Role of Saponins from Platycodon grandiflorum in Alzheimer’s Disease: DFT, Molecular Docking, and Simulation Studies in Key Enzymes
Abstract
1. Introduction
Saponins Against Age-Related Neurological Disorders
2. Results
2.1. Selection of the Best Inhibitors for Proteins
2.2. ADMET Results
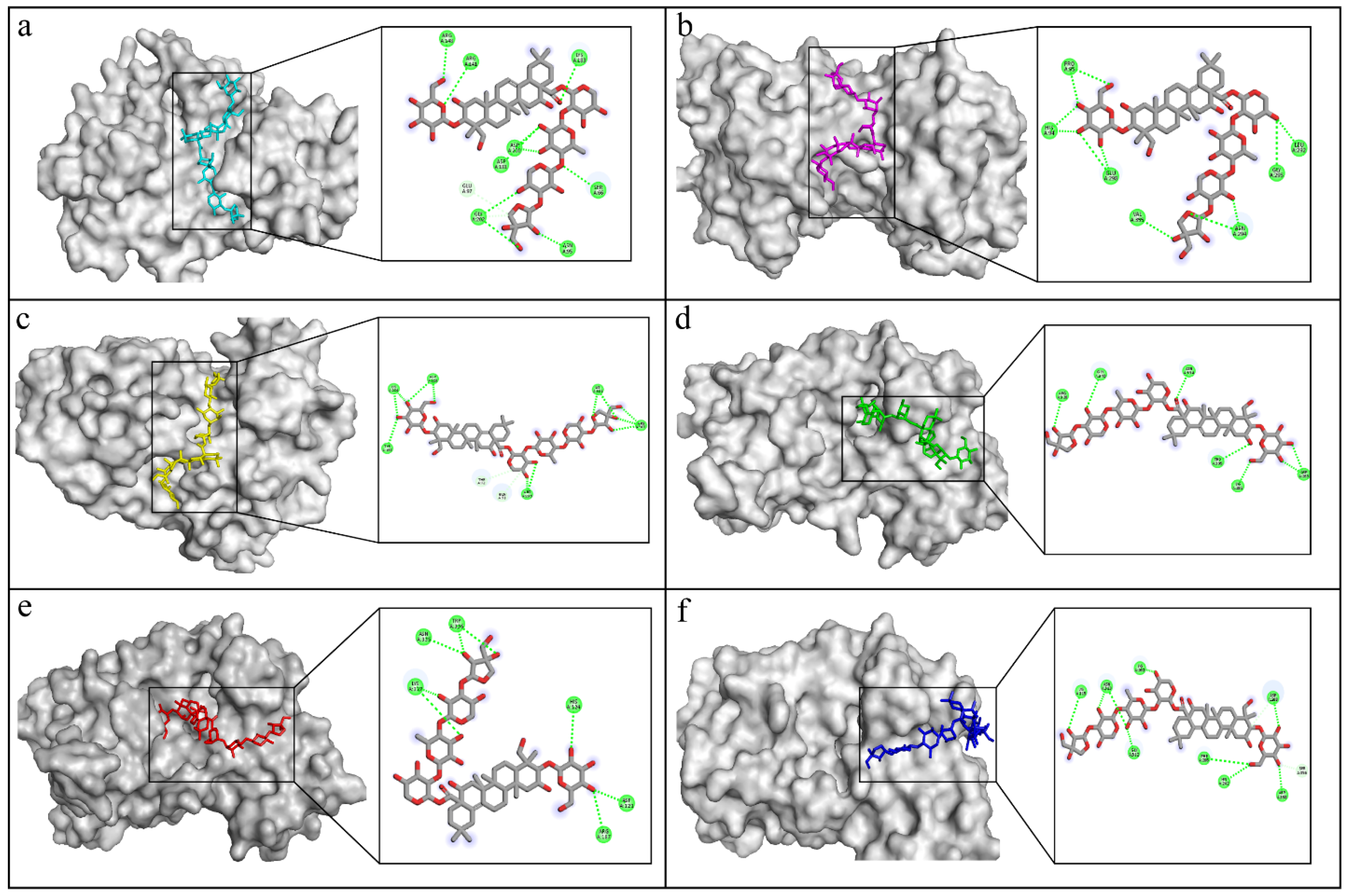
2.3. Molecular Docking
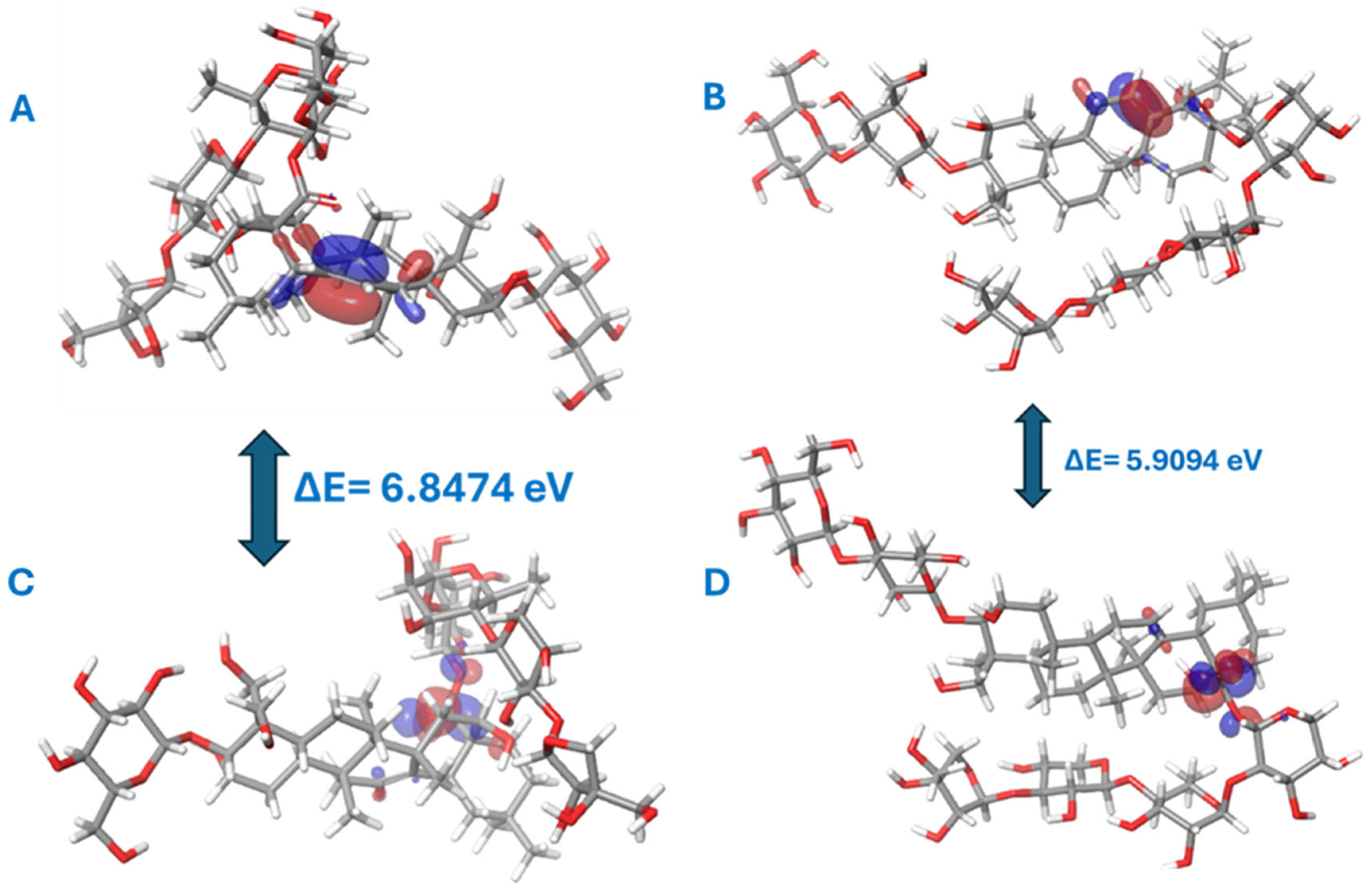
2.4. Molecular Quantum Calculations
2.5. Frontier Molecular Orbitals (FMOs)
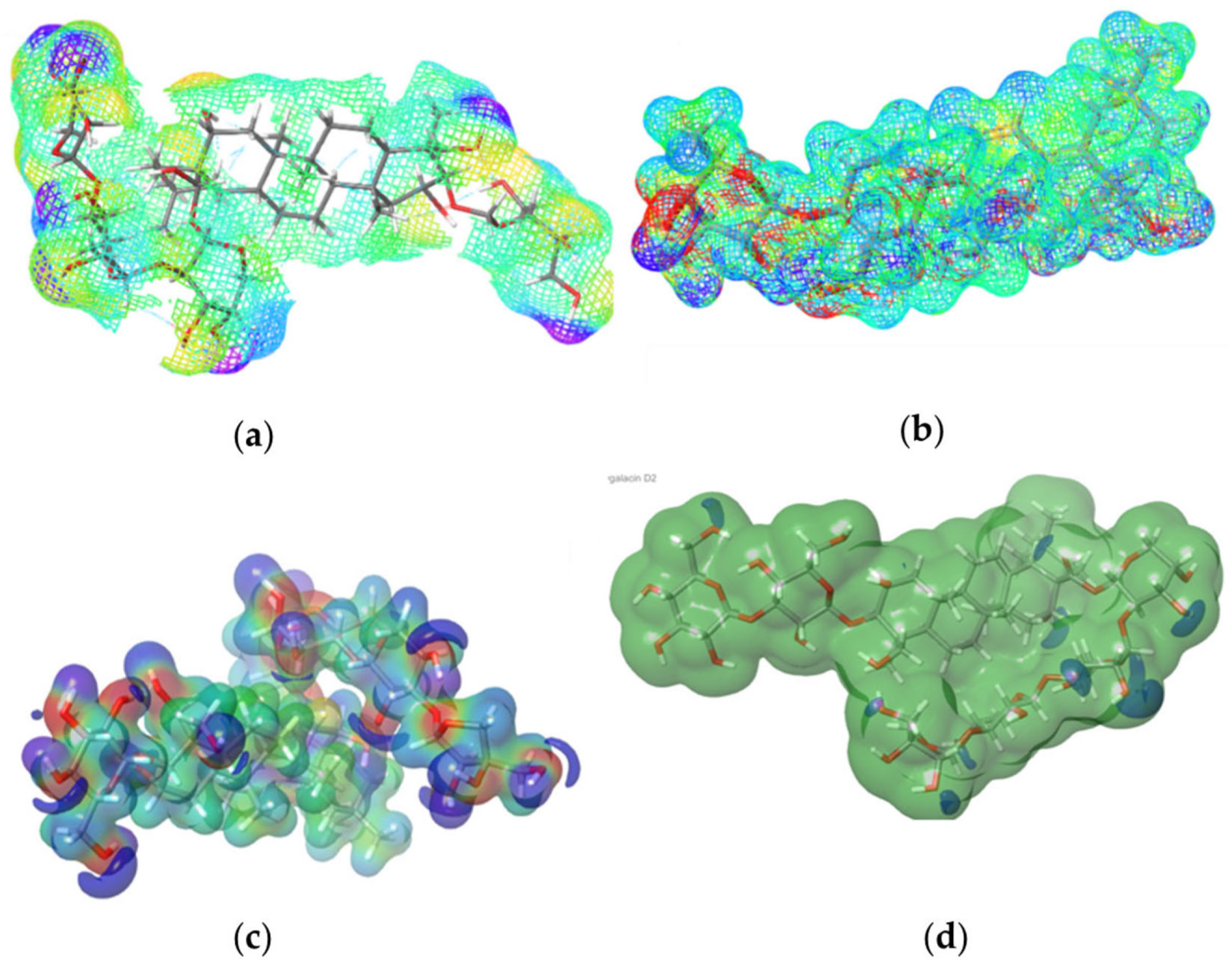
2.6. Molecular Electrostatic Potential (MEP)
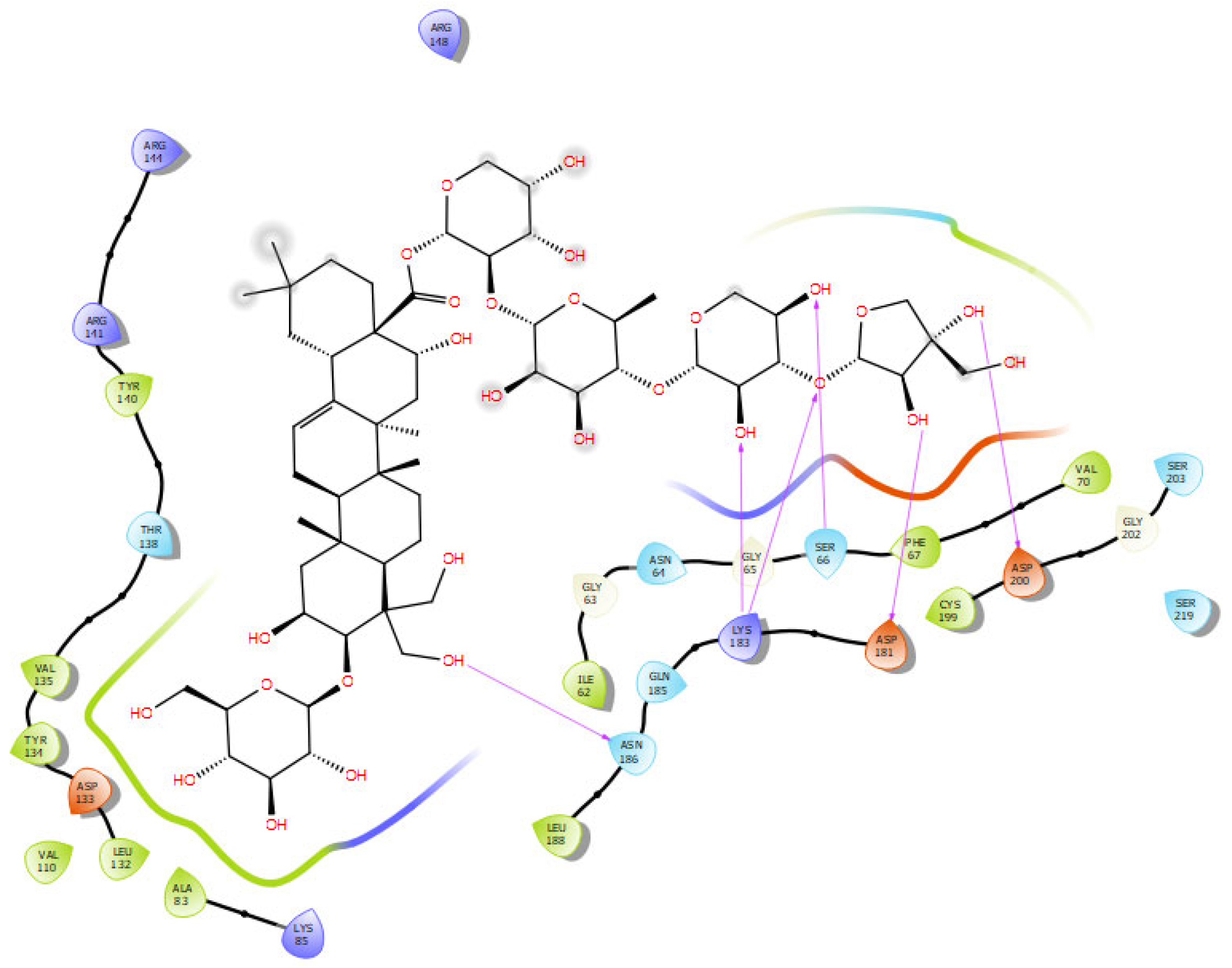
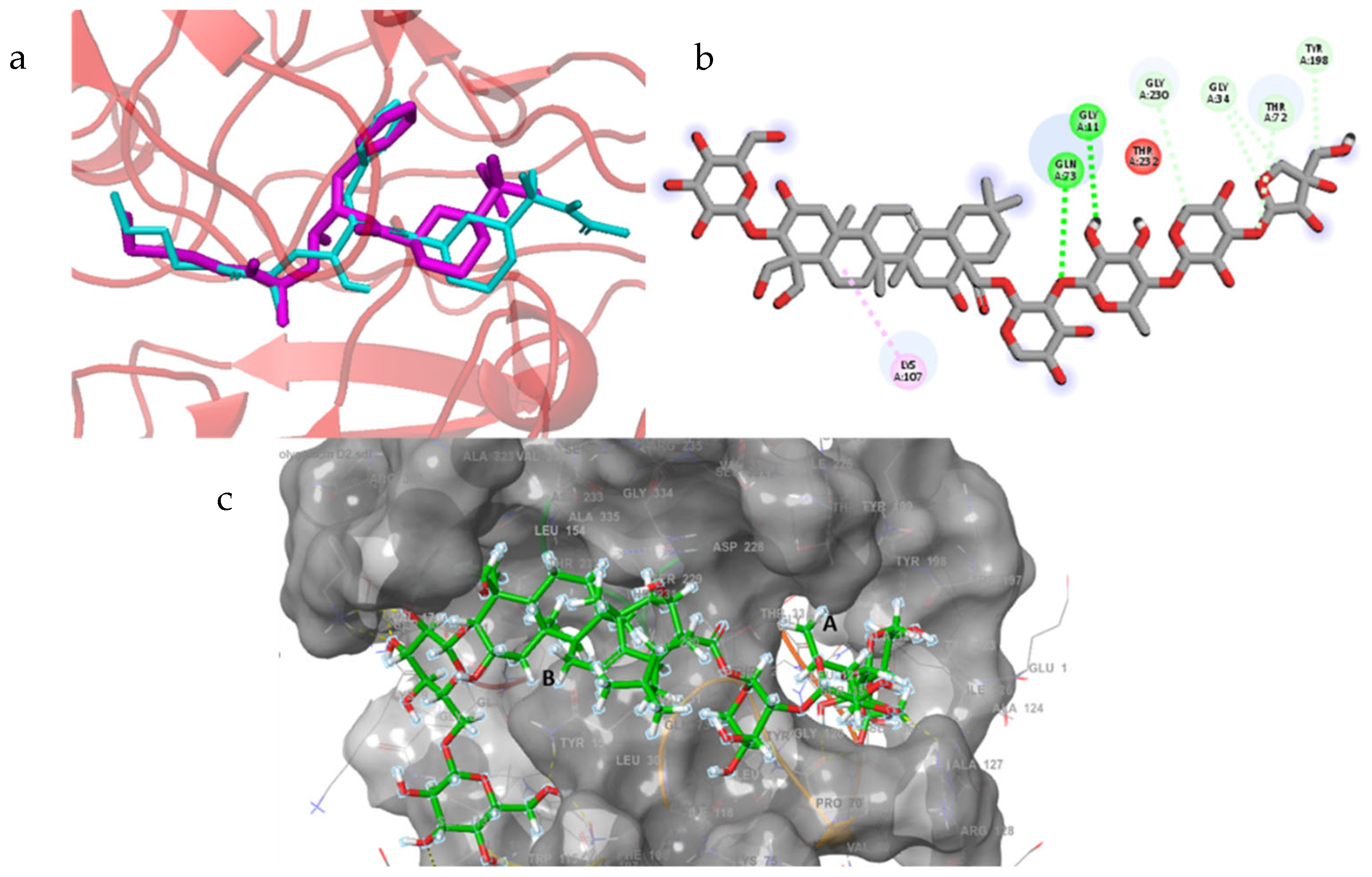
2.7. Docking Interactions in Selected Proteins
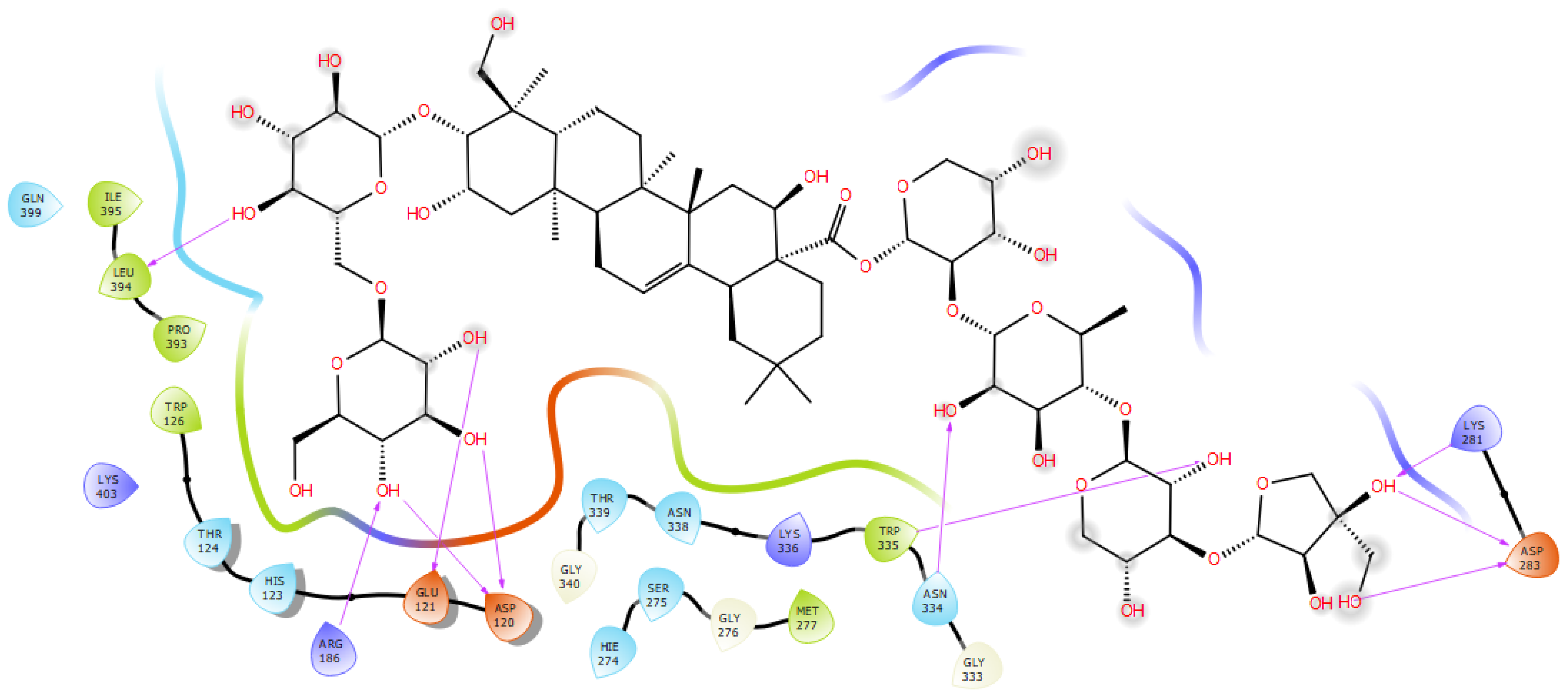
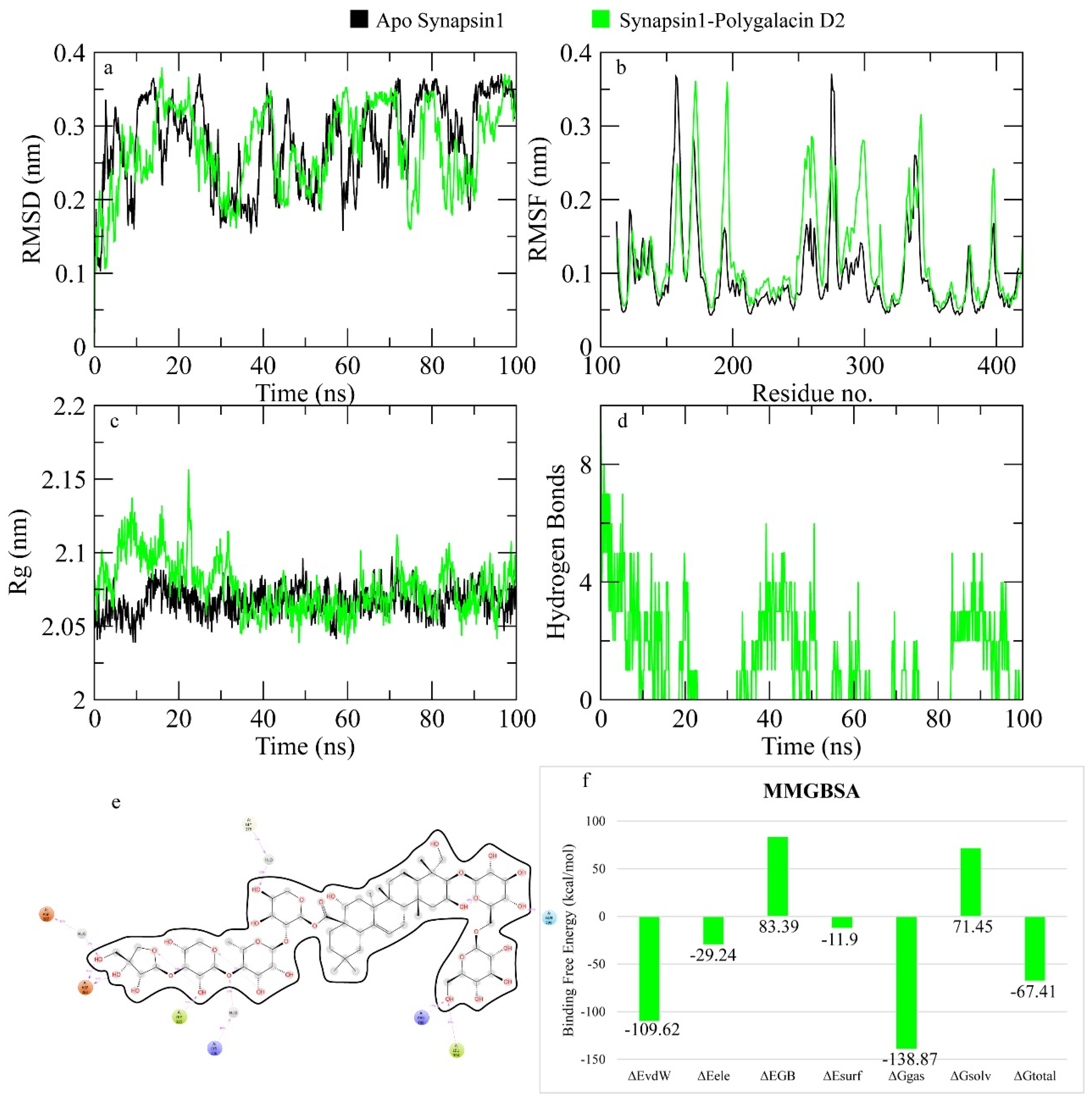
2.7.1. Synapsin I Receptor
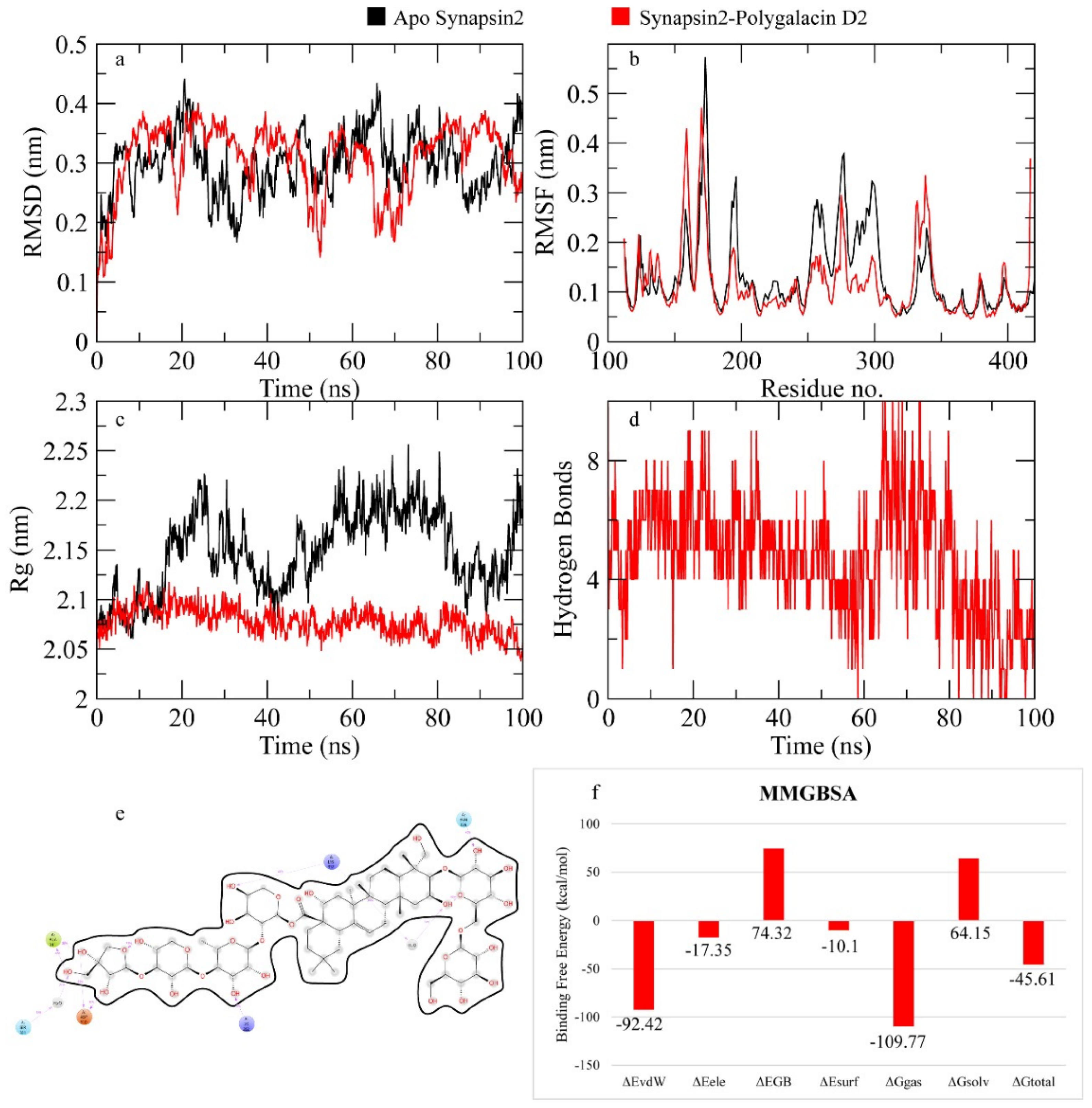
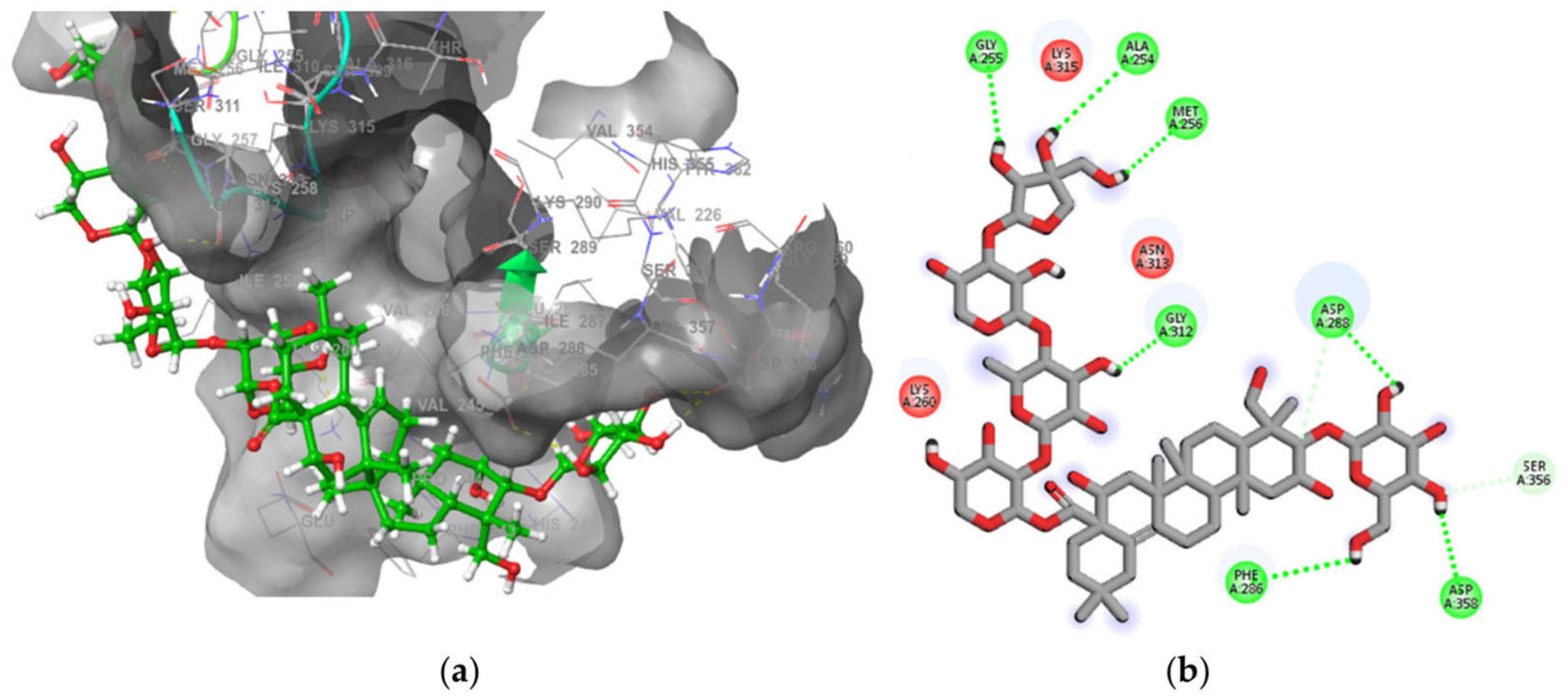
2.7.2. Synapsin II Receptor
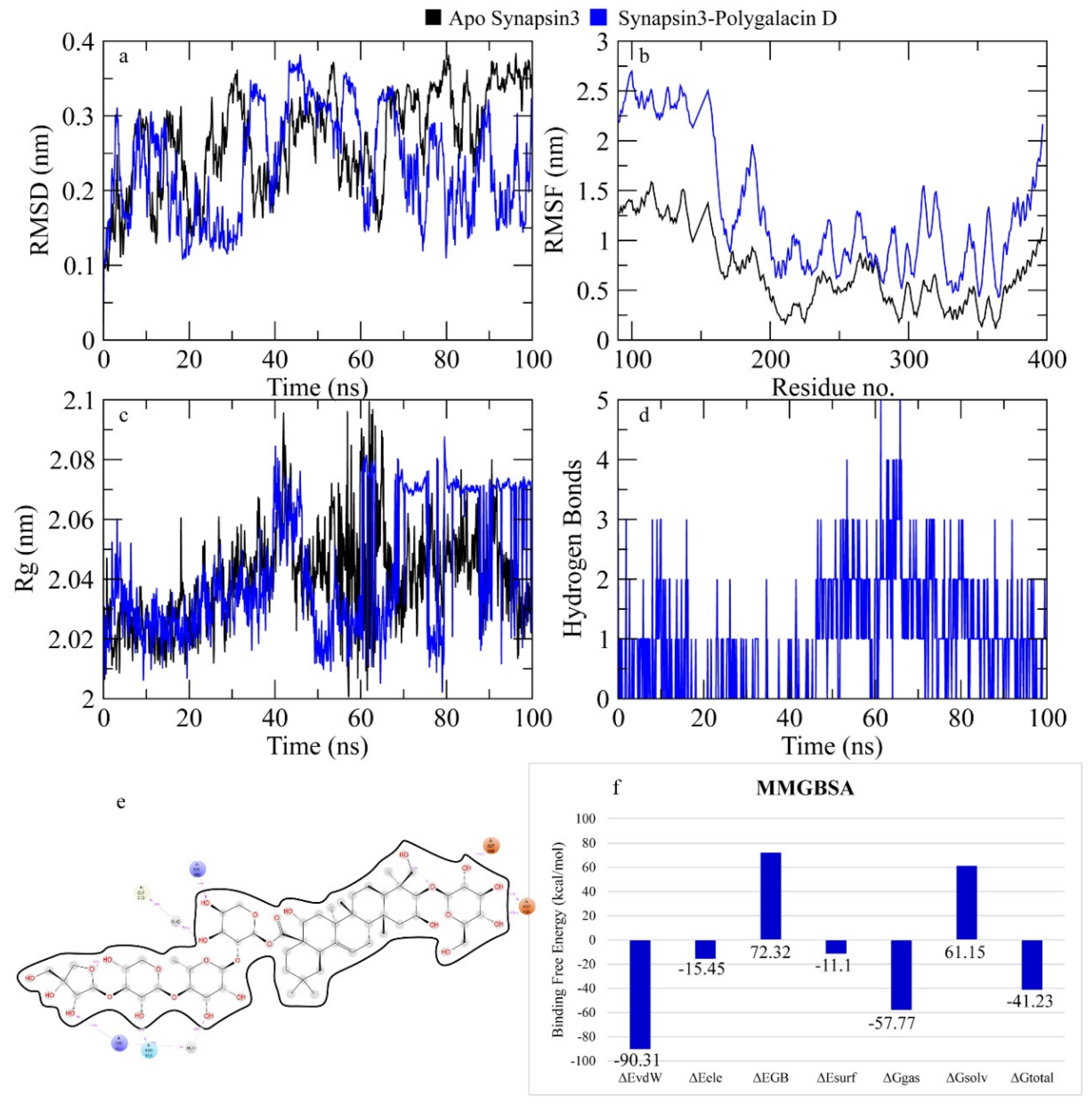
2.7.3. Synapsin III Receptor
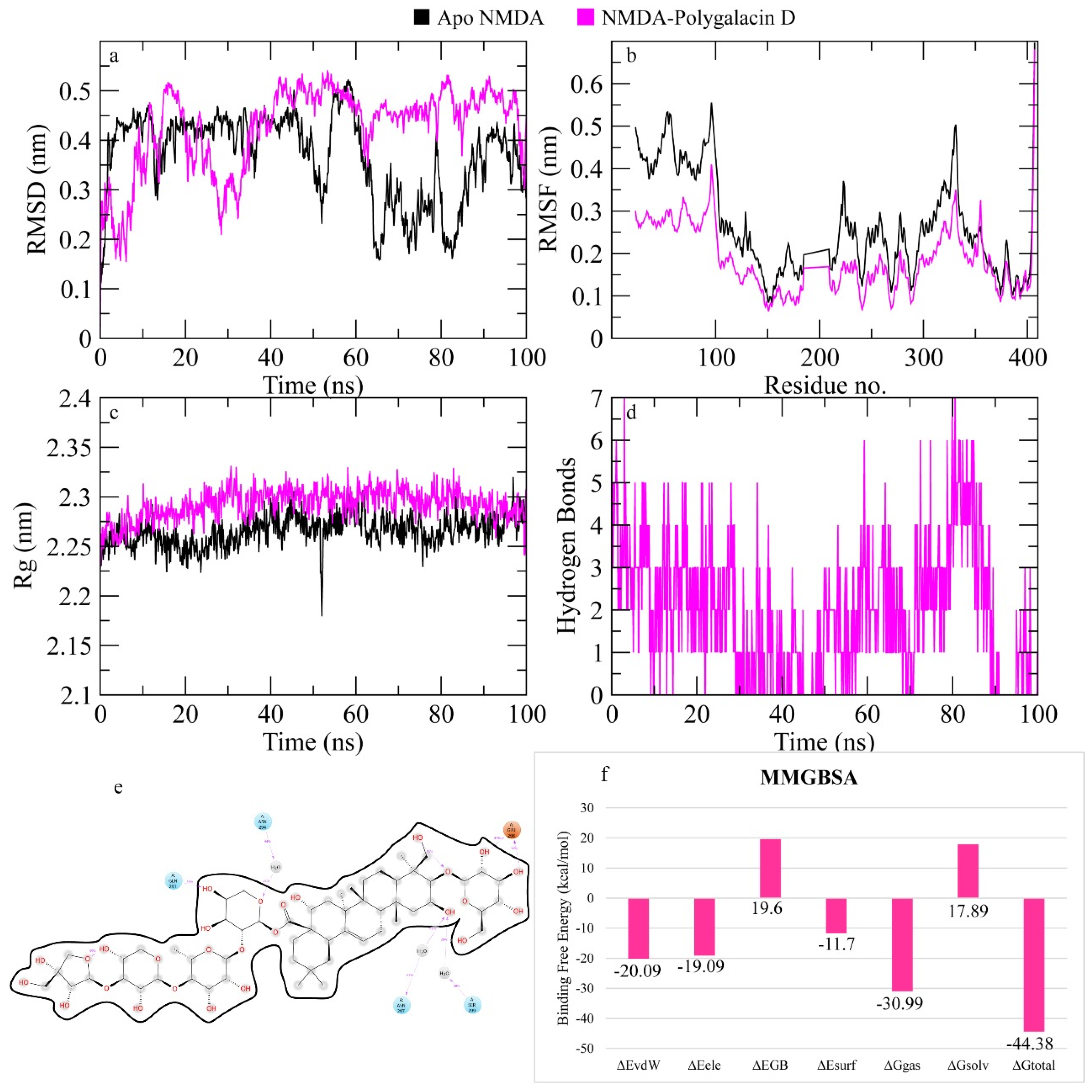
2.7.4. N-Methyl-D-Aspartate (NMDA) Receptor
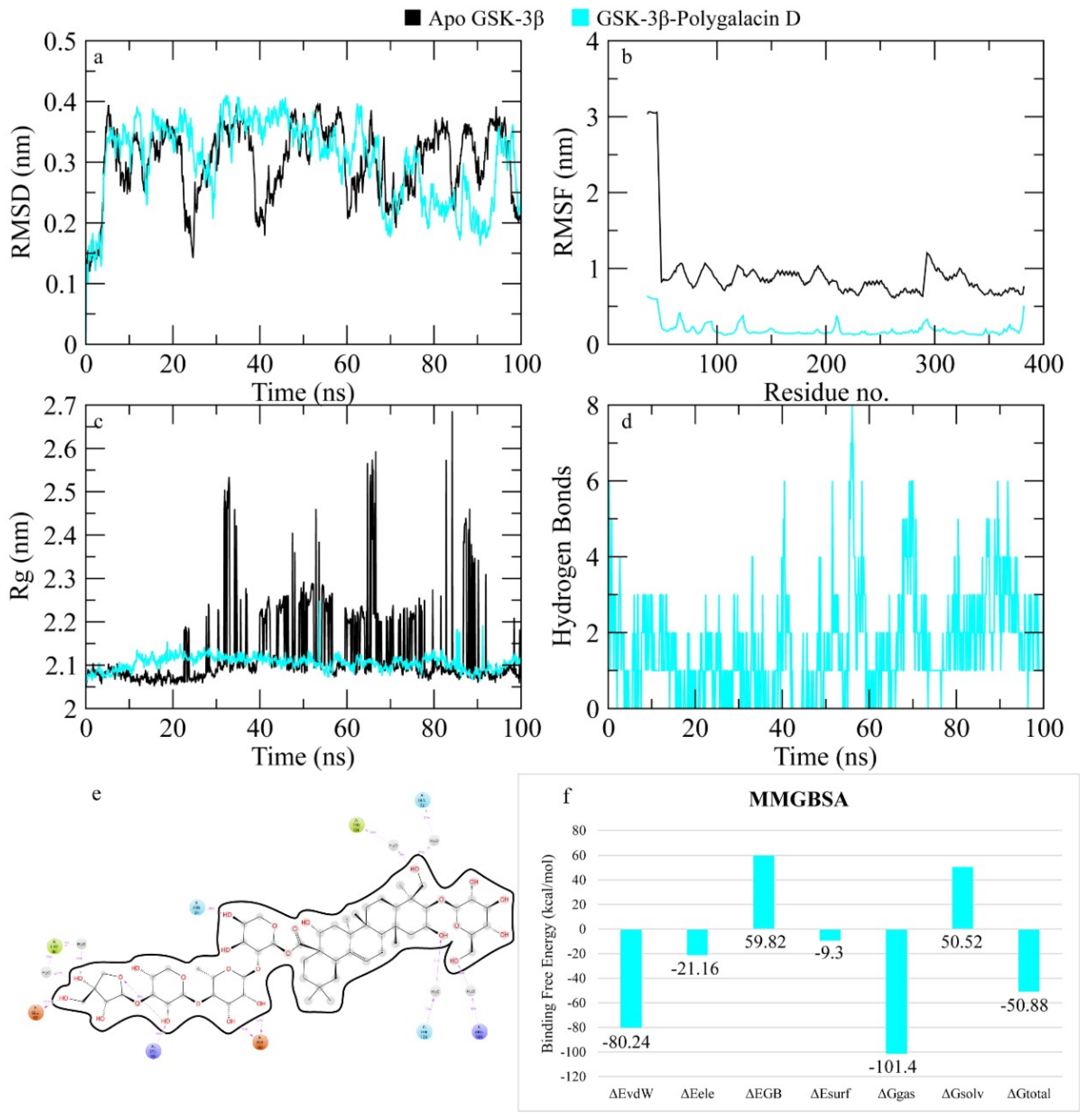
2.7.5. GSK-3β
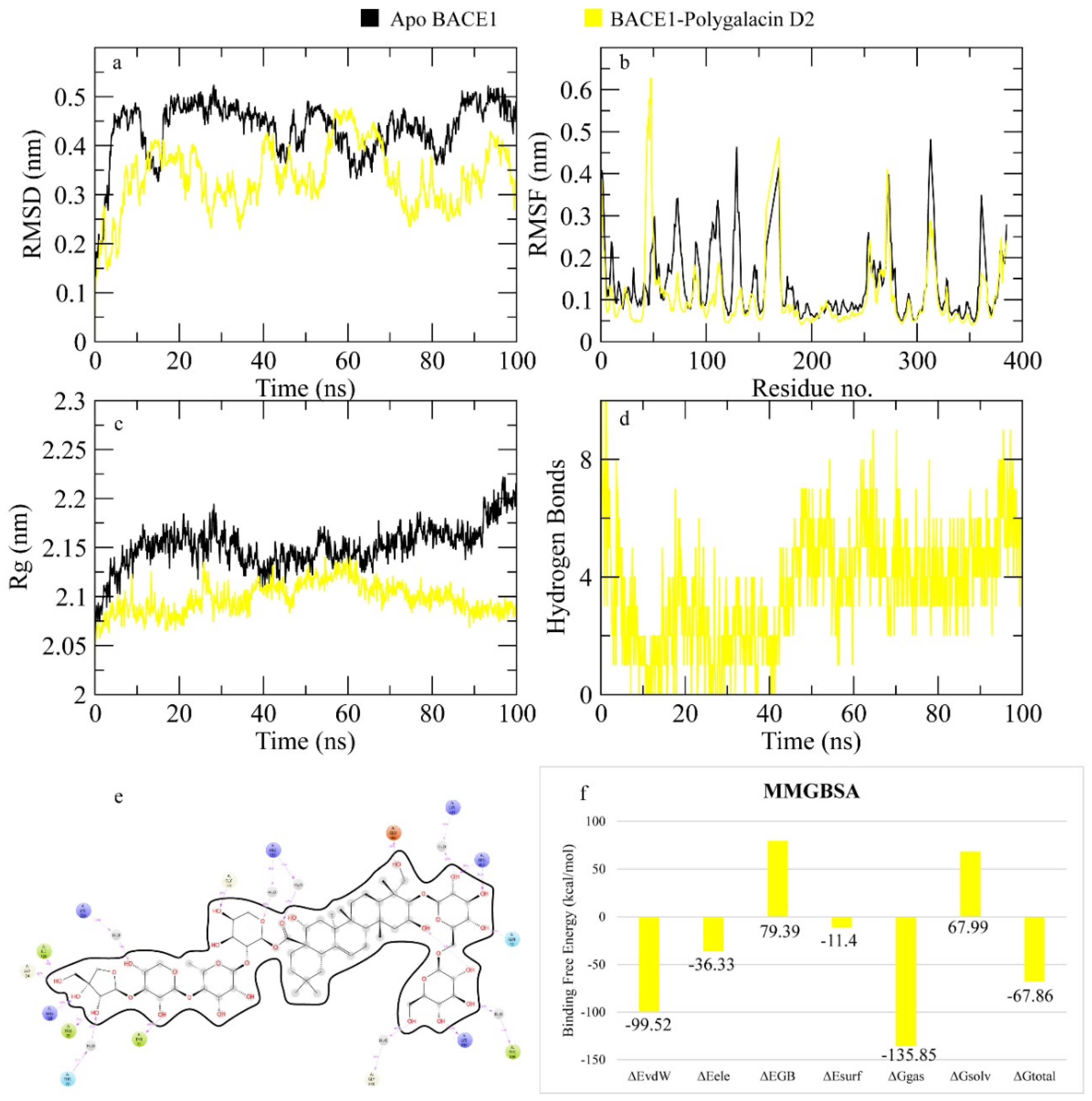
2.7.6. BACE1
3. Discussion
4. Materials and Methods
4.1. Protein and Ligand Structures Retrieval
4.2. Ligand Preparation
4.3. Molecular Docking
4.4. MD Simulation
4.5. Quantum Chemical Investigation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elhawary, E.A.; Moussa, A.Y.; Singab, A.N.B. Genus Curcuma: Chemical and ethnopharmacological role in aging process. BMC Complement. Med. Ther. 2024, 24, 31. [Google Scholar] [CrossRef] [PubMed]
- Better, M.A. 2023 Alzheimer’s disease facts and figures. Alzheimers Dement. 2023, 19, 1598–1695. [Google Scholar]
- Limantoro, J.; de Liyis, B.G.; Sutedja, J.C. Akt signaling pathway: A potential therapy for Alzheimer’s disease through glycogen synthase kinase 3 beta inhibition. Egypt. J. Neurol. Psychiatry Neurosurg. 2023, 59, 147. [Google Scholar] [CrossRef]
- Abeysinghe, A.A.D.T.; Deshapriya, R.D.U.S.; Udawatte, C. Alzheimer’s disease; a review of the pathophysiological basis and therapeutic interventions. Life Sci. 2020, 256, 117996. [Google Scholar] [CrossRef]
- Hampel, H.; Mesulam, M.M.; Cuello, A.C.; Farlow, M.R.; Giacobini, E.; Grossberg, G.T.; Khachaturian, A.S.; Vergallo, A.; Cavedo, E.; Snyder, P.J.; et al. The cholinergic system in the pathophysiology and treatment of Alzheimer’s disease. Brain 2018, 141, 1917–1933. [Google Scholar] [CrossRef]
- Liu, J.; Chang, L.; Song, Y.; Li, H.; Wu, Y. The role of NMDA receptors in Alzheimer’s disease. Front. Neurosci. 2019, 13, 43. [Google Scholar] [CrossRef]
- Dou, K.X.; Tan, M.S.; Tan, C.C.; Cao, X.-P.; Hou, X.-H.; Guo, Q.-H.; Tan, L.; Mok, V.; Yu, J.-T. Comparative safety and effectiveness of cholinesterase inhibitors and memantine for Alzheimer’s disease: A network meta-analysis of 41 randomized controlled trials. Alzheimers Res. Ther. 2018, 10, 126. [Google Scholar] [CrossRef]
- Iqbal, D.; Alsaweed, M.; Jamal, Q.M.S.; Asad, M.R.; Rizvi, S.M.D.; Rizvi, M.R.; Albadrani, H.M.; Hamed, M.; Jahan, S.; Alyenbaawi, H. Pharmacophore-based screening, molecular docking, and dynamic simulation of fungal metabolites as inhibitors of multi-targets in neurodegenerative disorders. Biomolecules 2023, 13, 1613. [Google Scholar] [CrossRef]
- Deng, M.; Yan, W.; Gu, Z.; Li, Y.; Chen, L.; He, B. Anti-neuroinflammatory potential of natural products in the treatment of Alzheimer’s disease. Molecules 2023, 28, 1486. [Google Scholar] [CrossRef]
- Back, M.K.; Ruggieri, S.; Jacobi, E.; von Engelhardt, J. Amyloid beta-mediated changes in synaptic function and spine number of neocortical neurons depend on NMDA receptors. Int. J. Mol. Sci. 2021, 22, 6298. [Google Scholar] [CrossRef]
