Substrate-Related Factors Affecting Cellulosome-Induced Hydrolysis for Lignocellulose Valorization
Abstract
:1. Introduction
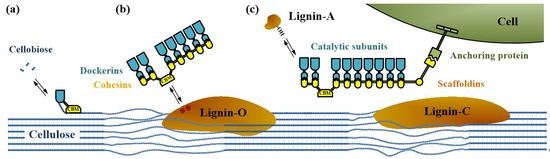
2. Cellulosome Composition and Assembly
2.1. Scaffoldin
2.2. Cohesin–Dockerin Interaction
2.3. CBMs (CBDs)
2.4. Cellulosomal Enzymes
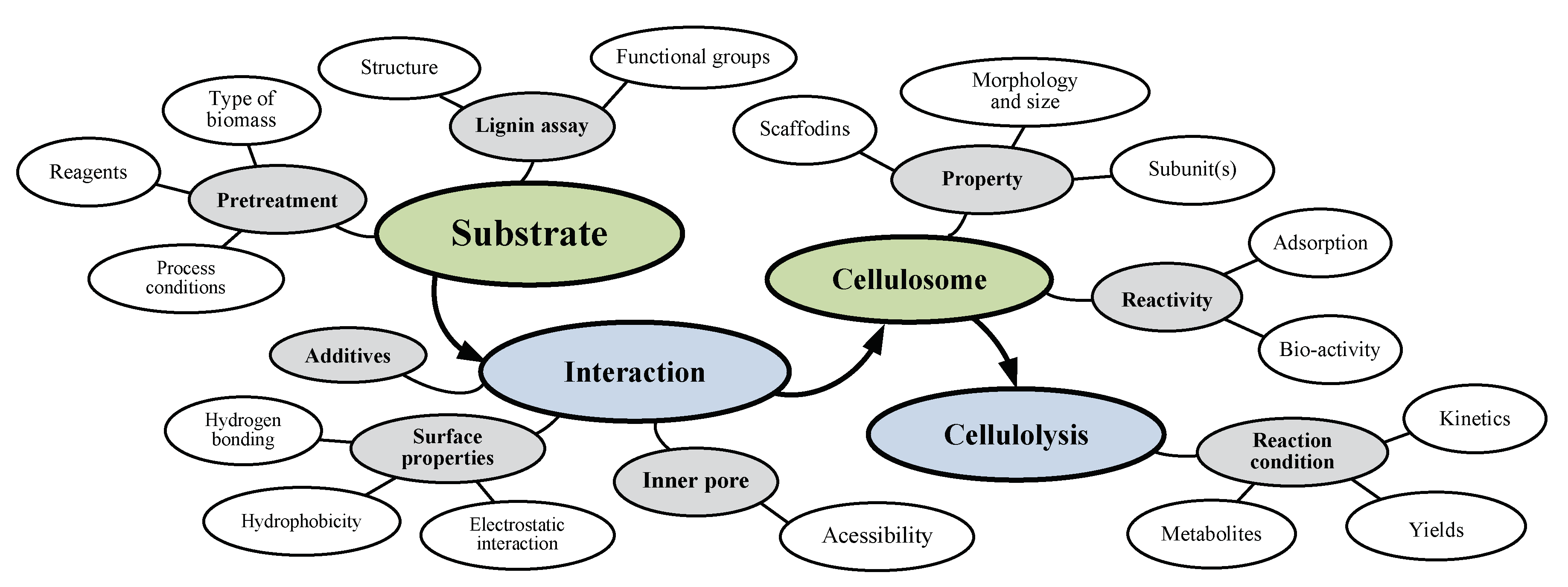
3. Effects of Several Substrate-Related Factors on Cellulosome-Induced Hydrolysis
3.1. Effects of Different Carbon Sources
3.2. Effects of Different Chemical Compounds
3.3. Effects of Pretreatment
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yamakawa, C.K.; Qin, F.; Mussatto, S.I. Advances and opportunities in biomass conversion technologies and biorefineries for the development of a bio-based economy. Biomass Bioenergy 2018, 119, 54–60. [Google Scholar] [CrossRef]
- Bauer, F.; Coenen, L.; Hansen, T.; McCormick, K.; Voytenko Palgan, Y. Technological innovation systems for biorefineries: A review of the literature. Biofuels Bioprod. Biorefin. 2017, 11, 534–548. [Google Scholar] [CrossRef]
- Periyasamy, K.; Santhalembi, L.; Mortha, G.; Aurousseau, M.; Boyer, A.; Subramanian, S. Bioconversion of lignocellulosic biomass to fermentable sugars by immobilized magnetic cellulolytic enzyme cocktails. Langmuir 2018, 34, 6546–6555. [Google Scholar] [CrossRef] [PubMed]
- Amezcua-Allieri, M.A.; Sánchez-Duran, T.; Aburto, J. Study of chemical and enzymatic hydrolysis of cellulosic material to obtain fermentable sugars. J. Chem. 2017, 2017, 5680105. [Google Scholar] [CrossRef]
- Chi, X.; Li, J.; Wang, X.; Zhang, Y.; Leu, S.; Wang, Y. Bioaugmentation with Clostridium tyrobutyricum to improve butyric acid production through direct rice straw bioconversion. Bioresour. Technol. 2018, 263, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.G.; McBride, J.E.; Shaw, A.J.; Lynd, L.R. Recent progress in consolidated bioprocessing. Curr. Opin. Biotechnol. 2012, 23, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tashiro, Y.; Sonomoto, K. Fermentative production of lactic acid from renewable materials: Recent achievements, prospects, and limits. J. Biosci. Bioeng. 2015, 119, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, S.P.; Henske, J.K.; O’Malley, M.A. Driving biomass breakdown through engineered cellulosomes. Bioengineered 2015, 6, 204–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, R.; Behera, S.; Kumar Sharma, N.; Kumar, S. Bioprospecting thermostable cellulosomes for efficient biofuel production from lignocellulosic biomass. Bioresour. Bioprocess. 2015, 2, 38. [Google Scholar] [CrossRef] [Green Version]
- Doi, R.H.; Kosugi, A.; Murashima, K.; Tamaru, Y.; Han, S.O. Cellulosomes from mesophilic bacteria. J. Bacteriol. 2003, 20, 5907–5914. [Google Scholar] [CrossRef]
- Lamed, R.; Setter, E.; Bayer, E.A. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 1983, 156, 828–836. [Google Scholar] [PubMed]
- Bayer, E.A.; Lamed, R.; White, B.A.; Flint, H.J. From cellulosomes to cellulosomics. Chem. Rec. 2008, 8, 364–377. [Google Scholar] [CrossRef] [PubMed]
- Desvaux, M. Clostridium cellulolyticum model organism of mesophilic cellulolytic clostridia. FEMS Microbiol. Rev. 2005, 29, 741–764. [Google Scholar] [CrossRef] [PubMed]
- Quiroz-Castañeda, R.E.; Folch-Mallol, J.L. Hydrolysis of biomass mediated by cellulases for the production of sugars. In Sustainable Degradation of Lignocellulosic Biomass Techniques, Applications and Commercialization; Chandel, A., Ed.; InTech: London, UK, 2013; pp. 119–155. [Google Scholar]
- Artzi, L.; Bayer, E.A.; Moraïs, S. Cellulosomes: Bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 2017, 15, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, G.A.L.; Mori, Y.; Kamiya, N. Biomolecular assembly strategies to develop potential artificial cellulosomes. Sustain. Chem. Process. 2014, 2, 19. [Google Scholar] [CrossRef] [Green Version]
- Moraïs, S.; Shterzer, N.; Lamed, R.; Bayer, E.A.; Mizrahi, I. A combined cell-consortium approach for lignocellulose degradation by specialized Lactobacillus plantarum cells. Biotechnol. Biofuel. 2014, 7, 112. [Google Scholar] [CrossRef]
- Béguin, P.; Lemaire, M. The Cellulosome: An exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 1996, 31, 201–236. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, R.; Lamberti, C.; Pessione, E. Engineering new metabolic capabilities in bacteria: Lessons from recombinant cellulolytic strategies. Trends Biotechnol. 2012, 30, 111–119. [Google Scholar] [CrossRef]
