Comprehensive Analysis of Stability and Variability of DNA Minimal I-Motif Structures
Abstract
:1. Introduction
2. Results and Discussion
2.1. Formation of I-Motifs with Base Tetrads
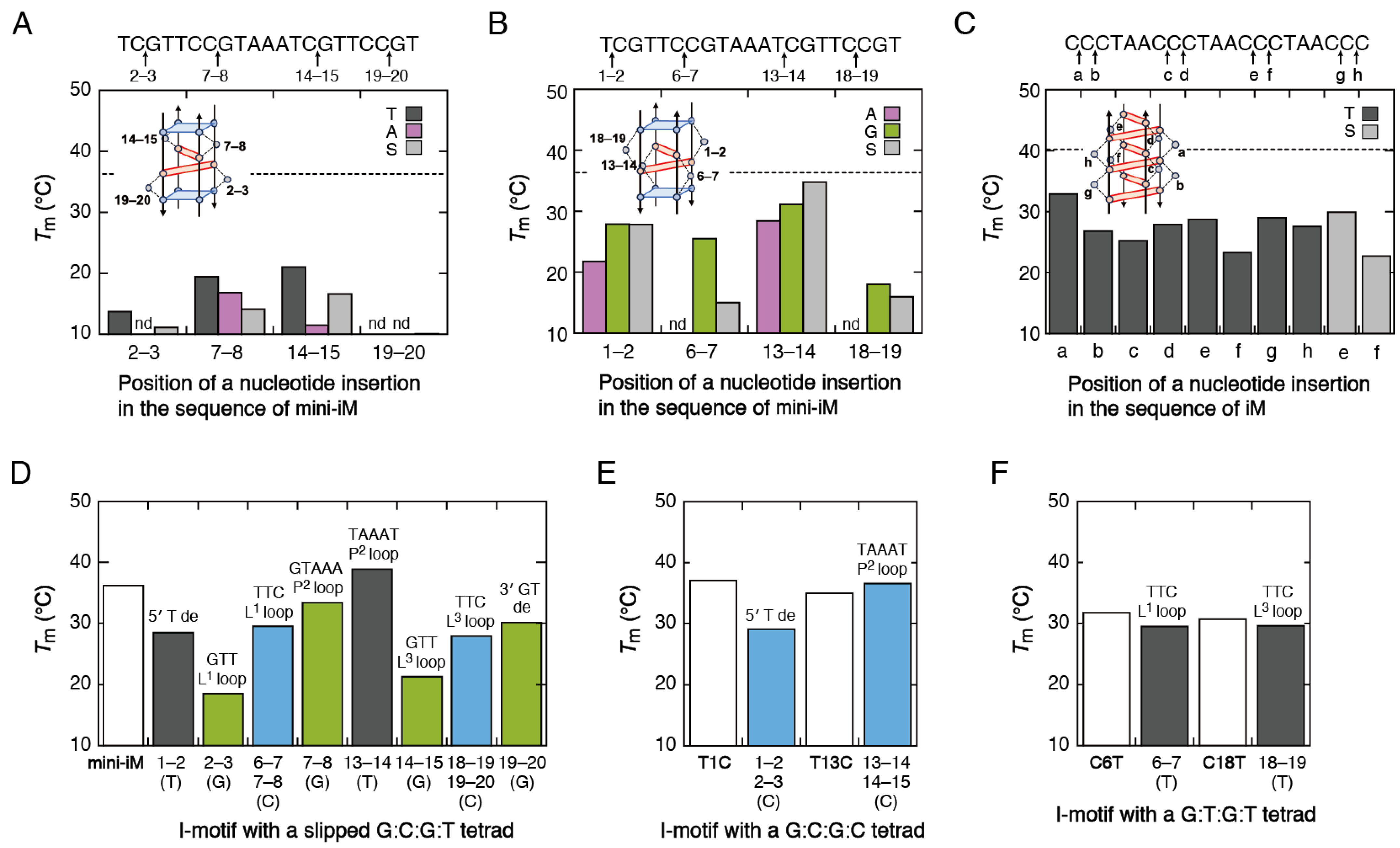
2.2. Variation in the Loop Sequences
2.3. I-Motifs Containing a Bulge
2.4. I-Motifs with a Slipped Base Tetrad
2.5. Influence of the Molecular Environment in the Presence of Spermine
2.6. Putative I-Motif-Forming Sequences Lacking C-Tracts
3. Materials and Methods
3.1. Preparation of Oligonucleotide and Buffer Solutions
3.2. Measurement of CD Spectra
3.3. Measurement and Analysis of DNA Thermal Melting
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Choi, J.; Majima, T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011, 40, 5893–5909. [Google Scholar] [CrossRef]
- Georgakopoulos-Soares, I.; Morganella, S.; Jain, N.; Hemberg, M.; Nik-Zainal, S. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 2018, 28, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Spiegel, J.; Marsico, G.; Tannahill, D.; Balasubramanian, S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018, 13, 551–564. [Google Scholar] [CrossRef]
- Mukundan, V.T.; Phan, A.T. Bulges in G-quadruplexes: Broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013, 135, 5017–5028. [Google Scholar] [CrossRef]
- Meier, M.; Moya-Torres, A.; Krahn, N.J.; McDougall, M.D.; Orriss, G.L.; McRae, E.K.S.; Booy, E.P.; McEleney, K.; Patel, T.R.; McKenna, S.A.; et al. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 2018, 46, 5319–5331. [Google Scholar] [CrossRef] [PubMed]