- Ugale, V.G.; Bari, S.B. Identification of potential Gly/NMDA receptor antagonists by cheminformatics approach: A combination of pharmacophore modelling, virtual screening and molecular docking studies. SAR QSAR Environ. Res. 2016, 27, 125–145. [Google Scholar] [CrossRef] [PubMed]
- Clyde, A. Ultrahigh throughput protein-ligand docking with deep learning. Methods Mol. Biol. 2022, 2390, 301–319. [Google Scholar] [PubMed]
- Moussa, A.Y.; Alanzi, A.R.; Riaz, M.; Fayez, S. Could mushrooms’ secondary metabolites ameliorate Alzheimer disease? A computational flexible docking investigation. J. Med. Food 2024, 27, 775–796. [Google Scholar] [CrossRef]
- Moussa, A.Y.; Alanzi, A.; Luo, J.; Chung, S.K.; Xu, B. Potential anti-obesity effect of saponin metabolites from adzuki beans: A computational approach. Food Sci. Nutr. 2024, 12, 3612–3627. [Google Scholar] [CrossRef] [PubMed]
- Moussa, A.Y.; Labib, R.M.; Ayoub, N.A. Isolation of chemical constituents and protective effect of Pistacia khinjuk against CCl4–induced damage on HepG2 cells. Phytopharmacology 2013, 2, 1–9. [Google Scholar]
- Torky, Z.A.; Moussa, A.Y.; Abdelghffar, E.A.; Abdel-Hameed, U.K.; Eldahshan, O.A. Chemical profiling, antiviral and antiproliferative activities of the essential oil of Phlomis aurea Decne grown in Egypt. Food Funct. 2021, 12, 4630–4643. [Google Scholar] [CrossRef]
- Sun, A.; Xu, X.; Lin, J.; Cui, X.; Xu, R. Neuroprotection by saponins. Phytother. Res. 2015, 29, 187–200. [Google Scholar] [CrossRef]
- Yang, L.; Hao, J.; Zhang, J.; Xia, W.; Dong, X.; Hu, X.; Kong, F.; Cui, X. Ginsenoside Rg3 promotes beta-amyloid peptide degradation by enhancing gene expression of neprilysin. J. Pharm. Pharmacol. 2009, 61, 375–380. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, W.; Lin, Z.; Wu, Y.; Li, Y.; Xia, X. Panax notoginseng saponins prevent dementia and oxidative stress in brains of SAMP8 mice by enhancing mitophagy. BMC Complement. Med. Ther. 2024, 24, 144. [Google Scholar] [CrossRef]
- Xu, T.Z.; Shen, X.Y.; Sun, L.L.; Chen, Y.; Zhang, B.; Huang, D.; Li, W. Ginsenoside Rg1 protects against H2O2-induced neuronal damage due to inhibition of the NLRP1 inflammasome signalling pathway in hippocampal neurons in vitro. Int. J. Mol. Med. 2019, 43, 717–726. [Google Scholar] [CrossRef]
- Shivakumar, D.; Harder, E.; Damm, W.; Friesner, R.A.; Sherman, W. Improving the prediction of absolute solvation free energies using the next generation OPLS force field. J. Chem. Theory Comput. 2012, 8, 2553–2558. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.H.; Zhang, J.T. Ginsenoside Rg1 promotes proliferation of hippocampal progenitor cells. Neurol. Res. 2004, 26, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.F.; Li, Q.; Li, Y. Long-term ginsenoside administration prevents memory loss in aged female C57BL/6J mice by modulating the redox status and up-regulating the plasticity-related proteins in hippocampus. Neuroscience 2011, 183, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liang, J.; Gao, C.; Wang, A.; Xia, J.; Hong, C.; Zhong, Z.; Zuo, Z.; Kim, J.; Ren, H.; et al. Multifunctional ginsenoside Rg3-based liposomes for glioma targeting therapy. J. Control. Release 2021, 330, 641–657. [Google Scholar] [CrossRef]
- Mao, Y.; Yuan, W.; Gai, J.; Zhang, Y.; Wu, S.; Xu, E.-Y.; Wang, L.; Zhang, X.; Guan, J.; Mao, S. Enhanced brain distribution of Ginsenoside F1 via intranasal administration in combination with absorption enhancers. Int. J. Pharm. 2024, 654, 123930. [Google Scholar] [CrossRef]
- Wang, C.; Schuller Levis, G.B.; Lee, E.B.; Levis, W.R.; Lee, D.W.; Kim, B.S.; Park, S.Y.; Park, E. Platycodin D and D3 isolated from the root of Platycodon grandiflorum modulate the production of nitric oxide and secretion of TNF-alpha in activated RAW 264.7 cells. Int. Immunopharmacol. 2004, 4, 1039–1049. [Google Scholar] [CrossRef]