- Pagès, S.; Bélaïch, A.; Fierobe, H.P.; Tardif, C.; Gaudin, C.; Bélaïch, J.P. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: Comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 1999, 181, 1801–1810. [Google Scholar]
- Erickson, S.J.; Duvall, S.W.; Fuller, J.; Schrader, R.; MacLean, P.; Lowe, J.R. Differential associations between maternal scaffolding and toddler emotion regulation in toddlers born preterm and full term. Early Hum. Dev. 2013, 89, 699–704. [Google Scholar] [CrossRef] [Green Version]
- Bayer, E.A.; Bélaïch, J.P.; Shoham, Y.; Lamed, R. The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 2004, 58, 521–554. [Google Scholar] [CrossRef] [PubMed]
- Shoham, Y.; Lamed, R.; Bayer, E.A. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 1999, 275, 275–281. [Google Scholar] [CrossRef]
- Bomble, Y.J.; Beckham, G.T.; Matthews, J.F.; Nimlos, M.R.; Himmel, M.E.; Crowley, M.F. Modeling the self-assembly of the cellulosome enzyme complex. J. Biol. Chem. 2011, 286, 5614–5623. [Google Scholar] [CrossRef] [PubMed]
- Koukiekolo, R.; Cho, H.Y.; Kosugi, A.; Inui, M.; Yukawa, H.; Doi, R.H. Degradation of corn fiber by Clostridium cellulovorans cellulases and hemicellulases and contribution of scaffolding protein CbpA. Appl. Environ. Microbiol. 2005, 71, 3504–3511. [Google Scholar] [CrossRef] [PubMed]
- Fierobe, H.P.; Mingardon, F.; Mechaly, A.; Belaich, A.; Rincon, M.T.; Pages, S.; Lamed, R.; Tardif, C.; Belaich, J.P.; Bayer, E.A. Action of designer cellulosomes on homogeneous versus complex substrates: Controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 2005, 280, 16325–16334. [Google Scholar] [CrossRef] [PubMed]
- Valbuena, A.; Oroz, J.; Hervas, R.; Manuel Vera, A.; Rodriguez, D.; Menendez, M.; Sulkowska, J.I.; Cieplak, M.; Carrion-Vazquez, M. On the remarkable mechanostability of scaffoldins and the mechanical clamp motif. Proc. Natl. Acad. Sci. USA 2009, 106, 13791–13796. [Google Scholar] [CrossRef] [Green Version]
- Gunnoo, M.; Cazade, P.A.; Galera-Prat, A.; Nash, M.A.; Czjzek, M.; Cieplak, M.; Alvarez, B.; Aguilar, M.; Karpol, A.; Gaub, H.; et al. Nanoscale engineering of designer cellulosomes. Adv. Mater. 2016, 28, 5619–5647. [Google Scholar] [CrossRef]
- Bras, J.L.A.; Alves, V.D.; Carvalho, A.L.; Najmudin, S.; Prates, J.A.M.; Ferreira, L.M.A.; Bolam, D.N.; Romao, M.J.; Gilbert, H.J.; Fontes, C.M.G.A. Novel Clostridium thermocellum Type I cohesin–dockerin complexes reveal a single binding mode. J. Biol. Chem. 2012, 287, 44394–44405. [Google Scholar] [CrossRef]
- Peer, A.; Smith, S.P.; Bayer, E.A.; Lamed, R.; Borovok, I. Noncellulosomal cohesin- and dockerin-like modules in the three domains of life. FEMS Microbiol. Lett. 2009, 291, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Voronov-Goldman, M.; Yaniv, O.; Gul, O.; Yoffe, H.; Salama-Alber, O.; Slutzki, M.; Levy-Assaraf, M.; Jindou, S.; Shimon, L.J.W.; Borovok, I.; et al. Standalone cohesin as a molecular shuttle in cellulosome assembly. FEBS Lett. 2015, 589, 1569–1576. [Google Scholar] [CrossRef]
- Resch, M.G.; Donohoe, B.S.; Baker, J.O.; Decker, S.R.; Bayer, E.A.; Beckham, G.T.; Himmel, M.E. Fungal cellulases and complexed cellulosomal enzymes exhibit synergistic mechanisms in cellulose deconstruction. Energ. Environ. Sci. 2013, 6, 1858–1867. [Google Scholar] [CrossRef]
- Ichikawa, S.; Karita, S.; Kondo, M.; Goto, M. Cellulosomal carbohydrate-binding module from Clostridium josui binds to crystalline and non-crystalline cellulose, and soluble polysaccharides. FEBS Lett. 2011, 588, 3886–3890. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, M.A.; Takagi, M.; Hashida, S.; Shoseyov, O.; Doi, R.H.; Segel, I.H. Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A (CbpA). J. Bacteriol. 1993, 175, 5762–5768. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, Y.; Karita, S.; Ibrahim, A.; Chan, H.; Doi, R.H. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 2000, 182, 5906–5910. [Google Scholar] [CrossRef] [PubMed]
- Caspi, J.; Irwin, D.; Lamed, R.; Li, Y.; Fierobe, H.P.; Wilson, D.B.; Bayer, E.A. Conversion of Thermobifida fusca free exoglucanases into cellulosomal components: Comparative impact on cellulose-degrading activity. J. Biotechnol. 2008, 135, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Gerngross, U.T.; Romaniec, M.P.; Kobayashi, T.; Huskisson, N.S.; Demain, A.L. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 1993, 10, 1155. [Google Scholar] [CrossRef]
