossRef] [PubMed]
- Sabaratnam, V.; Kah-hui, W.; Naidu, M.; Rosie David, P. Neuronal health-can culinary and medicinal mushrooms help? J. Tradit. Complement. Med. 2013, 3, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Heinzelmann, R.; Dutech, C.; Tsykun, T.; Labbé, F.; Soularue, J.P.; Prospero, S. Latest advances and future perspectives in Armillaria research. Can. J. Plant Pathol. 2019, 41, 1–23. [Google Scholar] [CrossRef]
- Lech, P.; Mychayliv, O.; Hildebrand, R.; Orman, O. Weather conditions drive the damage area caused by Armillaria root disease in coniferous forests across poland. Plant Pathol. J. 2023, 39, 548–565. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Gao, Y.; Li, H.; Ma, H.; Han, X.; Yang, Z.; Chen, W. Research status and application prospects of the medicinal mushroom Armillaria mellea. Appl. Biochem. Biotechnol. 2023, 195, 3491–3507. [Google Scholar] [CrossRef] [PubMed]
- Dörfer, M.; Gressler, M.; Hoffmeister, D. Diversity and bioactivity of Armillaria sesquiterpene aryl ester natural products. Mycol. Prog. 2019, 18, 1027–1037. [Google Scholar] [CrossRef]
- Liu, T.P.; Chen, C.C.; Shiao, P.Y.; Shieh, H.R.; Chen, Y.Y.; Chen, Y.J. Armillaridin, a honey medicinal mushroom, Armillaria mellea (higher basidiomycetes) component, inhibits differentiation and activation of human macrophages. Int. J. Med. Mushrooms 2015, 17, 161–168. [Google Scholar] [CrossRef]
- Leu, Y.S.; Chen, Y.J.; Chen, C.C.; Huang, H.L. Induction of autophagic death of human hepatocellular carcinoma cells by Armillaridin from Armillaria mellea. Am. J. Chin. Med. 2019, 47, 1365–1380. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.H.; Huang, H.L.; Huang, W.P.; Chen, C.C.; Chen, Y.J. Armillaridin induces autophagy-associated cell death in human chronic myelogenous leukemia K562 cells. Tumour Biol. 2016, 37, 14291–14300. [Google Scholar] [CrossRef]
- Huang, K.; Xu, B. Critical review of the phytochemical profiles and health-promoting effects of the edible mushroom Armillaria mellea. Food Funct. 2023, 14, 9518–9533. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. Swiss Target Prediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2022; pp. 27–56. [Google Scholar]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Pinero, J.; Queralt-Rosinach, N.; Bravo, A.; Deu-Pons, J.; Bauer-Mehren, A.; Baron, M.; Sanz, F.; Furlong, L.I. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database-J. Biol. Databases Curation 2015, 2015, bav028. [Google Scholar] [CrossRef]
- Whirl-Carrillo, M.; Huddart, R.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Whaley, R.; Klein, T.E. An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2021, 110, 563–572. [Google Scholar] [CrossRef]