- Kiranmayee, M.; Rajesh, N.; Vidya Vani, M.; Khadri, H.; Mohammed, A.; Chinni, S.V.; Ramachawolran, G.; Riazunnisa, K.; Moussa, A.Y. Green synthesis of Piper nigrum copper-based nanoparticles: In silico study and ADMET analysis to assess their antioxidant, antibacterial, and cytotoxic effects. Front. Chem. 2023, 11, 1218588. [Google Scholar] [CrossRef]
- Kaya, S.; Putz, M.V. Atoms-in-molecules’ faces of chemical hardness by conceptual density functional theory. Molecules 2022, 27, 8825. [Google Scholar] [CrossRef]
- Branches, A.D.S.; da Silva, J.N.; de Oliveira, M.D.L.; Bezerra, D.P.; Soares, M.B.P.; Costa, E.V.; Oliveira, K.M.T. DFT calculations, molecular docking, binding free energy analysis and cytotoxicity assay of 7,7-dimethylaporphine alkaloids with methylenedioxy ring in positions 1 and 2. Comput. Theor. Chem. 2024, 1233, 114483. [Google Scholar] [CrossRef]
- Yamari, I.; Abchir, O.; Nour, H.; Khedraoui, M.; Rossafi, B.; Errougui, A.; Talbi, M.; Samadi, A.; El Kouali, M.; Chtita, S. Unveiling Moroccan nature’s Arsenal: A computational molecular docking, density functional theory, and molecular dynamics study of natural compounds against drug-resistant fungal infections. Pharmaceuticals 2024, 17, 886. [Google Scholar] [CrossRef]
- Yovanno, R.A.; Chou, T.H.; Brantley, S.J.; Furukawa, H.; Lau, A.Y. Excitatory and inhibitory D-serine binding to the NMDA receptor. eLife 2022, 11, e77645. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.L. The challenge of interpreting glutamate-receptor ion-channel structures. Biophys. J. 2017, 113, 2143–2151. [Google Scholar] [CrossRef] [PubMed]
- Takashima, A. GSK-3 is essential in the pathogenesis of Alzheimer’s disease. J. Alzheimers Dis. 2006, 9 (Suppl. S3), 309–317. [Google Scholar] [CrossRef]
- Wang, C.; Cui, Y.; Xu, T.; Zhou, Y.; Yang, R.; Wang, T. New insights into glycogen synthase kinase-3: A common target for neurodegenerative diseases. Biochem. Pharmacol. 2023, 218, 115923. [Google Scholar] [CrossRef]
- Casiraghi, A.; Longhena, F.; Faustini, G.; Ribaudo, G.; Suigo, L.; Camacho-Hernandez, G.A.; Bono, F.; Brembati, V.; Newman, A.H.; Gianoncelli, A.; et al. Methylphenidate analogues as a new class of potential disease-modifying agents for Parkinson’s disease: Evidence from cell models and alpha-synuclein transgenic mice. Pharmaceutics 2022, 14, 1595. [Google Scholar] [CrossRef]
- Zhan, Q.; Zhang, F.; Sun, L.; Wu, Z.; Chen, W. Two new oleanane-type triterpenoids from Platycodi Radix and anti-proliferative activity in HSC-T6 cells. Molecules 2012, 17, 14899–14907. [Google Scholar] [CrossRef]
- Nan, F.; Nan, W.; Yu, Z.; Wang, H.; Cui, X.; Jiang, S.; Zhang, X.; Li, J.; Wang, Z.; Zhang, S.; et al. Polygalacin D inhibits the growth of hepatocellular carcinoma cells through BNIP3L-mediated mitophagy and endogenous apoptosis pathways. Chin. J. Nat. Med. 2023, 21, 346–358. [Google Scholar] [CrossRef]
- Seo, Y.S.; Kang, O.H.; Kong, R.; Zhou, T.; Kim, S.-A.; Ryu, S.; Kim, H.-R.; Kwon, D.-Y. Polygalacin D induces apoptosis and cell cycle arrest via the PI3K/Akt pathway in non-small cell lung cancer. Oncol. Rep. 2018, 39, 1702–1710. [Google Scholar] [CrossRef]
- Kim, M.; Hwang, I.G.; Kim, S.B.; Choi, A.J. Chemical characterization of balloon flower (Platycodon grandiflorum) sprout extracts and their regulation of inflammatory activity in lipopolysaccharide-stimulated RAW 264.7 murine macrophage cells. Food Sci. Nutr. 2019, 8, 246–256. [Google Scholar] [CrossRef]
- Maesako, M.; Zoltowska, K.M.; Berezovska, O. Synapsin 1 promotes Aβ generation via BACE1 modulation. PLoS ONE 2019, 14, e0226368. [Google Scholar] [CrossRef]