- Yaniv, O.; Jindou, S.; Frolow, F.; Lamed, R.; Bayer, E.A. A simple method for determining specificity of carbohydrate-binding modules for purified and crude insoluble polysaccharide substrates. Methods Mol. Biol. 2012, 908, 101–107. [Google Scholar] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Shoseyov, O.; Shani, Z.; Levy, I. Carbohydrate binding modules: Biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 2006, 70, 283–295. [Google Scholar] [CrossRef]
- Hazlewood, G.P.; Romaniec, M.P.M.; Davidson, K.; Grépinet, O.; Béguin, P.; Millet, J.; Raynaud, O.; Aubert, J.P. A catalogue of Clostridium thermocellum endoglucanase, β-glucosidase and xylanase genes cloned in Escherichia coli. FEMS Microbiol. Lett. 1988, 51, 231–236. [Google Scholar] [CrossRef]
- Vazana, Y.; Moraïs, S.; Barak, Y.; Lamed, R.; Bayer, E.A. Interplay between Clostridium thermocellum family 48 and family 9 cellulases in cellulosomal versus noncellulosomal states. Appl. Environ. Microbiol. 2010, 76, 3236–3243. [Google Scholar] [CrossRef] [PubMed]
- Bayer, E.A.; Chanzy, H.; Lamed, R.; Shoham, Y. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 1998, 8, 548–557. [Google Scholar] [CrossRef]
- Bayer, E.A.; Morag, E.; Lamed, R. The cellulosome—A treasure-trove for biotechnology. Trends Biotechnol. 1994, 12, 378–386. [Google Scholar] [CrossRef]
- Ding, S.Y.; Xu, Q.; Crowley, M.; Zeng, Y.; Nimlos, M.; Lamed, R.; Bayer, E.A.; Himmel, M.E. A biophysical perspective on the cellulosome: New opportunities for biomass conversion. Curr. Opin. Biotechnol. 2008, 19, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Morag, E.; Halevy, I.; Bayer, E.A.; Lamed, R. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J. Bacteriol. 1991, 173, 4155–4162. [Google Scholar] [CrossRef] [PubMed]
- Ravachol, J.; Borne, R.; Meynial-Salles, I.; Soucaille, P.; Pages, S.; Tardif, C. Combining free and aggregated cellulolytic systems in the cellulosome-producing bacterium Ruminiclostridium cellulolyticum. Biotechnol. Biofuels 2015, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Morag, E.; Bayer, E.A.; Lamed, R. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J. Bacteriol. 1993, 172, 6098–6105. [Google Scholar] [CrossRef] [PubMed]
- Mohand-Oussaid, O.; Payot, S.; Guedon, E.; Gelhaye, E.; Youyou, A.; Petitdemange, H. The extracellular xylan degradative system in Clostridium cellulolyticum cultivated on xylan: Evidence for cell-free cellulosome production. J. Bacteriol. 1999, 181, 4035–4040. [Google Scholar] [PubMed]
- Maki, M.; Leung, K.T.; Qin, W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 2009, 5, 500–516. [Google Scholar]
- Akinosho, H.; Yee, K.; Close, D.; Ragauskas, A. The emergence of Clostridium thermocellum as a high utility candidate for consolidated bioprocessing applications. Front. Chem. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Yuan, L. Production of multifunctional chimaeric enzymes in plants: A promising approach for degrading plant cell wall from within. Plant Biotechnol. J. 2010, 8, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Cragg, S.M.; Beckham, G.T.; Bruce, N.C.; Bugg, T.D.; Distel, D.L.; Dupree, P.; Etxabe, A.G.; Goodell, B.S.; Jellison, J.; McGeehan, J.E.; et al. Lignocellulose degradation mechanism sacross the tree of life. Curr. Opin. Chem. Biol. 2015, 29, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Fernandes, V.O.; Dias, F.M.V.; Prates, JA.M.; Ferreira, L.M.A.; Fontes, C.M.G.A.; Goyal, A.; Centeno, M.S.J. Role of pectinolytic enzymes identified in Clostridium thermocellum cellulosome. PLoS ONE 2015, 10, e0116787. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, Y.; Doi, R.H. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. Proc. Natl. Acad. Sci. USA 2001, 98, 4125–4129. [Google Scholar] [CrossRef] [PubMed]
- Perret, S.; Belaich, A.; Fierobe, H.P.; Belaich, J.P.; Tardif, C. Towards designer cellulosomes in Clostridia: Mannanase enrichment of the cellulosomes produced by Clostridium cellulolyticum. J. Bacteriol. 2004, 186, 6544–6552. [Google Scholar] [CrossRef] [PubMed]
- Sabathé, F.; Bélaïch, A.; Soucaille, P. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 2002, 217, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, J.; Hemjinda, E.; Arai, T.; Karita, S.; Kimura, T.; Sakka, K.; Ohmiya, K. Sequence of the Clostridium thermocellum mannanase gene man26B and characterization of the translated product. Biosci. Biotechnol. Biochem. 2001, 65, 548–554. [Google Scholar] [CrossRef]
