Genome Editing Using Mammalian Haploid Cells
Abstract
:1. Introduction

2. Derivation of Haploid ESCs
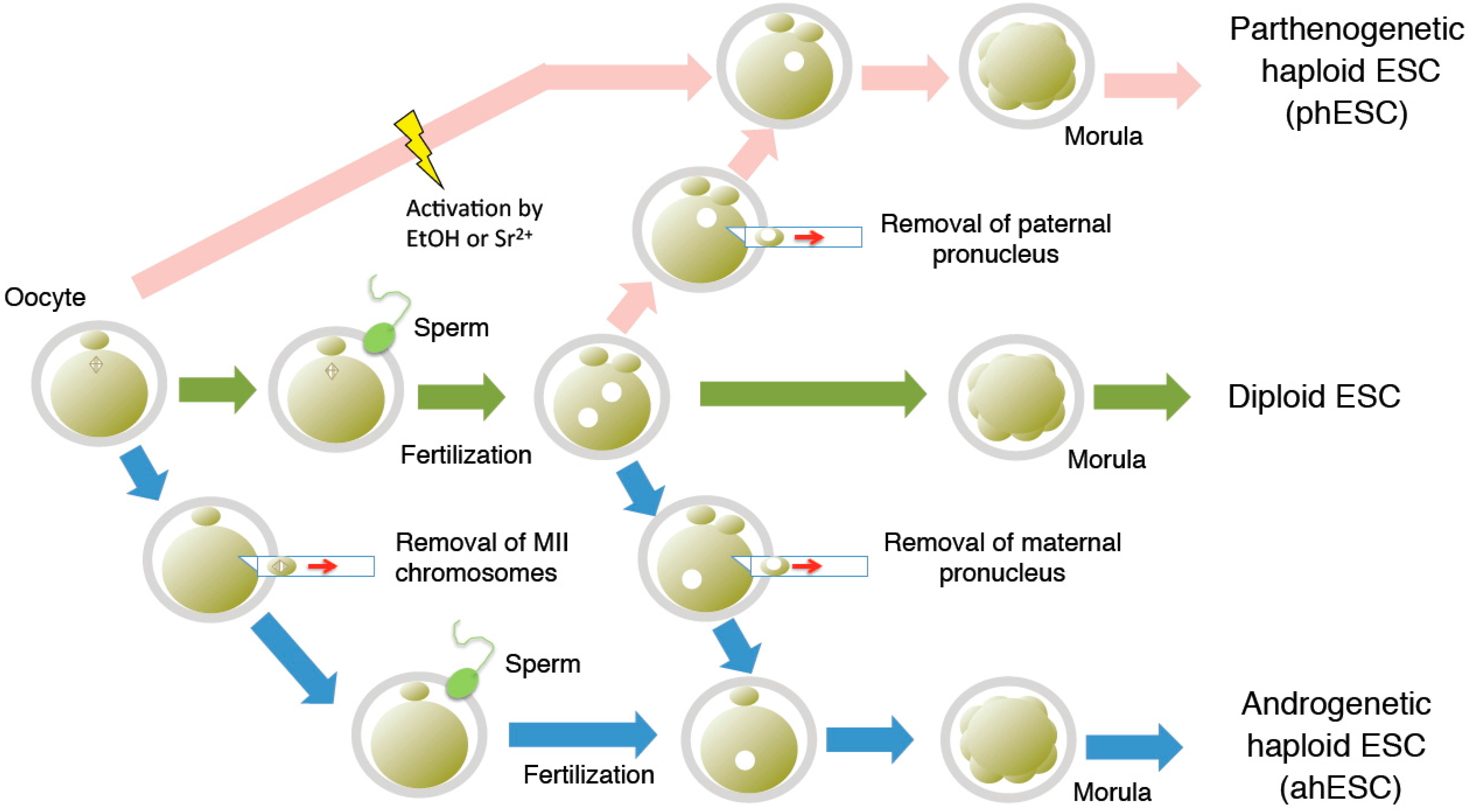
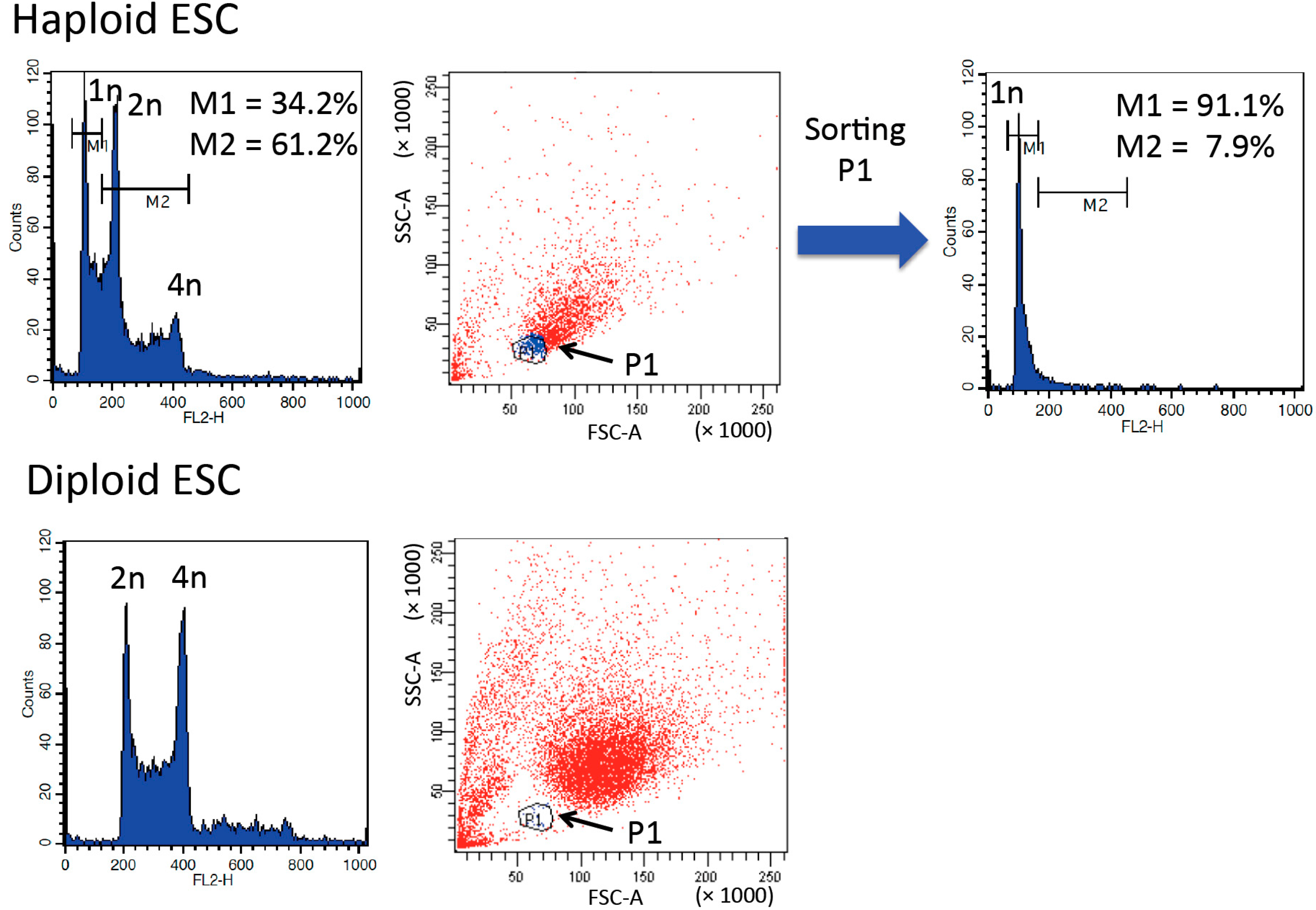
3. Purification and Maintenance of Haploid ESCs

4. Genome Editing in Haploid ESCs

5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mortensen, R.M.; Zubiaur, M.; Neer, E.J.; Seidman, J.G. Embryonic stem cells lacking a functional inhibitory G-protein subunit (α i2) produced by gene targeting of both alleles. Proc. Natl. Acad. Sci. USA 1991, 88, 7036–7040. [Google Scholar] [CrossRef] [PubMed]
- Milstone, D.S.; Bradwin, G.; Mortensen, R.M. Simultaneous Cre catalyzed recombination of two alleles to restore neomycin sensitivity and facilitate homozygous mutations. Nucleic Acids Res. 1999, 27, e10. [Google Scholar] [CrossRef]
- Freed, J.J.; Mezger-Freed, L. Stable haploid cultured cell lines from frog embryos. Proc. Natl. Acad. Sci. USA 1970, 65, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.F. The effect of cell size and DNA content on the cellular regulation of DNA synthesis in haploid and diploid embryos. Exp. Cell Res. 1966, 43, 13–19. [Google Scholar] [CrossRef]
- Modlinski, J.A. Haploidmouse embryos obtained by microsurgical removal of one pronucleus. J. Embryol. Exp. Morphol. 1975, 33, 897–905. [Google Scholar] [PubMed]
- Tarkowski, A.K.; Rossant, J. Haploidmouse blastocysts developed from bisected zygotes. Nature 1976, 259, 663–665. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, M.H. Chromosome analysis of early postimplantation presumptive haploid parthenogenetic mouse embryos. J. Embryol. Exp. Morphol. 1978, 45, 85–91. [Google Scholar] [PubMed]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, M.H.; Robertson, E.J.; Handyside, A.H.; Evans, M.J. Establishment of pluripotential cell-lines from haploid mouse embryos. J. Embryol. Exp. Morph. 1983, 73, 249–261. [Google Scholar] [PubMed]
- Yi, M.; Hong, N.; Hong, Y. Generation of medaka fish haploid embryonic stem cells. Science 2009, 326, 430–433. [Google Scholar] [CrossRef] [PubMed]
- Elling, U.; Taubenschmid, J.; Wirnsberger, G.; O’Malley, R.; Demers, S.P.; Vanhaelen, Q.; Shukalyuk, A.I.; Schmauss, G.; Schramek, D.; Schnuetgen, F.; et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell 2011, 9, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Leeb, M.; Wutz, A. Derivation of haploid embryonic stem cells from mouse embryos. Nature 2011, 479, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shuai, L.; Wan, H.; Dong, M.; Wang, M.; Sang, L.; Feng, C.; Luo, G.Z.; Li, T.; Li, X.; et al. Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature 2012, 490, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Shi, L.; Wang, B.A.; Liang, D.; Zhong, C.; Liu, W.; Nie, Y.; Liu, J.; Zhao, J.; Gao, X.; et al. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell 2012, 149, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; He, Z.; Dong, M.; Gu, T.; Luo, G.Z.; Teng, F.; Xia, B.; Li, W.; Feng, C.; Li, X.; et al. Parthenogenetic haploid embryonic stem cells produce fertile mice. Cell Res. 2013, 23, 1330–1333. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, X.; Li, T.; Jiang, M.G.; Wan, H.; Luo, G.Z.; Feng, C.; Cui, X.; Teng, F.; Yuan, Y.; et al. Genetic modification and screening in rat using haploid embryonic stem cells. Cell Stem Cell 2013, 14, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, Z.; Ma, Y.; Zhong, C.; Yin, Q.; Zhou, C.; Shi, L.; Cai, Y.; Zhao, H.; Wang, H.; et al. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Res. 2013, 23, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Kotecki, M.; Reddy, P.S.; Cochran, B.H. Isolation and characterization of a near-haploid human cell line. Exp. Cell Res. 1999, 252, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Carette, J.E.; Guimaraes, C.P.; Varadarajan, M.; Park, A.S.; Wuethrich, I.; Godarova, A.; Kotecki, M.; Cochran, B.H.; Spooner, E.; Ploegh, H.L.; et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science 2009, 326, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Carette, J.E.; Raaben, M.; Wong, A.C.; Herbert, A.S.; Obernosterer, G.; Mulherkar, N.; Kuehne, A.I.; Kranzusch, P.J.; Griffin, A.M.; Ruthel, G.; et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 2011, 477, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Rosmarin, D.M.; Carette, J.E.; Olive, A.J.; Starnbach, M.N.; Brummelkamp, T.R.; Ploegh, H.L. Attachment of Chlamydia trachomatis L2 to host cells requires sulfation. Proc. Natl. Acad. Sci. USA 2012, 109, 10059–10064. [Google Scholar] [CrossRef] [PubMed]
- Reiling, J.H.; Clish, C.B.; Carette, J.E.; Varadarajan, M.; Brummelkamp, T.R.; Sabatini, D.M. A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc. Natl. Acad. Sci. USA 2011, 108, 11756–11765. [Google Scholar] [CrossRef] [PubMed]