- Yan, R.; Fan, Q.; Zhou, J.; Vassar, R. Inhibiting BACE1 to reverse synaptic dysfunctions in Alzheimer’s disease. Neurosci. Biobehav. Rev. 2016, 65, 326–340. [Google Scholar] [CrossRef] [PubMed]
- Egbertson, M.; McGaughey, G.B.; Pitzenberger, S.M.; Stauffer, S.R.; Coburn, C.A.; Stachel, S.J.; Yang, W.; Barrow, J.C.; Neilson, L.A.; McWherter, M.; et al. Methyl-substitution of an iminohydantoin spiropiperidine β-secretase (BACE-1) inhibitor has a profound effect on its potency. Bioorg Med. Chem. Lett. 2015, 25, 4812–4819. [Google Scholar] [CrossRef] [PubMed]
- Joseph, O.A.; Babatomiwa, K.; Niyi, A.; Olaposi, O.; Olumide, I. molecular docking and 3D osar studies of C000000956 as a potent inhibitor of Bace-1. Drug Res. 2019, 69, 451–457. [Google Scholar]
- Han, B.; Luo, J.; Xu, B. Revealing molecular mechanisms of the bioactive saponins from edible root of Platycodon grandiflorum in combating obesity. Plants 2024, 13, 1123. [Google Scholar] [CrossRef]
- Ji, M.Y.; Bo, A.; Yang, M.; Xu, J.-F.; Jiang, L.-L.; Zhou, B.-C.; Li, M.-H. The pharmacological effects and health benefits of Platycodon grandifloras—A medicine food homology species. Foods 2020, 9, 142. [Google Scholar] [CrossRef]
- Moon, M.K.; Ahn, J.Y.; Kim, S.; Ryu, S.Y.; Kim, Y.S.; Ha, T.Y. Ethanol extract and saponin of Platycodon grandiflorum ameliorate scopolamine-induced amnesia in mice. J. Med. Food 2010, 13, 584–588. [Google Scholar] [CrossRef]
- Zhang, J.T.; Xie, L.Y.; Shen, Q.; Liu, W.; Li, M.-H.; Hu, R.-Y.; Hu, J.-N.; Wang, Z.; Chen, C. Platycodin D stimulates AMPK activity to inhibit the neurodegeneration caused by reactive oxygen species-induced inflammation and apoptosis. J. Ethnopharmacol. 2023, 308, 116294. [Google Scholar] [CrossRef]
- Choi, J.H.; Yoo, K.Y.; Park, O.K.; Lee, C.H.; Won, M.-H.; Hwang, I.K.; Ryu, S.Y.; Kim, Y.S.; Yi, J.-S.; Bae, Y.-S.; et al. Platycodin D and 2″-O-acetyl-polygalacin D2 isolated from Platycodon grandiflorum protect ischemia/reperfusion injury in the gerbil hippocampus. Brain Res. 2009, 1279, 197–208. [Google Scholar] [CrossRef]
- Choi, Y.; Kang, S.; Cha, S.H.; Kim, H.-S.; Song, K.; Lee, Y.J.; Kim, K.; Kim, Y.S.; Cho, S.; Park, Y. Platycodon saponins from Platycodi Radix (Platycodon grandiflorum) for the green synthesis of gold and silver nanoparticles. Nanoscale Res. Lett. 2018, 13, 23. [Google Scholar] [CrossRef]
- Ha, Y.W.; Kim, Y.S. Preparative isolation of six platycosides from Platycodi Radix by high-speed counter-current chromatography with evaporative light scattering detection. Planta Med. 2008, 74, PC89. [Google Scholar] [CrossRef]
- Shin, K.C.; Oh, D.K. Biotransformation of platycosides, saponins from balloon flower root, into bioactive deglycosylated platycosides. Antioxidants 2023, 12, 327. [Google Scholar] [CrossRef] [PubMed]
- Nyakudya, E.; Jeong, J.H.; Lee, N.K.; Jeong, Y.S. Platycosides from the roots of Platycodon grandiflorum and their health benefits. Prev. Nutr. Food Sci. 2014, 19, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Mirza, F.J.; Zahid, S.; Amber, S.; Sumera, J.H.; Asim, N.; Ali Shah, S.A. Multitargeted molecular docking and dynamic simulation studies of bioactive compounds from Rosmarinus officinalis against Alzheimer’s disease. Molecules 2022, 27, 7241. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.J.; Kang, M.H.; Kim, G.S.; Kim, H.D.; Jang, G.Y. Platycodon grandiflorum exhibits anti-neuroinflammatory potential against beta-amyloid-induced toxicity in microglia cells. Front. Nutr. 2024, 11, 1427121. [Google Scholar] [CrossRef] [PubMed]
- Pallas-Bazarra, N.; Draffin, J.; Cuadros, R.; Antonio Esteban, J.; Avila, J. Tau is required for the function of extrasynaptic NMDA receptors. Sci. Rep. 2019, 9, 9116. [Google Scholar] [CrossRef]