- Zverlov, V.V.; Fuchs, K.P.; Schwarz, W.H. Chi18A, the endochitinase in the cellulosome of the thermophilic, cellulolytic bacterium Clostridium thermocellum. Appl. Environ. Microbiol. 2002, 68, 3176–3179. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, Y.; Miyake, H.; Kuroda, K.; Ueda, M.; Doi, R.H. Comparative genomics of the mesophilic cellulosome-producing Clostridium cellulovorans and its application to biofuel production via consolidated bioprocessing. Envion. Technol. 2010, 31, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Vodovnik, M.; Logar, R.M. Cellulosomes-promising supramolecular machines of anaerobic cellulolytic microorganisms. Acta. Chim. Slov. 2010, 57, 767–774. [Google Scholar] [PubMed]
- Gold, N.D.; Martin, V.J.J. Global view of the Clostridium thermocellum cellulosome revealed by quantitative proteomic analysis. J. Bacteriol. 2007, 189, 6787–6795. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.; Jeon, S.D.; Shim, H.J.; Doi, R.H.; Han, S.O. Cellulosomic profiling produced by Clostridium cellulovorans during growth on different carbon sources explored by the cohesin marker. J. Biotechnol. 2010, 145, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.L.; Goyal, G.; Chen, W. Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 2010, 76, 7514–7520. [Google Scholar] [CrossRef] [PubMed]
- Artzi, L.; Morag, E.; Barak, Y.; Lamed, R.; Bayer, E.A. Clostridium clariflavum: Key cellulosome players are revealed by proteomic analysis. mBio 2015, 6, e00411-15. [Google Scholar] [CrossRef] [PubMed]
- Leu, S.Y.; Zhu, J.Y. Substrate-related factors affecting enzymatic saccharification of lignocelluloses: Our recent understanding. Bioenergy Res. 2013, 6, 405–415. [Google Scholar] [CrossRef]
- Chan, K.L.; Dong, C.; Wong, M.S.; Kim, L.H.; Leu, S.Y. Plant chemistry associated dynamic modelling to enhance urban vegetation carbon sequestration potential via bioenergy harvesting. J. Clean. Prod. 2018, 197, 1084–1094. [Google Scholar] [CrossRef]
- Sosnowski, P.; Wieczorek, A.; Ledakowicz, S. Anaerobic co-digestion of sewage sludge and organic fraction of municipal solid wastes. Adv. Environ. Res. 2003, 7, 609–616. [Google Scholar] [CrossRef]
- Carrère, H.; Dumas, C.; Battimelli, A.; Batstone, D.J.; Delgenès, J.P.; Steyer, J.P.; Ferrer, I. Pretreatment methods to improve sludge anaerobic degradability: A review. J. Hazard. Mater. 2010, 183, 1–15. [Google Scholar]
- Zhen, G.; Lu, X.; Kato, H.; Zhao, Y.; Li, Y.Y. Overview of pretreatment strategies for enhancing sewage sludge disintegration and subsequent anaerobic digestion: Current advances, full-scale application and future perspectives. Renew. Sustain. Energy Rev. 2017, 69, 559–577. [Google Scholar] [CrossRef]
- Liu, H.; Sun, J.; Leu, S.Y.; Chen, S. Toward a fundamental understanding of cellulase-lignin interactions in the whole slurry enzymatic saccharification process. Biofuels Bioprod. Biorefin. 2016, 10, 648–663. [Google Scholar] [CrossRef]
- Pan, X.; Gilkes, N.; Kadla, J.; Pye, K.; Saka, S.; Gregg, D.; Ehara, K.; Xie, D.; Lam, D.; Saddler, J. Bioconversion of hybrid poplar to ethanol and co-products using an organosolv fractionation process: Optimization of process yields. Biotechnol. Bioeng. 2006, 94, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.; Goodenough, P.W.; Owen, E.; Bhat, M.K. Cellobiose: A true inducer of cellulosome in different strains of Clostridium thermocellum. FEMS Microbiol. Lett. 1993, 111, 73–78. [Google Scholar] [CrossRef]
- Bae, J.; Morisaka, H.; Kuroda, K.; Ueda, M. Cellulosome complexes: Natural biocatalysts as arming microcompartments of enzymes. J. Mol. Microbiol. Biotechnol. 2013, 23, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Han, S.O.; Yukawa, H.; Inui, M.; Doi, R.H. Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulose genes. J. Bacteriol. 2003, 185, 2520–2527. [Google Scholar] [CrossRef] [PubMed]
- Raman, B.; Pan, C.; Hurst, G.B.; Rodriguez, M.J.; McKeown, C.K.; Lankford, P.K.; Samatova, N.F.; Mielenz, J.R. Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: A quantitative proteomic analysis. PLoS ONE 2009, 4, e5271. [Google Scholar] [CrossRef] [PubMed]
- Morisaka, H.; Matsui, K.; Tatsukami, Y.; Kuroda, K.; Miyake, H.; Tamaru, Y.; Ueda, M. Profile of native cellulosomal proteins of Clostridium cellulovorans adapted to various carbon sources. AMB Express 2012, 2, 37. [Google Scholar] [CrossRef] [PubMed]
- Han, S.O.; Yukawa, H.; Inui, M.; Doi, R.H. Effect of carbon source on the cellulosomal subpopulations of Clostridium cellulovorans. Microbiology 2005, 151, 1491–1497. [Google Scholar] [CrossRef]
- Yoav, S.; Barak, Y.; Shamshoum, M.; Borovok, I.; Lamed, R.; Dassa, B.; Hadar, Y.; Morag, E.; Bayer, E.A. How does cellulosome composition influence deconstruction of lignocellulosic substrates in Clostridium (Ruminiclostridium) thermocellum DSM 1313? Biotechnol. Biofuels 2017, 10, 222. [Google Scholar] [CrossRef]