- Reiling, J.H.; Olive, A.J.; Sanyal, S.; Carette, J.E.; Brummelkamp, T.R.; Ploegh, H.L.; Starnbach, M.N.; Sabatini, D.M. ACREB3-ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat. Cell Biol. 2013, 15, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Birsoy, K.; Wang, T.; Possemato, R.; Yilmaz, O.H.; Koch, C.E.; Chen, W.W.; Hutchins, A.W.; Gultekin, Y.; Peterson, T.R.; Carette, J.E.; et al. MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors. Nat. Genet. 2013, 45, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Leeb, M.; Perry, A.C.; Wutz, A. Establishment and use of mouse haploid ES cells. Curr. Protoc. Mouse Biol. 2015, 5, 155–185. [Google Scholar] [PubMed]
- Leeb, M.; Walker, R.; Mansfield, B.; Nichols, J.; Smith, A.; Wutz, A. Germline potential of parthenogenetic haploid mouse embryonic stem cells. Development 2012, 139, 3301–3305. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Yang, J.; Nichols, J.; Hall, J.S.; Eyres, I.; Mansfield, W.; Smith, A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 2009, 136, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Lee, J.; Kohda, T.; Matsuzawa, A.; Kawasumi, M.; Kanai-Azuma, M.; Kaneko-Ishino, T.; Ishino, F. Induction of the G2/M transition stabilizes haploid embryonic stem cells. Development 2014, 141, 3842–3847. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. eLife 2013, 2, e00471. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Koike-Yusa, H.; Li, Y.; Tan, E.P.; Velasco-Herrera, M.D.C.; Yusa, K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2014, 32, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014, 343, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Horii, T.; Tamura, D.; Morita, S.; Kimura, M.; Hatada, I. Generation of an ICF syndrome model by efficient genome editing of human induced pluripotent stem cells using the CRISPR system. Int. J. Mol. Sci. 2013, 14, 19774–19781. [Google Scholar] [CrossRef] [PubMed]
- An, M.C.; O’Brien, R.N.; Zhang, N.; Patra, B.N.; de La Cruz, M.; Ray, A.; Ellerby, L.M. Polyglutamine disease modeling: Epitope based screen for homologous recombination using CRISPR/Cas9 system. PLoS Curr. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Ye, L.; Chang, J.C.; Beyer, A.I.; Wang, J.; Muench, M.O.; Kan, Y.W. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyback. Genome Res. 2014, 24, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Fujimoto, N.; Sasakawa, N.; Shirai, S.; Ohkame, T.; Sakuma, T.; Tanaka, M.; Amano, N.; Watanabe, A.; Sakurai, H.; et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep. 2015, 4, 143–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ousterout, D.G.; Kabadi, A.M.; Thakore, P.I.; Majoros, W.H.; Reddy, T.E.; Gersbach, C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015, 6, 6244. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Fan, Y.; He, W.; Zhu, D.; Niu, X.; Wang, D.; Ou, Z.; Luo, M.; Sun, X. Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev. 2015, 24, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, Y.; Yan, W.; Smith, C.; Ye, Z.; Wang, J.; Gao, Y.; Mendelsohn, L.; Cheng, L. Production of gene-corrected adult β globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells 2015, 33, 1470–1479. [Google Scholar] [CrossRef] [PubMed]
- Horii, T.; Morita, S.; Kimura, M.; Kobayashi, R.; Tamura, D.; Takahashi, R.U.; Kimura, H.; Suetake, I.; Ohata, H.; Okamoto, K.; et al. Genome engineering of mammalian haploid embryonic stem cells using the Cas9/RNA system. PeerJ 2013, 1, e230. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Canver, M.C.; Bauer, D.E.; Dass, A.; Yien, Y.Y.; Chung, J.; Masuda, T.; Maeda, T.; Paw, B.H.; Orkin, S.H. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 2014, 289, 21312–21324. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Oda, M.; Nakatani, T.; Sekita, Y.; Monfort, A.; Wutz, A.; Mochizuki, H.; Nakano, T. CRISPR/Cas9-mediated reporter knock-in in mouse haploid embryonic stem cells. Sci. Rep. 2015, 5, 10710. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Yin, Q.; Xie, Z.; Bai, M.; Dong, R.; Tang, W.; Xing, Y.H.; Zhang, H.; Yang, S.; Chen, L.L.; et al. CRISPR-Cas9-Mediated Genetic Screening in Mice with Haploid Embryonic Stem Cells Carrying a Guide RNA Library. Cell Stem Cell 2015, 17, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, V.; Lin, S.; Guilinger, J.P.; Ma, E.; Doudna, J.A.; Liu, D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013, 31, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. One-Step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; Zhang, F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horii, T.; Hatada, I. Genome Editing Using Mammalian Haploid Cells. Int. J. Mol. Sci. 2015, 16, 23604-23614. https://doi.org/10.3390/ijms161023604
Horii T, Hatada I. Genome Editing Using Mammalian Haploid Cells. International Journal of Molecular Sciences. 2015; 16(10):23604-23614. https://doi.org/10.3390/ijms161023604
Chicago/Turabian StyleHorii, Takuro, and Izuho Hatada. 2015. "Genome Editing Using Mammalian Haploid Cells" International Journal of Molecular Sciences 16, no. 10: 23604-23614. https://doi.org/10.3390/ijms161023604
APA StyleHorii, T., & Hatada, I. (2015). Genome Editing Using Mammalian Haploid Cells. International Journal of Molecular Sciences, 16(10), 23604-23614. https://doi.org/10.3390/ijms161023604