- Pluta, R.; Ułamek-Kozioł, M. Tau protein-targeted therapies in Alzheimer’s disease: Current state and future perspectives. In Alzheimer’s Disease: Drug Discovery; Huang, X., Ed.; Exon Publications: Brisbane, Australia, 2020. [Google Scholar]
- Vashisth, M.K.; Hu, J.; Liu, M.; Basha, S.H.; Yu, C.; Huang, W. In-Silico discovery of 17alpha-hydroxywithanolide-D as potential neuroprotective allosteric modulator of NMDA receptor targeting Alzheimer’s disease. Sci. Rep. 2024, 14, 27908. [Google Scholar] [CrossRef]
- Dajani, R.; Fraser, E.; Roe, S.M.; Young, N.; Good, V.; Dale, T.C.; Pearl, L.H. Crystal structure of glycogen synthase kinase 3 beta: Structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 2001, 105, 721–732. [Google Scholar] [CrossRef]
- Sayas, C.L.; Ávila, J. GSK-3 and tau: A key duet in Alzheimer’s disease. Cells 2021, 10, 721. [Google Scholar] [CrossRef]
- Iwaloye, O.; Elekofehinti, O.O.; Oluwarotimi, E.A.; Kikiowo, B.I.; Fadipe, T.M. Insight into glycogen synthase kinase-3β inhibitory activity of phyto-constituents from Melissa officinalis: In silico studies. In Silico Pharmacol. 2020, 8, 2. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2021, 2199, 239–255. [Google Scholar]
- Kim, M.O.; Nichols, S.E.; Wang, Y.; McCammon, J.A. Effects of histidine protonation and rotameric states on virtual screening of M. tuberculosis RmlC. J. Comput. Aided Mol. Des. 2013, 27, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Alanzi, A.; Moussa, A.Y.; Mothana, R.A.; Abbas, M.; Ali, I. In silico exploration of PD-L1 binding compounds: Structure-based virtual screening, molecular docking, and MD simulation. PLoS ONE 2024, 19, e0306804. [Google Scholar] [CrossRef]
- Yang, Y.; Yao, K.; Repasky, M.P.; Leswing, K.; Abel, R.; Shoichet, B.K.; Jerome, S.V. Efficient exploration of chemical space with docking and deep learning. J. Chem. Theory Comput. 2021, 17, 7106–7119. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Liu, Y.; Grimm, M.; Dai, W.T.; Hou, M.C.; Xiao, Z.X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein-ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef]
- Qureshi, K.A.; Al Nasr, I.; Koko, W.S.; Khan, T.A.; Fatmi, M.Q.; Imtiaz, M.; Khan, R.A.; Mohammed, H.A.; Jaremko, M.; Emwas, A.-H.; et al. In vitro and in silico approaches for the antileishmanial activity evaluations of actinomycins isolated from novel Streptomyces smyrnaeus strain UKAQ_23. Antibiotics 2021, 10, 887. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Grubmüller, H.; Heller, H.; Windemuth, A.; Schulten, K. Generalized verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol. Simul. 1991, 6, 121–142. [Google Scholar] [CrossRef]
- Kholmurodov, K.; Smith, W.; Yasuoka, K.; Darden, T.; Ebisuzaki, T. A smooth-particle mesh Ewald method for DL_POLY molecular dynamics simulation package on the Fujitsu VPP700. J. Comput. Chem. 2000, 21, 1187–1191. [Google Scholar] [CrossRef]
- Grant, B.J.; Skjaerven, L.; Yao, X.Q. The Bio3D packages for structural bioinformatics. Protein Sci. 2021, 30, 20–30. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Frisch, R.; Gary, M.J.; Trucks, G.; Trucks, S.H.B. Gaussian 6.0; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]



| No. | PubChem CID | Compounds | MW (g/mol) | HBD | HBA | logP | Drug-like |
|---|---|---|---|---|---|---|---|
| 1 | 101596922 | Prosapogenin D | 603.817 | 6 | 8 | 3.62969 | TRUE |
| 2 | 162859 | Platycodin D | 1225.335 | 17 | 28 | −5.3782 | TRUE |
| 3 | 385678065 | Polygalacin D2 | 1371.477 | 19 | 32 | −6.5264 | FALSE |
| 4 | 385678073 | Polygalacin D3 | 1371.477 | 19 | 32 | −6.5264 | FALSE |
| 5 | 46173919 | Platycodin C | 1267.372 | 16 | 29 | −4.8074 | TRUE |
| 6 | 50900852 | Platyconic acid B lactone | 1383.444 | 18 | 33 | −6.9818 | FALSE |