- Papp, C.; Mukundan, V.T.; Jenjaroenpun, P.; Winnerdy, F.R.; Ow, G.S.; Phan, A.T.; Kuznetsov, V.A. Stable bulged G-quadruplexes in the human genome: Identification, experimental validation and functionalization. Nucleic Acids Res. 2023, 51, 4148–4177. [Google Scholar] [CrossRef]
- Skolakova, P.; Renciuk, D.; Palacky, J.; Krafcik, D.; Dvorakova, Z.; Kejnovska, I.; Bednarova, K.; Vorlickova, M. Systematic investigation of sequence requirements for DNA i-motif formation. Nucleic Acids Res. 2019, 47, 2177–2189. [Google Scholar] [CrossRef]
- Zanin, I.; Ruggiero, E.; Nicoletto, G.; Lago, S.; Maurizio, I.; Gallina, I.; Richter, S.N. Genome-wide mapping of i-motifs reveals their association with transcription regulation in live human cells. Nucleic Acids Res. 2023, 51, 8309–8321. [Google Scholar] [CrossRef]
- Roxo, C.; Pasternak, A. Switching off cancer—An overview of G-quadruplex and i-motif functional role in oncogene expression. Bioorg. Med. Chem. Lett. 2024, 116, 130038. [Google Scholar] [CrossRef]
- Pena Martinez, C.D.; Zeraati, M.; Rouet, R.; Mazigi, O.; Henry, J.Y.; Gloss, B.; Kretzmann, J.A.; Evans, C.W.; Ruggiero, E.; Zanin, I.; et al. Human genomic DNA is widely interspersed with i-motif structures. EMBO J. 2024, 43, 4786–4804. [Google Scholar] [CrossRef]