- Han, S.O.; Cho, H.Y.; Yukawa, H.; Inui, M.; Doi, R.H. Regulation of expression of cellulosomes and noncellulosomal (hemi)cellulolytic enzymes in Clostridium cellulovorans during growth on different carbon sources. J. Bacteriol. 2004, 188, 4218–4227. [Google Scholar] [CrossRef]
- Fierobe, H.B.; Bayer, E.A.; Tardif, C.; Czjzek, M.; Mechaly, A.; Belaich, A.; Lamed, R.; Shoham, Y.; Belaich, J.P. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J. Biol. Chem. 2002, 227, 49621–49630. [Google Scholar] [CrossRef]
- Artzi, L.; Dadosh, T.; Milrot, E.; Moraïs, S.; Levin-Zaidman, S.; Morag, E.; Bayer, E.A. Colocalization and disposition of cellulosomes in Clostridium clariflavum as revealed by correlative superresolution imaging. mBio 2018, 9, e00012-18. [Google Scholar] [CrossRef] [PubMed]
- Han, S.O.; Yukawa, H.; Inui, M.; Doi, R.H. Regulation of expression of cellulosomal cellulase and hemicellulase genes in Clostridium cellulovorans. J. Bacteriol. 2003, 185, 6067–6075. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, A.; Murashima, K.; Doi, R.H. Characterization of xylanolytic enzymes in Clostridium cellulovorans: Expression of xylanase activity dependent on growth substrates. J. Bacteriol. 2001, 183, 7037–7043. [Google Scholar] [CrossRef] [PubMed]
- Murashima, K.; Kosugi, A.; Doi, R.H. Determination of subunit composition of Clostridium cellulovorans cellulosomes that degrade plant cell walls. Appl. Environ. Microbiol. 2002, 68, 1610–1615. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Matano, Y.; Doi, R.H. Cohesin–dockerin interactions of cellulosomal subunits of Clostridium cellulovorans. J. Bacteriol. 2001, 183, 5431–5435. [Google Scholar] [CrossRef] [PubMed]
- Nataf, Y.; Bahari, L.; Kahei-Raifer, H.; Borovok, I.; Lamed, R.; Bayer, E.A.; Sonenshein, A.L.; Shoham, Y. Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18646–18651. [Google Scholar] [CrossRef]
- Qing, Q.; Yang, B.; Wyman, C.E. Xylooligomers are strong inhibitors of cellulose hydrolysis by enzymes. Bioresour. Technol. 2010, 101, 9624–9630. [Google Scholar] [CrossRef]
- Teugjas, H.; Valjamae, P. Product inhibition of cellulases studied with 14C-labeled cellulose substrates. Biotechnol. Biofuels 2013, 6, 104. [Google Scholar] [CrossRef]
- Carere, C.R.; Sparling, R.; Cicek, N.; Levin, D.B. Third generation biofuels via direct cellulose fermentation. Int. J. Mol. Sci. 2008, 9, 1342–1360. [Google Scholar] [CrossRef]
- Lin, C.C.; Kan, S.C.; Yeh, C.W.; Chen, C.I.; Shieh, C.J.; Liu, Y.C. Kinetics Study for the Recombinant Cellulosome to the Degradation of Chlorella Cell Residuals; International Journal of Biological, Biomolecular, Agricultural, Food and Biotechnological Engineering; World Academy of Science Engineering and Technology: Paris, France, 2015; Volume 9, pp. 782–785. [Google Scholar]
- Zhang, P.; Wang, B.; Xiao, Q.; Wu, S. A kinetics modeling study on the inhibition of glucose on cellulosome of Clostridium thermocellum. Bioresour. Technol. 2015, 190, 36–43. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, Y.H.P.; Lynd, L.R. Enzyme–microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc. Natl. Acad. Sci. USA 2006, 103, 16165–16169. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Wang, Y.; Zhang, H.; Leu, S.Y. Feasibility of high-concentration cellulosic bioethanol production from undetoxified whole Monterey pine slurry. Bioresour. Technol. 2018, 250, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Weil, J.R.; Dien, B.; Bothast, R.; Hendrickson, R.; Mosier, N.S.; Ladisch, M.R. Removal of fermentation inhibitors formed during pretreatment of biomass by polymeric adsorbents. Ind. Eng. Chem. Res. 2002, 41, 6132–6138. [Google Scholar] [CrossRef]
- Min, S.; Kim, O.J.; Bae, J.; Chung, T.N. Effect of pretreatment with the NADPH xxidase inhibitor Apocynin on the therapeutic efficacy of human placenta-derived mesenchymal stem cells in intracerebral hemorrhage. Intl. J. Mol. Sci. 2018, 19, 3679. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Qin, Y.; Li, Y.; Ji, Y.; Huang, J.; Song, H.; Xu, J. Factors influencing cellulosome activity in consolidated bioprocessing of cellulosic ethanol. Bioresour. Technol. 2010, 101, 9560–9569. [Google Scholar] [CrossRef] [PubMed]
- Duarte, L.; Carvalheiro, F.; Neves, I.; Girio, F. Effects of aliphatic acids, furfural, and phenolic compounds on Debaryomyces hansenii CCMI 941. Appl. Biochem. Biotechnol. 2005, 121, 413–425. [Google Scholar] [CrossRef]