| 7 | 53317652 | Platycodin D2 | 1387.476 | 20 | 33 | −7.554 | FALSE |
| 8 | 70698202 | Platycoside E | 1549.617 | 23 | 38 | −9.7298 | FALSE |
| 9 | 70698266 | Deapioplatycodin D | 1093.22 | 15 | 24 | −3.8431 | TRUE |
| 10 | 70698289 | Deapioplatycoside E | 1417.502 | 21 | 34 | −8.1947 | FALSE |
| 11 | 75251137 | Platycodin D3 | 1387.476 | 20 | 33 | −7.554 | FALSE |
| 12 | 96023791 | Polygalacin D | 1209.336 | 16 | 27 | −4.3506 | TRUE |
| 13 | 70698190 | Deapi-platycodin D3 | 1255.361 | 18 | 29 | −6.0189 | TRUE |
| 14 | 46173910 | Platycodin A | 1267.4 g | 16 | 29 | −3.1 | TRUE |
| Synapsin I | Binding Free Energy (Kcal/mol) | H-Bonds | Hydrophobic and VDW |
|---|---|---|---|
| polygalacin D2 | −6.742 | leu394, Arg186, Asp120, Glu121, Asn 334, Trp335, Asp283, Lys281 (10) | ile395, Pro393, Trp126, Met277, Gly276, Met297, Ser275. His123, Thr124, Asn338 |
| 3′-O-acetyl platyconic acid | −6.361 | Asp309, Lys281, Gly333, Asn334, Asn 338, Thr339 (8) | Phe307, Pro265, Lys378, Trp335, Lys336, Gly340, Met277, Gly276, Ser275 |
| LY281137 | −6.916 | Trp335 | Arg328, Lys336, Thr337, Asn 338, Gly276, Ile385, Glu386, Lys279, Lys269, Leu375, Val267, Phe307, Ile308, Ala310, Asp313 |
| PF-0480 | −6.703 | Lys269, Glu373, Ile308 | Ala310, Leu375, Trp335, Asp309, Asn338, Phe307, Gly276 |
| donepezil | −5.951 | Lys279, Gly276, Trp335 | Val388, Ile385, Leu375, Ala310, Ile308, Phe307, Pro306, Ile247, Val267 |
| ifenoprodil | −4.672 | Lys279 | Trp335, Leu375, Ala310, Met392, Pro393, Val388, Ile385, Val267 |
| synapsin II | |||
| polygalacin D | −6.101 | Lys337, trp336, asn335, glu122, asp121, arg187 (7) | ile396, leu395, pro304, ser392, met278, val281, lys282, gly279, met278, gly277, asn339, ser276 |
| Polygalacin D2 | −6.011 | asp310(1.85, 2.50), lys379(2.05), lys282(1.89, 1.85, 2.08), asn339, ser276(1.88), glu197(1.65, 1.72) (11) | phe308, ala311, gly314, trp336, val281, met278, gly341, gly277, ala194, lys337, lys280, gly277, gly341 |
| PF-0480 | −6.839 | ile309, phe386 | val 376, trp 336, pro307, phe308, ile248, ala311, val268, lys379, lys374, lys337, lys280, ser276, asn335, glu306, asp310, asp314, ser331, arg329, lys270 |
| LY-2811376 | −5.727 | phe386, val268, ile248, pro307, phe308, ile309, ala311, val376, trp336, lys280, lys270, lys337, lys374, arg329, gly277, ser276, asn339, thr338, glu306, asp314 | |
| ifenoprodil | −5.187 | lys374, glu306, phe386, asp314 | ile248, val376, pro307, phe308, ile309, val268, phe386, trp336, met389, pro394, lys270, lys337, arg329, arg316, thr338, asn339, ser276, gly277 |
| donepezil | −4.879 | lys374, glu306, phe386, asp34 | ile248, val376, pro307, phe308, ile309, val268, phe386, trp336, met389 |
| synapsin III | |||
| polygalacin D | −6.145 | ala254, asn313, gly312, lys260, phe286, asp288, asp358 (8) | met256, trp314, pro244, ala285, ile287, lys357, ser356 |
| polygalacin D2 | −5.882 | ala254, asn313, lys260, asp288, asp358 (8) | met256, trp314, gly312, pro244,lys260, phe286, ile287, ser356, lys357, arg360, phe243 |
| pf-0480 | −6.361 | lys248, ala254, lys352, trp314, ile287 | met367, ile364, ala252, pro372, ala310, val354, phe286, ala285, val226, val246 |
| ifenoprodil | −5.345 | trp314 | ala252, met367, ala254, ile364, val354, pro372, ala316, val246, ala285, phe286, ile287, val354 |
| donepezil | −3.513 | lys352, ser289 | ala254, phe286, ile287, tyr291, ala316, trp314, val354, tyr291, val246, ile364 |
| GSK-3B | |||
| platycodin D | −7.147 | asp200, asp181, lys183, ser66, asn186 (6) | val70, phe67, cys199, ile62, leu188, ala83, leu132, val110, tyr134, val135, tyr140 |
| polygalacin D | −6.873 | arg148, lys183, ile62, asp200, gly202, asn95 (9) | Tyr140, tyr222, ile217, val87, lys85, arg96, ser203, phe67, ser66, gly 65, gly63 |