- Hamosh, A.; Amberger, J.S.; Bocchini, C.; Scott, A.F.; Rasmussen, S.A. Online Mendelian Inheritance in Man (OMIM ®): Victor McKusick’s magnum opus. Am. J. Med. Genet. Part A 2021, 185, 3259–3265. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.T.; Lian, X.C.; Li, F.C.; Zhu, F.; Qiu, Y.Q.; Chen, Y.Z. Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022, 50, D1398–D1407. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Ochagavia, M.E.; Rabasa, L.C.; Miranda, J.; Fernandez-de-Cossio, J.; Bringas, R. BisoGenet: A new tool for gene network building, visualization and analysis. BMC Bioinform. 2010, 11, 91. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Peng, X.; Wang, J.; Wang, J.; Wu, F.X.; Pan, Y. Rechecking the centrality-lethality rule in the scope of protein subcellular localization interaction networks. PLoS ONE 2015, 10, e0130743. [Google Scholar] [CrossRef]
- Li, M.; Wang, J.; Chen, X.; Wang, H.; Pan, Y. A local average connectivity-based method for identifying essential proteins from the network level. Comput. Biol. Chem. 2011, 35, 143–150. [Google Scholar] [CrossRef]
- Wang, J.; Li, M.; Wang, H.; Pan, Y. Identification of essential proteins based on edge clustering coefficient. IEEE-ACM Trans. Comput. Biol. Bioinform. 2012, 9, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.; Hurley, R.; Almutlaqah, N.; O’Keeffe, G.; Keane, T.M.; Fitzpatrick, D.A.; Owens, R.A. Proteomic characterization of Armillaria mellea reveals oxidative stress response mechanisms and altered secondary metabolism profiles. Microorganisms 2017, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Mrabti, H.N.; Hachlafi, N.E.; Al-Mijalli, S.H.; Jeddi, M.; Elbouzidi, A.; Abdallah, E.M.; Flouchi, R.; Assaggaf, H.; Qasem, A.; Zengin, G.; et al. Phytochemical profile, assessment of antimicrobial and antioxidant properties of essential oils of Artemisia herba-alba Asso., and Artemisia dracunculus L.: Experimental and computational approaches. J. Mol. Struct. 2023, 1294, 136479. [Google Scholar] [CrossRef]
- Wang, F.; Feng, J.C.; Yang, Y.; Liu, J.G.; Liu, M.X.; Wang, Z.Y.; Pei, H.; Wei, Y.; Li, H. The Chinese herbal formula Fuzheng Quxie Decoction attenuates cognitive impairment and protects cerebrovascular function in SAMP8 mice. Neuropsychiatr. Dis. Treat. 2018, 14, 3037–3051. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.Y.; Wu, J.; Xie, C.Q.; Wu, L.; Gu, Z.; Hu, J.W.; Xiong, W. Antioxidant and anti-inflammatory activities of Gynura procumbens flowers extract through suppressing LPS-induced MAPK/NF-kappa B signalling pathways. Food Agric. Immunol. 2022, 33, 511–529. [Google Scholar] [CrossRef]
- Luo, Y.; Yang, Z.; Su, L.; Shan, J.J.; Xu, H.L.; Xu, Y.M.; Liu, L.M.; Zhu, W.; Chen, X.J.; Liu, C.G.; et al. Non-CSCs nourish CSCs through interleukin-17E-mediated activation of NF-kappa B and JAK/STAT3 signaling in human hepatocellular carcinoma. Cancer Lett. 2016, 375, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, S.; Starz-Gaiano, M. Drosophila Jak/STAT Signaling: Regulation and relevance in human cancer and metastasis. Int. J. Mol. Sci. 2018, 19, 4056. [Google Scholar] [CrossRef] [PubMed]