- Dzatko, S.; Krafcikova, M.; Hansel-Hertsch, R.; Fessl, T.; Fiala, R.; Loja, T.; Krafcik, D.; Mergny, J.L.; Foldynova-Trantirkova, S.; Trantirek, L. Evaluation of the stability of DNA i-motifs in the nuclei of living mammalian cells. Angew. Chem. Int. Ed. Engl. 2018, 57, 2165–2169. [Google Scholar] [CrossRef] [PubMed]
- Zeraati, M.; Langley, D.B.; Schofield, P.; Moye, A.L.; Rouet, R.; Hughes, W.E.; Bryan, T.M.; Dinger, M.E.; Christ, D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018, 10, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Qiu, D.; Tamon, L.; Istvankova, E.; Viskova, P.; Amrane, S.; Guedin, A.; Chen, J.; Lacroix, L.; Ju, H.; et al. Thermal and pH stabilities of i-DNA: Confronting in vitro experiments with models and in-cell NMR data. Angew. Chem. Int. Ed. Engl. 2021, 60, 10286–10294. [Google Scholar] [CrossRef] [PubMed]
- Viskova, P.; Istvankova, E.; Rynes, J.; Dzatko, S.; Loja, T.; Zivkovic, M.L.; Rigo, R.; El-Khoury, R.; Serrano-Chacon, I.; Damha, M.J.; et al. In-cell NMR suggests that DNA i-motif levels are strongly depleted in living human cells. Nat. Commun. 2024, 15, 1992. [Google Scholar] [CrossRef]
- Wright, E.P.; Huppert, J.L.; Waller, Z.A.E. Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 2017, 45, 2951–2959. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Rogers, R.A.; Zhu, J.; Zhu, J.; Burton, A.D.; Carlisle, C.B.; Burrows, C.J. 4n-1 Is a “sweet spot” in DNA i-motif folding of 2′-deoxycytidine homopolymers. J. Am. Chem. Soc. 2017, 139, 4682–4689. [Google Scholar] [CrossRef]