- Guarnieri, M.T.; Franden, M.A.; Johnson, C.W.; Beckham, G.T. Conversion and assimilation of furfural and 5-(hydroxymethyl)furfural by Pseudomonas putida KT2440. Metabol. Eng. Commun. 2017, 4, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Parsiegla, G.; Juy, M.; Reverbel-Leroy, C.; Tardif, C.; Velaich, J.P.; Driguez, H.; Haser, R. The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 Å resolution. EMBO J. 1998, 17, 5551–5562. [Google Scholar] [CrossRef] [PubMed]
- You, C.; Zhang, X.Z.; Sathitsukssanoh, N.; Lynd, L.R.; Zhang, Y.H.P. Enhanced microbial utilization of recalcitrant cellulose by an ex vivo cellulosome-microbe complex. Appl. Environ. Microbiol. 2012, 78, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.P. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol. Adv. 2011, 29, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Alonso, S.; Rendueles, M.; Diaz, M. Microbial production of specialty organic acids from renewable and waste materials. Crit. Rev. Biotechol. 2013, 35, 497–513. [Google Scholar] [CrossRef] [PubMed]
- Wackett, L.P. Microbial acid fermentation products: An annotated selection of world wide web sites relevant to the topics in microbial biotechnology. Microb. Biotechnol. 2018, 11, 268–269. [Google Scholar] [CrossRef] [PubMed]
- Zverlov, V.V.; Velikodvorskaya, G.A.; Schwarz, W.H. Two new cellulosome components encoded downstream of celI in the genome of Clostridium thermocellum: The non-processive endoglucanase CelN and the possibly structural protein CseP. Microbiology 2003, 149, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Bras, J.L.; Pinheiro, B.A.; Cameron, K.; Cuskin, F.; Viegas, A.; Najmudin, S.; Bule, P.; Pires, V.M.R.; Romao, M.J.; Bayer, E.A.; et al. Diverse specificity of cellulosome attachment to the bacterial cell surface. Sci. Rep. 2016, 6, 38292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leis, B.; Held, C.; Andreeen, B.; Liebl, W.; Graubner, S.; Schulte, L.P.; Schwarz, W.H.; Zverlov, V.V. Optimizing the composition of a synthetic cellulosome complex for the hydrolysis of softwood pulp: Identification of the enzymatic core functions and biochemical complex characterization. Biotechnol. Biofuels 2018, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Correia, M.A.; Prates, J.A.; Bras, J.; Fontes, C.M.; Newman, J.A.; Lewis, R.J.; Gilbert, H.J.; Flint, J.E. Crystal structure of a cellulosomal family 3 carbohydrate esterase from Clostridium thermocellum provides insights into the mechanism of substrate recognition. J. Mol. Biol. 2008, 379, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Taherzadeh, M.; Karimi, K. Pretreatment of lignocellulosic wastes to improve ethanol and biogas production: A review. Int. J. Mol. Sci. 2008, 9, 1621–1651. [Google Scholar] [CrossRef]
- Zabed, H.; Sahu, J.N.; Boyce, A.N.; Faruq, C. Fuel ethanol production from lignocellulosic biomass: An overview on feedstocks and technological approaches. Renew. Sustain. Energy Rev. 2016, 66, 751–774. [Google Scholar] [CrossRef]
- Ding, S.Y.; Liu, Y.S.; Zeng, Y.; Himmel, M.E.; Baker, J.O.; Bayer, E.A. How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science 2012, 338, 1055–1060. [Google Scholar] [CrossRef]
- Siqueira, G.; Várnai, A.; Ferraz, A.; Milagres, A.M.F. Enhancement of cellulose hydrolysis in sugarcane bagasse by the selective removal of lignin with sodium chlorite. Appl. Energy 2013, 102, 399–402. [Google Scholar] [CrossRef]
- Wallace, J.; Brienzo, M.; García-Aparicio, M.P.; Görgens, J.F. Lignin enrichment and enzyme deactivation as the root cause of enzymatic hydrolysis slowdown of steam pretreated sugarcane bagasse. New Biotechnol. 2016, 33, 361–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zhang, L.; Zhou, T.; Wu, Y.; Xu, F. The dual effects of lignin content on enzymatic hydrolysis using film composed of cellulose and lignin as a structure model. Bioresour. Technol. 2016, 200, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Shi, W.; Sun, W.; Li, X.; Wang, F.; Zhao, J.; Qu, Y. Differences in the adsorption of enzymes onto lignins from diverse types of lignocellulosic biomass and the underlying mechanism. Biotechnol. Biofuels 2014, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Nakagame, S.; Chandra, R.P.; Kadla, J.F.; Saddler, J.N. Enhancing the enzymatic hydrolysis of lignocellulosic biomass by increasing the carboxylic acid content of the associated lignin. Biotechnol. Bioeng. 2011, 108, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Huang, Y.; Sun, R.; Tu, M. The strong association of condensed phenolic moieties in isolated lignins with their inhibition of enzymatic hydrolysis. Green Chem. 2016, 18, 4276–4286. [Google Scholar] [CrossRef]