- Owen, K.L.; Brockwell, N.K.; Parker, B.S. JAK-STAT signaling: A double-edged sword of immune regulation and cancer progression. Cancers 2019, 11, 2002. [Google Scholar] [CrossRef]
- Bose, S.; Banerjee, S.; Mondal, A.; Chakraborty, U.; Pumarol, J.; Croley, C.R.; Bishayee, A. Targeting the JAK/STAT signaling pathway using phytocompounds for cancer prevention and therapy. Cells 2020, 9, 1451. [Google Scholar] [CrossRef]
- Chen, J.; Wang, X.Y.; Zhang, J.L.; Chang, J.W.; Han, C.J.; Xu, Z.W.; Yu, H.Z. Effects of the wnt/beta-catenin signaling pathway on proliferation and apoptosis of gastric cancer cells. Contrast Media Mol. Imaging 2022, 2022, 5132691. [Google Scholar] [CrossRef] [PubMed]
- Ba, M.; Long, H.; Yan, Z.F.; Wang, S.; Wu, Y.B.; Tu, Y.N.; Gong, Y.F.; Cui, S.Z. BRD4 promotes gastric cancer progression through the transcriptional and epigenetic regulation of c-MYC. J. Cell. Biochem. 2018, 119, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yan, S.P.; Chu, D.X.; Xie, Y.; Wang, C.F.; Zhang, J.Y.; Li, W.C.; Guo, R.X. Silencing of long non-coding RNA RP1-93H18.6 acts as a tumor suppressor in cervical cancer through the blockade of the PI3K/Akt axis. Mol. Ther.-Nucleic Acids 2020, 19, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Dai, Q.J.; Su, X.R.; Fu, J.J.; Feng, X.C.; Peng, J. Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Mol. Biol. Rep. 2020, 47, 4587–4629. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]



| Ingredient ID | Ingredient Name | Ingredient ID | Ingredient Name | Ingredient ID | Ingredient Name |
|---|---|---|---|---|---|
| AO1 | α-Eleostearic acid | AO2 | ValylIsoleucine | AO3 | Tretinoin |
| AO4 | Suberic acid | AO5 | Spinochalcone A | AO6 | Saccharin |
| AO7 | Propofol | AO8 | Promethazine sulfoxide | AO9 | Preclamol |
| AO10 | Penbutolol | AO11 | Pazelliptine | AO12 | Parthenolide |
| AO13 | Palmatine | AO14 | Olodaterol | AO15 | N-benzyl-4-(isobutylamino)-2-[4-(tetrahydropyran-3-ylmethyl) piperazin-1-yl] pyrimidine-5-carboxamide |
| AO16 | N-acetyl-2-phenylethylamine | AO17 | N-{[(4E)-2-(Hydroxymethyl)-4-(3-isobutyl-5-methylhexylidene)-5-oxotetrahydro-2-furanyl] methyl}-2,2-dimethylpropanamide | AO18 | N-[2-(3,4-Dimethoxyphenyl) ethyl]-3-(2-methoxyphenyl)-5-methylpyrazolo[1,5-a] pyrimidin-7-amine |
| AO19 | N-[(9Z)-9-Octadecenoyl] glycine | AO20 | N,N-Dimethylarginine | AO21 | N,N-Dibutylethanolamine |
| AO22 | Momelotinib | AO23 | Metirosine | AO24 | Lonchocarpol E |
| AO25 | Linoleic acid | AO26 | L-Glutamic acid | AO27 | Leucylleucine |
| AO28 | Jasmonic acid | AO29 | Isopropyl 4-[3-(3-chloro-4-methoxyphenyl)-1-phenyl-1H-pyrazol-4-yl]-6-methyl-2-oxo-1,2,3,4-tetrahydro-5-pyrimidinecarboxylate | AO30 | Isophorone |