| platycodin D3 | −6.514 | ser66, asn64, gln185, arg141, thr138, pro136, tyr134 (7) | ile217, cys218, phe67, leu188, tyr140, pro136, val135, tyr134, ser219, asp181, lys183, phe67, gly65, gly62, ile62, lys60, gln72 |
| platyconic acid B-lactone | −6.418 | arg148, arg144, arg141, tyr140, glu137, tyr221, ser203, asp200, asp181, lys183 (13) | arg223, tyr222, tyr221, arg220, ser219, cys218, ser66, phe67, gly202, lys85, glu185, asn186 |
| −6.297 | asp200, lys183, thr138, arg141, ser66, lys60, gln72 (9) | asp181, gln185, asn186, tyr140, phe67, gly65, asn64, gly63, ile62, val61, arg 148 | |
| PF-04820 | −8.373 | lys85, asp200, val135, arg141 | phe67, val70, ala83, cys199, leu132, leu188, val110, tyr134, pro126, ile62 |
| ifenprodil | −5.956 | asn186 | cys199, val110, ala83, leu132, ile62, tyr134, val135, pro134, phe67, val70, leu188 |
| donepezil | −5.918 | lys85 | phe67, val70, ile62, phe201, cys199, met101, ala83, leu130, val110, leu132, tyr134, leu188, val135, pro136 |
| BACE1 | |||
| polygalacin D2 | −7.298 | phe109, lys107, asn111, arg307, lys321, gly264, glu265, gln73, pro70, ile126, ser36, tyr198 (15) | phe47, phe108, ile110, leu263, tyr71, val69, ala127, gly34 |
| polygalacin D | −5.607 | Arg307, lys321, arg235, asp223, lys224 (9 bonds) | val309, tyr71, pro70, tyr198, Gly11, Gln12, Ser10, Lys107, Glu265, Thr72, Gln73, Ser327, Ser328, Thr329 |
| 3′-O-acetyl platyconic acid | −7.716 | tyr198, tyr71, lys75, asn233, thr232, thr 231, gly230 (7) | trp197, lys224, arg128, pro70, thr72, gln73, gly74, thr329, ser328, gln326, er325, arg236, leu263, gly264, leu30, ile110, arg307, gln12 |
| platyconic acid -B lactone | −6.93 | lys75, tyr71, thr72, gln326, glu265, gly264, lys321, arg307, asn111 (9) | gly74, pro70, leu263, val309, phe47, ile110, phe109, val309, thr329, ser328, ser327, ser325, arg235, asn233, ser10, gly11, gln12, lys107 |
| platycodin D3 | −6.891 | asp106, lys107, lys75, gly264, gln73, gly11, thr232, gly230 (10) | phe108, ile110, leu263, gly74, arg307, lys321, ser325, glu265, arg235, asn233, thr231, gln12, gly13, leu30, trp115 |
| platycodin D | −6.758 | gly11, thr232, the231, thr72, asp106 (5) | pro46, phe47, lys107, phe108, phe109, ile110, asn111, ile118, val332, ile226, tyr198, tyr71, gln73,gln12, arg235, lys321, arg307, glu265 |
| ifenoprodil | −5.726 | pro70, trp115, asp228 | tyr71, val69, ile118, phe108, ile110, tyr198, ile226, leu30, val332 |
| PF-04820 | −5.147 | tyr198, tyr71, asp228, | ile126, val332, phe108, ile110, trp115, ile118, pro70, val69, ile226, leu30 |
| donepezil | −5.052 | thr72 | ile126, tyr198, ile226, val332, leu30, pro70, tyr71, ile118, phe108, ile110, trp115, arg128, gly34, ser35, asp228, asp32 |
| LY-2811376 | −5.018 | gly11, thr72 | trp115, ile118, leu30, tyr198, tyr71, phe108, ile110, asp228, ser35, asp32 |
| NMDA | |||
| polygalacin D2 | −7.755 | Gln48, tyr351, asp353, lys296, gly354, gln291, arg380 (10) | leu280, leu377, met375, ile275, leu292, ile293, val355, ala307 |
| platycodin D | −7.296 | leu382, gln371, gly354, asp353, tyr351, ser303 (7) | val383, ile293, Met375, val355, ala307, gln384, gln291, ser373, asn297, thr356, arg52, asp304 |
| 3′-O-acetyl platyconic acid | −7.214 | pro95, thr122, lys37,glu298, lys296, asn294 (10) | pro96, pro95, ile41, ile293, ser93, ser34, thr35, ile293, ser299, asn297, gln291 |
| platycodin D3 | −7.183 | glu246, tyr144, lys296, leu292, ser373, gln384 (10) | ser245, arg273, ser276, gly277, met125, arg124, thr123, thr122, asn297, gly295, asn294, ile293, gln291 |
| polygalacin D | −6.736 | pro95, glu298, lys296, leu292, asn294, val355 (8) | pro96, ser93, thr 122, ser34, asn297, gly295, ile293, gln 291, glu272, gln384, ser373 |
| ifenoprodil | −5.29 | ser393, gln291 (2) | leu292, ile293 |