- Yang, Q.; Pan, X. Correlation between lignin physicochemical properties and inhibition to enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 2016, 113, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Pu, Y.; Ragauskas, A.J. Current understanding of the correlation of lignin structure with biomass recalcitrance. Front. Chem. 2016, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Pareek, N.; Gillgren, T.; Jönsson, L.J. Adsorption of proteins involved in hydrolysis of lignocellulose on lignins and hemicelluloses. Bioresour. Technol. 2013, 148, 70–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lancefield, C.S.; Panovic, I.; Deuss, P.J.; Bartac, K.; Westwood, N.J. Pre-treatment of lignocellulosic feedstocks using biorenewable alcohols: Towards complete biomass valorisation. Green Chem. 2017, 19, 202–214. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, Y.Y.; Kim, T.H. A review on alkaline pretreatment technology for bioconversion of lignocellulosic biomass. Bioresour. Technol. 2016, 199, 42–48. [Google Scholar] [CrossRef]
- Sipponen, M.H.; Pihlajaniemi, V.; Pastinen, O.; Laakso, S. Reduction of surface area of lignin improves enzymatic hydrolysis of cellulose from hydrothermally pretreated wheat straw. RSC Adv. 2014, 4, 36591–36596. [Google Scholar] [CrossRef] [Green Version]
- Sipponen, M.H.; Rahikainen, J.; Leskinen, T.; Pihlajaniemi, V.; Mattinen, M.L.; Lange, H.; Crestini, C.; Österberg, Ö.M. Structural changes of lignin in biorefinery pretreatments and consequences to enzyme-lignin interactions. Nord. Pulp Pap. Res. J. 2017, 32, 550–571. [Google Scholar] [CrossRef]
- Huang, C.; He, J.; Min, D.; Lai, C.; Yong, Q. Understanding the nonproductive enzyme adsorption and physicochemical properties of residual lignins in moso bamboo pretreated with sulfuric acid and kraft pulping. Appl. Biochem. Biotechnol. 2016, 180, 1508–1523. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Gwak, K.S.; Treasure, T.; Jameel, H.; Chang, H.M.; Park, S. Effect of lignin chemistry on the enzymatic hydrolysis of woody biomass. ChemSusChem 2014, 7, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Hu, F.; Sannigrahi, P.; Jung, S.; Ragauskas, A.J.; Wyman, C.E. Carbohydrate derived-pseudo-lignin can retard cellulose biological conversion. Biotechnol. Bioeng. 2013, 110, 737–753. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, H.; Tanner, D.; Sørensen, H.R.; Meyer, A.S. New degradation compounds from lignocellulosic biomass pretreatment: Routes for formation of potent oligophenolic enzyme inhibitors. Green Chem. 2017, 19, 464–473. [Google Scholar] [CrossRef]
- Taherzadeh, M.J.; Karimi, K. Enzyme-based hydrolysis processes for ethanol from lignocellulosic materials: A review. Bioresources 2007, 2, 707–738. [Google Scholar]
- Bubner, P.; Dohr, J.; Plank, H.; Mayrhofer, C.; Nidetzky, B. Cellulases dig deep: In Situ observation of the mesoscopic structural dynamics of enzymatic cellulose degradation. J. Biol. Chem. 2012, 287, 2759–2765. [Google Scholar] [CrossRef] [PubMed]
- Karimi, K.; Taherzadeh, M.J. A critical review on analysis in pretreatment of lignocelluloses: Degree of polymerization, adsorption/desorption, and accessibility. Bioresour. Technol. 2016, 203, 348–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafiei, M.; Kumar, R.; Karimi, K. Pretreatment of lignocellulosic biomass. In Lignocellulose-Based Bioproducts; Karimi, K., Ed.; Springer International Publishing: Berlin, Germany, 2015; Volume 1, pp. 85–154. [Google Scholar]
- Neuman, R.P.; Walker, L.P. Solute exclusion from cellulose in packed columns: Experimental investigation and pore volume measurements. Biotechnol. Bioeng. 1992, 40, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Resch, M.G.; Podkaminer, K.; Yang, S.; Baker, J.O.; Donohoe, B.S.; Wilson, C.; Klingeman, D.M.; Olson, D.G.; Decker, S.R.; et al. Dramatic performance of Clostridium thermocellum explained by its wide range of cellulase modalities. Sci. Adv. 2016, 2, e1501254. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.B. Demonstration of the importance for cellulose hydrolysis of CelS, the most abundant cellulosomal cellulase in Clostridium thermocellum. Proc. Natl. Acad. Sci. USA 2010, 107, 17855–17856. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, G.; Li, Y.; Yang, L.; Liang, Y.; Jin, H.; Han, W.; Feng, Y.; Zhang, Z.; Wang, J. Cloning, expression, and characterization of a thermophilic endoglucanase, AcCel12B from Acidothermus cellulolyticus 11B. Int. J. Mol. Sci. 2015, 16, 25080–25095. [Google Scholar] [CrossRef] [PubMed]