| AO31 | Isoliquiritigenin | AO32 | hypophyllanthin | AO33 | Histamine |
| AO34 | Heteroflavanone B | AO35 | goralatide | AO36 | Flavone |
| AO37 | Etiproston | AO38 | Estriol | AO39 | Embelin |
| AO40 | DMH4 | AO41 | DL-Arginine | AO42 | Decanamide |
| AO43 | Cortisol | AO44 | Catalposide | AO45 | Capsaicin |
| AO46 | Boldione | AO47 | Azelaic acid | AO48 | AUDA |
| AO49 | Arachidonic acid | AO50 | Anacardic acid | AO51 | Ambrosic acid |
| AO52 | Ajugarin I | AO53 | 8-{3-Oxo-2-[(2E)-2-penten-1-yl]-1-cyclopenten-1-yl} octanoic acid | AO54 | 7-Hydroxycoumarine |
| AO55 | 5α-Dihydrotestosterone | AO56 | 5-[4-(6-Methyl-4-phenylquinazolin-2-yl) piperazin-1-yl]-5-oxopentanoic acid | AO57 | 5-[(8aS)-2,5,5,8a-tetramethyl-3-oxo-4a,6,7,8-tetrahydro-4H-naphthalen-1-yl]-3-methylpentanoic acid |
| AO58 | 5-(3,4-Dimethoxyphenyl)-4-[(2E)-3-(2-furyl)-2-propenoyl]-3-hydroxy-1-[2-(4-morpholinyl) ethyl]-1,5-dihydro-2H-pyrrol-2-one | AO59 | 4-Phenylbutyric acid | AO60 | 4-Hydroxy-N’-[(E)-(4-isopropylphenyl) methylene] benzohydrazide |
| AO61 | 4-Hydroxyderricin | AO62 | 4-(Dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl] benzamide | AO63 | 3-Hydroxypregnane-11,20-dione |
| AO64 | 3-Formyl-2a-hydroxy-6,6,7b-trimethyl-2,2a,4a,5,6,7,7a,7b-octahydro-1H-cyclobuta[e]inden-2-yl 3-chloro-6-hydroxy-4-methoxy-2-methylbenzoate | AO65 | 3-BHA | AO66 | 2-Cyclododecyl-1-(4-morpholinyl) ethanone |
| AO67 | 2,2’-(Dodecylimino) diethanol | AO68 | 2-(4-Morpholinyl)-8-phenyl-4H-chromen-4-one | AO69 | 1-Amino-3-cyano-4,6-dimethylpyridine-2-one |
| AO70 | 13S-hydroxyoctadecadienoic acid | AO71 | 12-HHTrE | AO72 | 1-{2-Chloro-4-[(6,7-dimethoxy-4-quinazolinyl) oxy] phenyl}-3-propylurea |
| AO73 | 1-[2-(Dimethylamino) ethyl]-3-hydroxy-5-(3-methoxyphenyl)-4-[(5-methyl-1-phenyl-1H-pyrazol-4-yl) carbonyl]-1,5-dihydro-2H-pyrrol-2-one | AO74 | 1-(3-Ethoxypropyl)-2-imino-N-(5-methyl-1,2-oxazol-3-yl)-5-oxo-1,5-dihydro-2H-dipyrido[1,2-a:2’,3’-d] pyrimidine-3-carboxamide | AO75 | 1-(1,3-Benzodioxol-5-yl)-2-(1-pyrrolidinyl)-1-butanone |
| AO76 | [(2R,4S,4aR,7aS,7bR)-3-formyl-4-hydroxy-6,6,7b-trimethyl-2,4,4a,5,7,7a-hexahydro-1H-cyclobuta[e]inden-2-yl] 3-chloro-6-hydroxy-4-methoxy-2-methyl-benzoate | AO77 | (E)-3-(4-methoxyphenyl)-1-[2,4,6-trimethoxy-3-(3-methylbut-2-enyl) phenyl] prop-2-en-1-one | AO78 | (3S)-5-[(1R,2R,8aS)-2-hydroxy-2,5,5,8a-tetramethyl-3,4,4a,6,7,8-hexahydro-1H-naphthalen-1-yl]-3-methylpentanoic acid |
| AO79 | (3beta,5alpha,23beta)-17,23-Epoxy-5,6-dihydro-3-hydroxyveratraman-11-one | AO80 | (11Z)-17-chloro-16-[2-(morpholin-4-yl) ethoxy]-2-oxo-2,3,10,13-tetrahydro-1H-8,4-(azeno)-9,14,1,3,6-benzodioxatriazacyclohexadecine-7-carbonitrile | AO81 | (+)-Carpaine |
| AO82 | (+)-Aphidicolin |
| PubChem CID | Compound | IL6 | EGFR | SRC | |||
|---|---|---|---|---|---|---|---|