- Berger, I.; Egli, M.; Rich, A. Inter-strand C-H···O hydrogen bonds stabilizing four-stranded intercalated molecules: Stereoelectronic effects of O4′ in cytosine-rich DNA. Proc. Natl. Acad. Sci. USA 1996, 93, 12116–12121. [Google Scholar] [CrossRef]
- Gurung, S.P.; Schwarz, C.; Hall, J.P.; Cardin, C.J.; Brazier, J.A. The importance of loop length on the stability of i-motif structures. Chem. Commun. 2015, 51, 5630–5632. [Google Scholar] [CrossRef]
- McKim, M.; Buxton, A.; Johnson, C.; Metz, A.; Sheardy, R.D. Loop sequence context influences the formation and stability of the i-motif for DNA oligomers of sequence (CCCXXX)4, where X = A and/or T, under slightly acidic conditions. J. Phys. Chem. B 2016, 120, 7652–7661. [Google Scholar] [CrossRef]
- Abou Assi, H.; Garavis, M.; Gonzalez, C.; Damha, M.J. I-motif DNA: Structural features and significance to cell biology. Nucleic Acids Res. 2018, 46, 8038–8056. [Google Scholar] [CrossRef] [PubMed]
- Escaja, N.; Viladoms, J.; Garavis, M.; Villasante, A.; Pedroso, E.; Gonzalez, C. A minimal i-motif stabilized by minor groove G:T:G:T tetrads. Nucleic Acids Res. 2012, 40, 11737–11747. [Google Scholar] [CrossRef]
- Escaja, N.; Mir, B.; Garavis, M.; Gonzalez, C. Non-G base tetrads. Molecules 2022, 27, 5287. [Google Scholar] [CrossRef]
- Mir, B.; Serrano, I.; Buitrago, D.; Orozco, M.; Escaja, N.; Gonzalez, C. Prevalent sequences in the human genome can form mini i-motif structures at physiological pH. J. Am. Chem. Soc. 2017, 139, 13985–13988. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L.; Han, X.; Leroy, J.-L.; Hélène, C. Intramolecular folding of pyrimidine oligodeoxynucleotides into an i-DNA motif. J. Am. Chem. Soc. 1995, 117, 8887–8898. [Google Scholar] [CrossRef]