| prosapogenin D | −4.415 | lys296, gln291(4) | ser299, glu298, asn297, asn294, ile293, ile387, gln384, ser373, met375 |
| donepezil | −4.354 | lys37, ser299 (3) | pro95, pro96, ile41, gly295 |
| ly-2811375 | −4.269 | lys37, ser303 (2) | val355, ala45, ile41 |
| pf-04820 | −4.193 | lys296 (2) | ile41, pro95, pro96 |
| Compound | Polygalacin D | Prosapogenin D | Platycodin D | Polygalacin D2 |
|---|---|---|---|---|
| ELUMO (eV) | 1.1064155 | −0.02387 | −0.026395057 | 0.122995523 |
| EHOMO (eV) | −6.7048887 | −0.21197 | −6.11385713 | −6.03249504 |
| ΔE | 5.5984732 | 0.18826 | 6.087462073 | 5.909499517 |
| χ | 3.9056521 | 0.23538 | 3.070126094 | 2.954749759 |
| η | 3.9056521 | −0.09405 | 3.043731037 | 3.077745282 |
| σ | 1.716714271 | −2.25188569 | 2.008671941 | 0.324913178 |
| ω | 7.840523073 | −124.3888 | 8.516100697 | 10.58547613 |
| dipole moment (DEBYE) | 7.8259 | 4.8371 | 7.2001 | 8.0721 |
| chemical potential (µ) | 2.7992366 | 0.11792 | 3.070126094 | 2.954749759 |
| electronic energy | −17,794.64455 | −7977.932898 | −22,130.38172 | −25,782.40283 |
| Compounds | GSK-3β | NMDA Receptor | BACE1 | Synapsin I | Synapsin II | Synapsin III |
|---|---|---|---|---|---|---|
| Platycodin A | 0 | 0 | 0 | 0 | 0 | 0 |
| Platycodin C | 0 | 0 | 0 | 0 | 0 | 0 |
| Platycodin D | −7.147 | −7.296 | −6.758 | −4.898 | −3.712 | −3.202 |
| Deapioplatycodin D | 0 | 0 | 0 | 0 | 0 | 0 |
| Platycodin D2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Platycodin D3 | −6.514 | −7.183 | −6.891 | −5.995 | −5.513 | 0 |
| Deapioplatycodin D3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Deapioplatycoside E | 0 | 0 | 0 | 0 | 0 | 0 |
| Platyconic acid B lactone | −6.418 | −6.623 | −6.93 | −6.058 | −5.973 | −4.801 |
| 3″-O-acetylplatyconic acid A | −6.297 | −7.214 | −7.716 | −6.361 | −6.078 | −6.018 |
| Prosapogenin D | −5.456 | −4.415 | −4.407 | 0 | 0 | 0 |
| Polygalacin D | −6.873 | −6.736 | −5.607 | −4.841 | −6.101 | −6.145 |
| Polygalacin D3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Platycoside E | 0 | 0 | 0 | 0 | 0 | 0 |
| Polygalacin D2 | −5.532 | −7.755 | −7.298 | −6.742 | −6.011 | −5.882 |
| Dimethyl 2-O-methyl-3-O-a-D-glucopyranosyl platycogenate A | −4.79 | −3.918 | −4.282 | 0 | 0 | 0 |
| Platycodin V | 0 | 0 | 0 | 0 | 0 | 0 |
| PF-04802367 | −8.373 | / | / | / | / | / |
| Ifenprodil | / | −5.29 | / | / | / | / |
| LY2811376 | / | / | −5.018 | / | / | / |
| Staurosporine | / | / | / | −3.52 | / | / |
| Donzepil | / | / | / | / | −4.879 | / |
| Methylphenidate (MPH) | / | / | / | / | / | −3.942 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moussa, A.Y.; Alanzi, A.R.; Luo, J.; Wang, J.; Cheang, W.S.; Xu, B. Role of Saponins from Platycodon grandiflorum in Alzheimer’s Disease: DFT, Molecular Docking, and Simulation Studies in Key Enzymes. Molecules 2025, 30, 1812. https://doi.org/10.3390/molecules30081812
Moussa AY, Alanzi AR, Luo J, Wang J, Cheang WS, Xu B. Role of Saponins from Platycodon grandiflorum in Alzheimer’s Disease: DFT, Molecular Docking, and Simulation Studies in Key Enzymes. Molecules. 2025; 30(8):1812. https://doi.org/10.3390/molecules30081812
Chicago/Turabian StyleMoussa, Ashaimaa Y., Abdulah R. Alanzi, Jinhai Luo, Jingwen Wang, Wai San Cheang, and Baojun Xu. 2025. "Role of Saponins from Platycodon grandiflorum in Alzheimer’s Disease: DFT, Molecular Docking, and Simulation Studies in Key Enzymes" Molecules 30, no. 8: 1812. https://doi.org/10.3390/molecules30081812
APA StyleMoussa, A. Y., Alanzi, A. R., Luo, J., Wang, J., Cheang, W. S., & Xu, B. (2025). Role of Saponins from Platycodon grandiflorum in Alzheimer’s Disease: DFT, Molecular Docking, and Simulation Studies in Key Enzymes. Molecules, 30(8), 1812. https://doi.org/10.3390/molecules30081812