| XP G | MM-GBSA | XP G | MM-GBSA | XP G | MM-GBSA | ||
| 638278 | Isoliquiritigenin | −6.426 | −26.91 | −6.151 | −13.27 | −6.691 | −8.76 |
| 44557095 | [(2R,4S,4aR,7aS,7bR)-3-formyl-4-hydroxy-6,6,7b-trimethyl-2,4,4a,5,7,7a-hexahydro-1H-cyclobuta[e]inden-2-yl] 3-chloro-6-hydroxy-4-methoxy-2-methyl-benzoate | −4.141 | −37.16 | −3.718 | −23.42 | −3.663 | −29.57 |
| 126031 | 3-Formyl-2a-hydroxy-6,6,7b-trimethyl-2,2a,4a,5,6,7,7a,7b-octahydro-1H-cyclobuta[e]inden-2-yl 3-chloro-6-hydroxy-4-methoxy-2-methylbenzoate | −4.129 | −27.21 | −5.111 | −27.82 | −4.176 | −22.54 |
| 1548943 | Capsaicin | −3.933 | −41.97 | −4.730 | −46.93 | −4.803 | −33.98 |
| 3994 | 4-(Dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl] benzamide | −3.912 | −32.72 | −6.092 | −43.84 | −5.047 | −30.78 |
| 8456 | 3-BHA | −2.703 | −27.16 | −3.911 | −31.26 | −4.092 | −22.90 |
| 7251185 | Parthenolide | −2.376 | −25.27 | −2.421 | −10.37 | −2.767 | −0.63 |
| 5754 | Cortisol | −2.232 | −21.24 | −7.043 | −15.80 | −3.848 | −22.66 |
| 5281426 | 7-Hydroxycoumarine | −1.375 | −23.64 | −5.274 | −27.29 | −6.065 | −31.66 |
| 442630 | (+)-Carpaine | −1.212 | −6.72 | −3.072 | −29.27 | 0.886 | −27.36 |
| Group | CDI | Mean | Standard Deviation | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| Combination drug low concentration | 0.809 | 0.832 | 0.827 | 0.823 | 0.012 |
| Combination drug medium concentration | 0.299 | 0.341 | 0.364 | 0.335 | 0.033 |
| Combination drug high concentration | 0.249 | 0.301 | 0.293 | 0.281 | 0.028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Wang, R.; Na, Z.; Liang, S.; Wu, F.; Xie, H.; Zhang, X.; Xu, W.; Wang, X. Network Pharmacology Analysis of Liquid-Cultured Armillaria ostoyae Mycelial Metabolites and Their Molecular Mechanism of Action against Gastric Cancer. Molecules 2024, 29, 1668. https://doi.org/10.3390/molecules29071668
Wang Z, Wang R, Na Z, Liang S, Wu F, Xie H, Zhang X, Xu W, Wang X. Network Pharmacology Analysis of Liquid-Cultured Armillaria ostoyae Mycelial Metabolites and Their Molecular Mechanism of Action against Gastric Cancer. Molecules. 2024; 29(7):1668. https://doi.org/10.3390/molecules29071668
Chicago/Turabian StyleWang, Zhishuo, Ruiqi Wang, Zhiguo Na, Shanshan Liang, Fan Wu, Hongyao Xie, Xue Zhang, Wei Xu, and Xin Wang. 2024. "Network Pharmacology Analysis of Liquid-Cultured Armillaria ostoyae Mycelial Metabolites and Their Molecular Mechanism of Action against Gastric Cancer" Molecules 29, no. 7: 1668. https://doi.org/10.3390/molecules29071668
APA StyleWang, Z., Wang, R., Na, Z., Liang, S., Wu, F., Xie, H., Zhang, X., Xu, W., & Wang, X. (2024). Network Pharmacology Analysis of Liquid-Cultured Armillaria ostoyae Mycelial Metabolites and Their Molecular Mechanism of Action against Gastric Cancer. Molecules, 29(7), 1668. https://doi.org/10.3390/molecules29071668