- Benabou, S.; Aviñó, A.; Eritja, R.; González, C.; Gargallo, R. Fundamental aspects of the nucleic acid i-motif structures. RSC Adv. 2014, 4, 26956–26980. [Google Scholar] [CrossRef]
- Mir, B.; Soles, X.; Gonzalez, C.; Escaja, N. The effect of the neutral cytidine protonated analogue pseudoisocytidine on the stability of i-motif structures. Sci. Rep. 2017, 7, 2772. [Google Scholar] [CrossRef]
- Gao, B.; Hou, X.M. Opposite effects of potassium ions on the thermal stability of i-motif DNA in different buffer systems. ACS Omega 2021, 6, 8976–8985. [Google Scholar] [CrossRef]
- Phan, A.T.; Gueron, M.; Leroy, J.L. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000, 299, 123–144. [Google Scholar] [CrossRef]
- Childs, A.C.; Mehta, D.J.; Gerner, E.W. Polyamine-dependent gene expression. Cell Mol. Life Sci. 2003, 60, 1394–1406. [Google Scholar] [CrossRef]
- Lightfoot, H.L.; Hall, J. Endogenous polyamine function − the RNA perspective. Nucleic Acids Res. 2014, 42, 11275–11290. [Google Scholar] [CrossRef] [PubMed]
- Molnar, M.M.; Liddell, S.C.; Wadkins, R.M. Effects of polyamine binding on the stability of DNA i-motif structures. ACS Omega 2019, 4, 8967–8973. [Google Scholar] [CrossRef] [PubMed]