- Khodaverdi, M.; Jeihanipour, A.; Karimi, K.; Taherzadeh, M.J. Kinetic modeling of rapid enzymatic hydrolysis of crystalline cellulose after pretreatment by NMMO. J. Ind. Microbiol. Biotechnol. 2012, 39, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Moraïs, S.; Morag, E.; Barak, Y.; Goldman, D.; Hadar, Y.; Lamed, R.; Shoham, Y.; Wilson, D.B.; Bayer, E.A. Deconstruction of lignocellulose into soluble sugars by native and designer cellulosomes. mBio 2012, 3, e00508-12. [Google Scholar] [CrossRef] [PubMed]
- Keller, F.A.; Hamilton, J.E.; Nguyen, Q.A. Microbial pretreatment of biomass: Potential for reducing severity of thermochemical biomass pretreatment. Appl. Biochem. Biotechnol. 2003, 105, 27–41. [Google Scholar] [CrossRef]
- Shi, J.; Sharma-Shivappa, R.; Chinn, M.; Howell, N. Effect of microbial pretreatment on enzymatic hydrolysis and fermentation of cotton stalks for ethanol production. Biomass Bioenergy 2009, 33, 88–96. [Google Scholar] [CrossRef]
- Jönsson, L.J.; Martín, C. Pretreatment of lignocellulose: Formation of inhibitory byproducts and strategies for minimizing their effects. Bioresour. Technol. 2016, 199, 103–112. [Google Scholar] [CrossRef]
- Davidi, L.; Moraïs, S.; Artzi, L.; Knop, D.; Hadar, Y.; Arfi, Y.; Bayer, E.A. Toward combined delignification and saccharification of wheat straw by a laccase-containing designer cellulosome. Proc. Natl. Acad. Sci. USA 2016, 113, 10854–10859. [Google Scholar] [CrossRef] [Green Version]
- Glazunova, O.A.; Moiseenko, K.V.; Kamenihina, I.A.; Isaykina, T.U.; Yaropolov, A.I.; Fedorova, T.V. Laccases with variable properties from different strains of Steccherinum ochraceum: Does glycosylation matter? Int. J. Mol. Sci. 2019, 20, 2008. [Google Scholar] [CrossRef]

| Changes after Pretreatment | Effect of Pretreatment | Enzyme Efficiency | Feedstocks | Reference |
|---|---|---|---|---|
| Depletion of lignin content | Increase accessible surface area and porosity of substrate | Enhanced | Corn stover, sugarcane bagasse, Eucalyptus globulus | [111,112,113,114] |
| Formation of COOH | Reduce surface tension and increase electrostatic repulsion between lignin and enzymes | Enhanced | Aspen, corn stover, poplar, lodgepole pine | [115,116,117,118] |
| Sulphonation | Reduce surface tension and non-productive enzyme binding | Enhanced | Poplar, lodgepole pine, Norway spruce, black cottonwood | [118,119,120] |
| Alkoxylation of aliphatic side chains | Block lignin condensation | Enhanced | Beech | [121] |
| Reduced surface coverage by lignin | Increase porosity and surface area | Enhanced | Wheat straw, several wood and grass species | [122,123,124] |
| Formation of condensed units | Adsorb more enzymes due to hydrophobicity | Reduced | Eucalyptus globulus, red maple, loblolly pine, mixed hardwood, aspen, bamboo | [117,124,125,126] |
| Formation of phenolic OH | Hydrophobicity and hydrogen bonding | Reduced | Technical lignins, aspen, poplar, pine, bamboo, mixed hardwood, barley straw | [115,117,118,125] |
| Removal of aliphatic OH | Form more condense lignin | Reduced | Eucalyptus globulus, red maple, loblolly pine, mixed hardwood | [115,126] |
| Increased hydrophobicity | Adsorb more enzymes | Reduced | Poplar, lodgepole pine, bamboo | [118,125] |
| Formation of resinous products | Adsorb more enzymes | Reduced | Wheat straw | [127,128] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Leng, L.; Islam, M.K.; Liu, F.; Lin, C.S.K.; Leu, S.-Y. Substrate-Related Factors Affecting Cellulosome-Induced Hydrolysis for Lignocellulose Valorization. Int. J. Mol. Sci. 2019, 20, 3354. https://doi.org/10.3390/ijms20133354
Wang Y, Leng L, Islam MK, Liu F, Lin CSK, Leu S-Y. Substrate-Related Factors Affecting Cellulosome-Induced Hydrolysis for Lignocellulose Valorization. International Journal of Molecular Sciences. 2019; 20(13):3354. https://doi.org/10.3390/ijms20133354
Chicago/Turabian StyleWang, Ying, Ling Leng, Md Khairul Islam, Fanghua Liu, Carol Sze Ki Lin, and Shao-Yuan Leu. 2019. "Substrate-Related Factors Affecting Cellulosome-Induced Hydrolysis for Lignocellulose Valorization" International Journal of Molecular Sciences 20, no. 13: 3354. https://doi.org/10.3390/ijms20133354
APA StyleWang, Y., Leng, L., Islam, M. K., Liu, F., Lin, C. S. K., & Leu, S.-Y. (2019). Substrate-Related Factors Affecting Cellulosome-Induced Hydrolysis for Lignocellulose Valorization. International Journal of Molecular Sciences, 20(13), 3354. https://doi.org/10.3390/ijms20133354