- Raspaud, E.; Chaperon, I.; Leforestier, A.; Livolant, F. Spermine-induced aggregation of DNA, nucleosome, and chromatin. Biophys. J. 1999, 77, 1547–1555. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.M.; Poudyal, R.R.; Bevilacqua, P.C. Long tracts of guanines drive aggregation of RNA G-quadruplexes in the presence of spermine. Biochemistry 2021, 60, 2715–2726. [Google Scholar] [CrossRef]
- Ngoc Nguyen, T.Q.; Lim, K.W.; Phan, A.T. Duplex formation in a G-quadruplex bulge. Nucleic Acids Res. 2020, 48, 10567–10575. [Google Scholar] [CrossRef]
- Murray-Stewart, T.R.; Woster, P.M.; Casero, R.A., Jr. Targeting polyamine metabolism for cancer therapy and prevention. Biochem. J. 2016, 473, 2937–2953. [Google Scholar] [CrossRef]
- Novita Sari, I.; Setiawan, T.; Seock Kim, K.; Toni Wijaya, Y.; Won Cho, K.; Young Kwon, H. Metabolism and function of polyamines in cancer progression. Cancer Lett. 2021, 519, 91–104. [Google Scholar] [CrossRef]
- Richard, R.G. Handbook of Biochemistry and Molecular Biology: Nucleic Acids, 3rd ed.; CRC Press: Cleveland, OH, USA, 1975; Volume 1, p. 597. [Google Scholar]
- Puglisi, J.D.; Tinoco, I., Jr. Absorbance melting curves of RNA. Methods Enzymol. 1989, 180, 304–325. [Google Scholar] [CrossRef]
- Mergny, J.L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
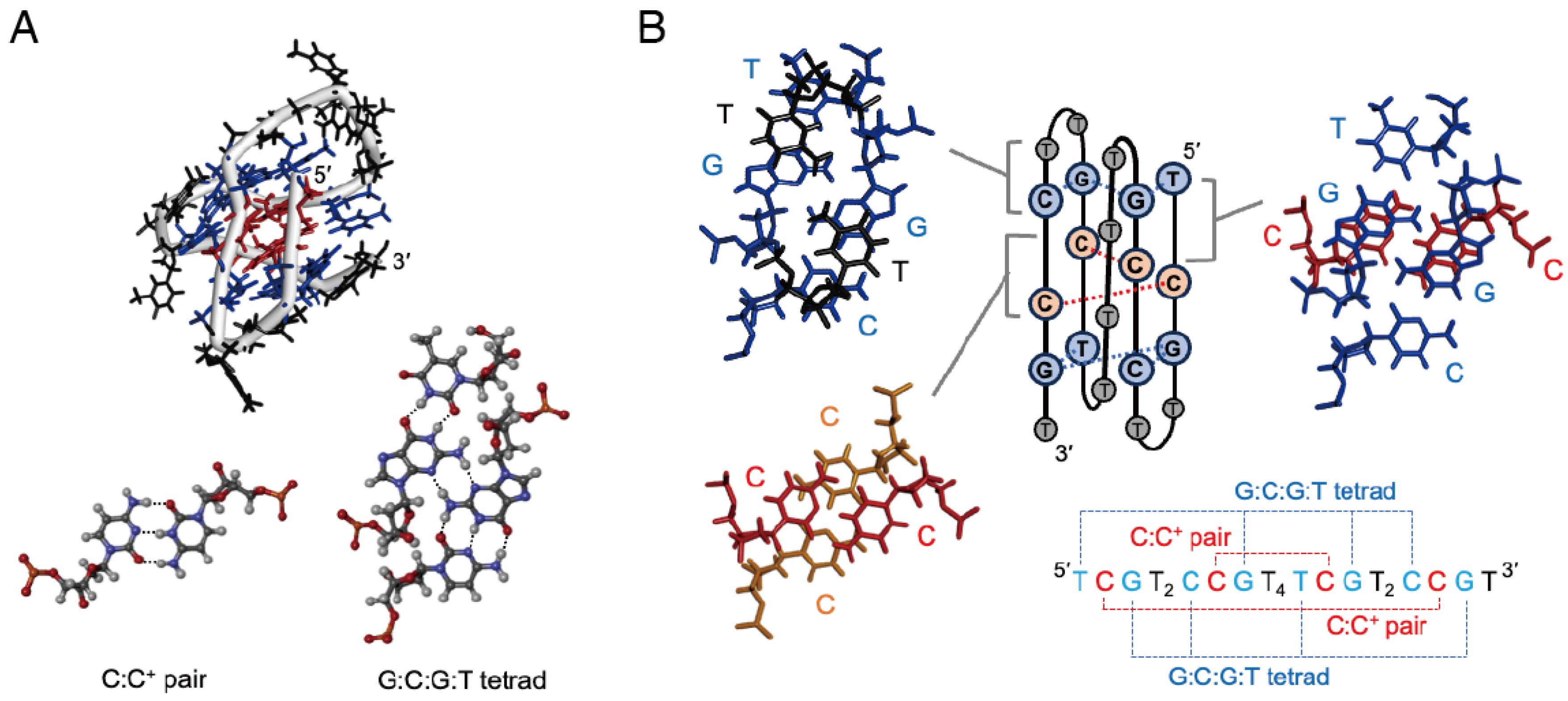
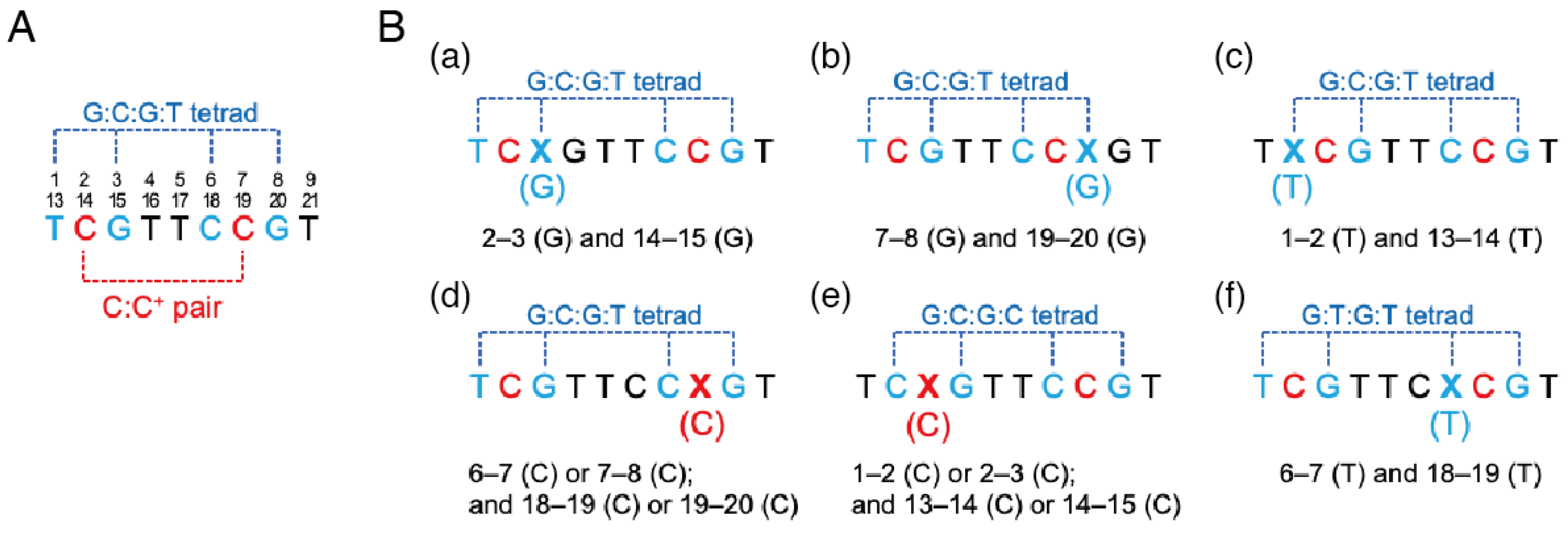
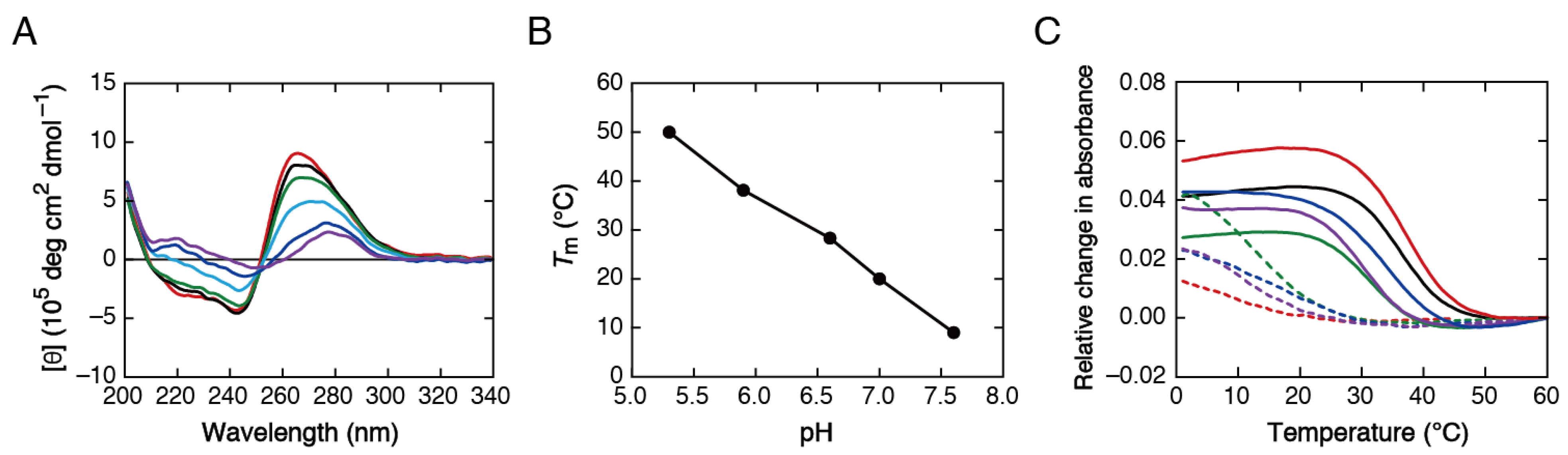
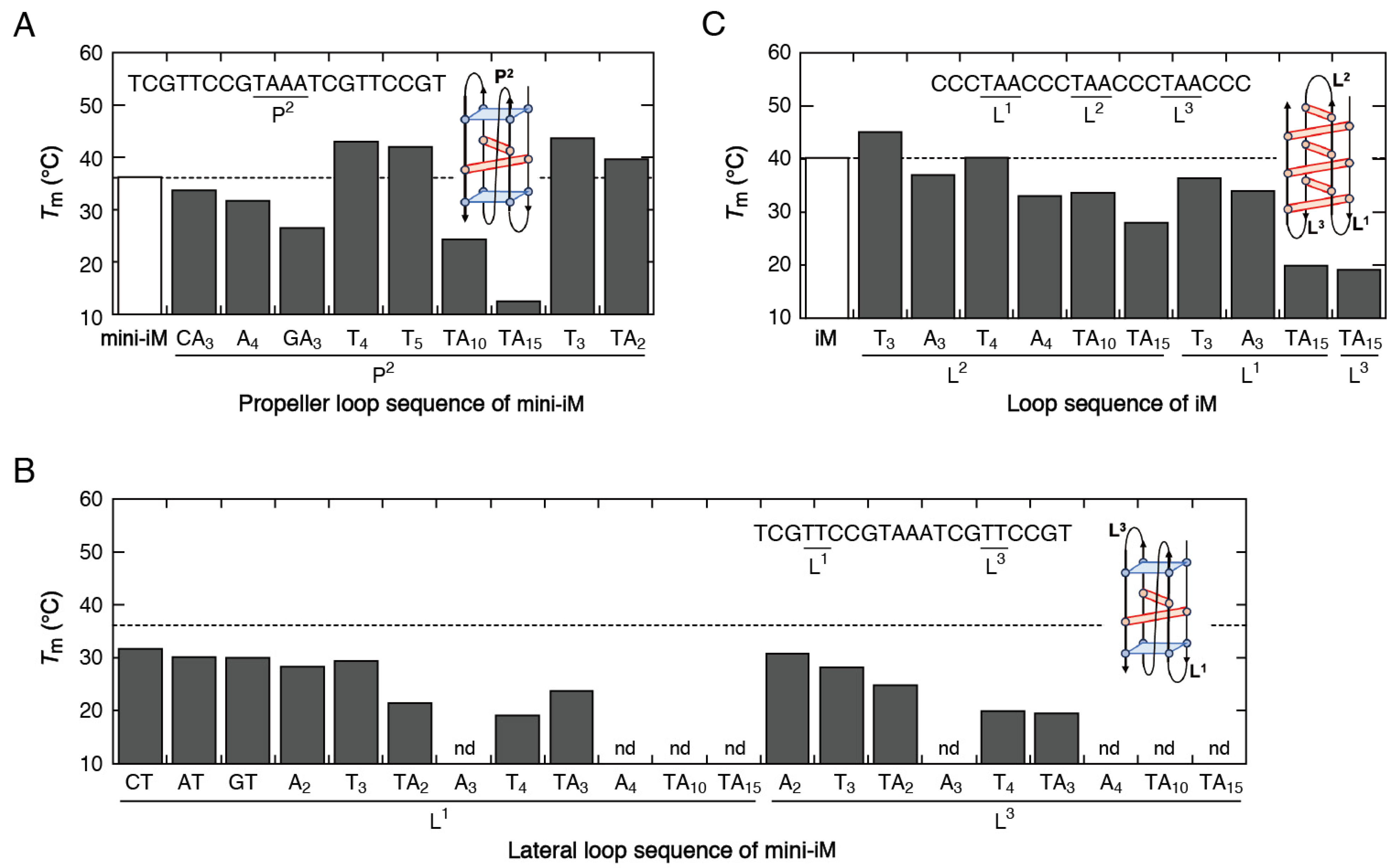

| DNA Sequence (from 5′ to 3′) | Tm (°C) | ∆G° (kcal mol−1) | ∆H° (kcal mol−1) | ∆S° (cal mol−1 K−1) | |
|---|---|---|---|---|---|
| mini-iM | TCGTTCCGTAAATCGTTCCGT | 36.2 ± 0.5 | 0.12 ± 0.08 | −47.7 ± 2.1 | −154 ± 7 |
| (two G:C:G:T tetrads) | |||||
| T1C | CCGTTCCGTAAATCGTTCCGT | 37.0 ± 0.5 | −0.01 ± 0.07 | −43.4 ± 1.3 | −140 ± 4 |
| (G:C:G:C and G:C:G:T tetrads) | |||||
| T13C | TCGTTCCGTAAACCGTTCCGT | 35.0 ± 0.3 | 0.24 ± 0.04 | −37.4 ± 0.8 | −121 ± 3 |
| (G:C:G:C and G:C:G:T tetrads) | |||||
| C6T | TCGTTTCGTAAATCGTTCCGT | 31.7 ± 0.3 | 0.77 ± 0.07 | −44.5 ± 2.1 | −146 ± 7 |
| (G:T:G:T and G:C:G:T tetrads) | |||||
| C18T | TCGTTCCGTAAATCGTTTCGT | 30.7 ± 0.5 | 0.92 ± 0.13 | −45.1 ± 3.8 | −148 ± 12 |
| (G:T:G:T and G:C:G:T tetrads) | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashida, K.; Kitabayashi, A.; Nishiyama, K.; Nakano, S.-i. Comprehensive Analysis of Stability and Variability of DNA Minimal I-Motif Structures. Molecules 2025, 30, 1831. https://doi.org/10.3390/molecules30081831
Ashida K, Kitabayashi A, Nishiyama K, Nakano S-i. Comprehensive Analysis of Stability and Variability of DNA Minimal I-Motif Structures. Molecules. 2025; 30(8):1831. https://doi.org/10.3390/molecules30081831
Chicago/Turabian StyleAshida, Koudai, Ayumi Kitabayashi, Kazuki Nishiyama, and Shu-ichi Nakano. 2025. "Comprehensive Analysis of Stability and Variability of DNA Minimal I-Motif Structures" Molecules 30, no. 8: 1831. https://doi.org/10.3390/molecules30081831
APA StyleAshida, K., Kitabayashi, A., Nishiyama, K., & Nakano, S.-i. (2025). Comprehensive Analysis of Stability and Variability of DNA Minimal I-Motif Structures. Molecules, 30(8), 1831. https://doi.org/10.3390/molecules30081831






