Screening of Cellular Stress Responses Induced by Ambient Aerosol Ultrafine Particle Fraction PM0.5 in A549 Cells
Abstract
1. Introduction
2. Results
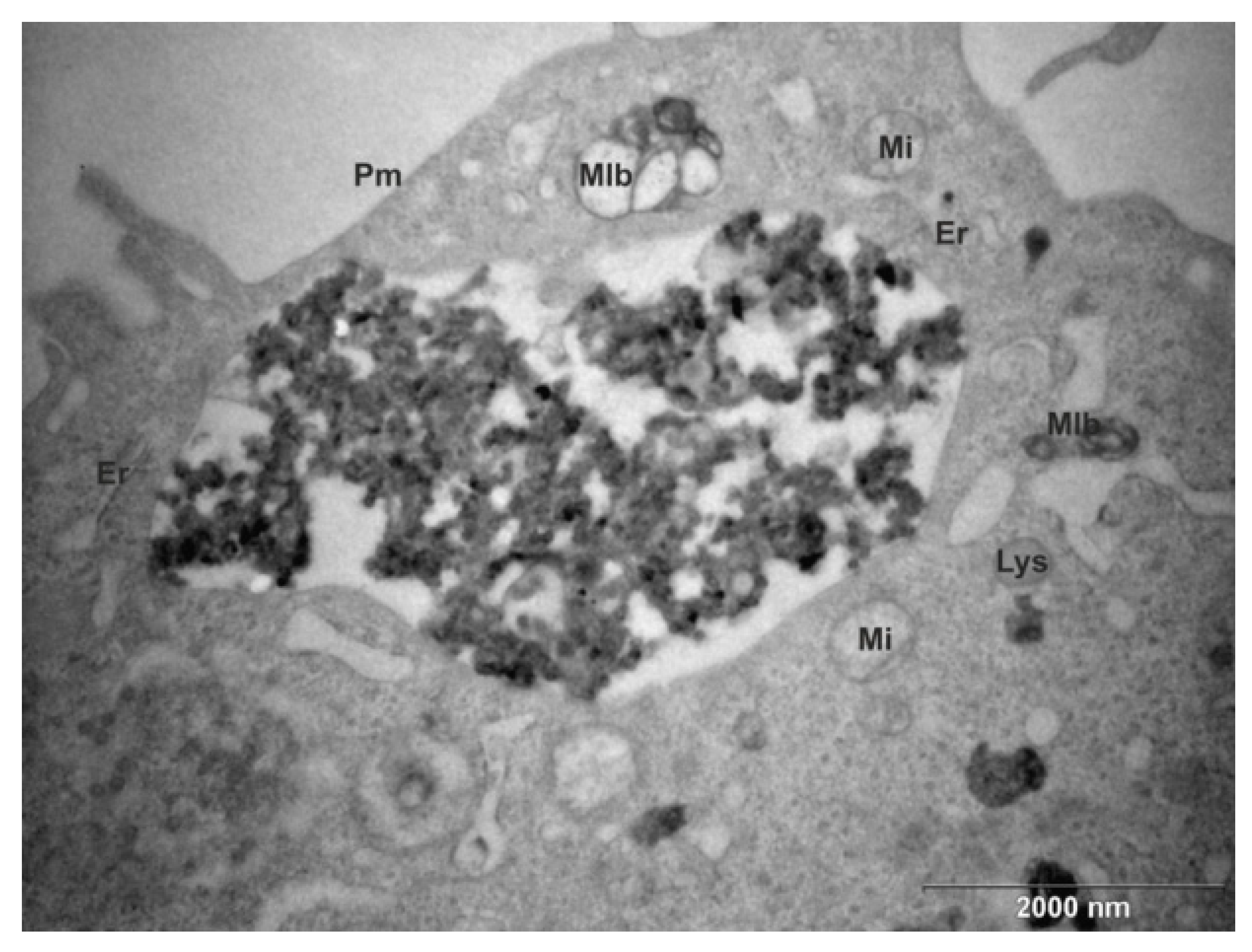
2.1. Intracellular Localization
2.2. Expression Analysis of a New Set of Genes Involved in Various Signaling Pathways Including AhR and p53
2.3. Induction of Novel Toxicity Markers—Early Stress Response Genes and Unfolded Protein Response
2.4. Autophagy
2.5. Arachidonic and Linoleic Acid Metabolites
3. Discussion
4. Materials and Methods
4.1. Aerosol Collection
4.2. Particle Separation
4.3. Cell Culture
4.4. Transmission Electron Microscopy
4.5. Real-Time Quantitative RT-PCR
4.6. Overrepresented Transcription Factor Binding Site (OTFBS) Analysis
4.7. Western Blotting
4.8. Flow Cytometry
4.9. Liquid Chromatography—Tandem Mass Spectrometry (LC-MS/MS)
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | arachidonic acid |
| AhR | Aryl hydrocarbon receptor |
| ATF3 | activating transcription factor 3 |
| BaP | benzo[a]pyrene |
| BiP | binding immunoglobulin protein |
| CDKN1A | cyclin dependent kinase inhibitor 1A |
| CYP1A1 | cytochrome P450 1A1 |
| dae | aerodynamic diameter |
| DDIT3 | DNA damage inducible transcript 3 |
| DNA | deoxyribonucleic acid |
| EGR1 | early growth response 1 |
| ER | endoplasmic reticulum |
| GDF15 | growth differentiation factor 15 |
| GREM1 | gremlin 1 |
| γH2AX | phosphorylated histone 2A family member X |
| HETE | hydroxyeicosatetranoic acid |
| HODE | hydroxyoctadecadienoic acid |
| HSPA5 | heat shock protein family A member 5 |
| IARC | International Agency for Research on Cancer |
| LAF | lower accumulation fraction |
| LC–MS/MS | liquid chromatography–tandem mass spectrometry |
| LC3B | microtubule associated protein 1 light chain 3 beta |
| OTFBS | overrepresented transcription factor binding sites |
| PARP | poly (ADP-ribose) polymerase-1 |
| PG | prostaglandin |
| PM | particulate matter |
| qRT-PCR | quantitative reverse transcription polymerase chain reaction |
| SQSTM1 | sequestosome 1 |
| TCDD | 2,3,7,8–tetrachlorodibenzo–p–dioxin |
| Tx | thromboxane |
| UPR | unfolded protein response |
| WHO | World Health Organization |
| XBP1s | spliced form of X–box binding protein 1 |
References
- IARC. Monographs on the evaluation of carcinogenic risks to humans. In Outdoor Air Pollution; IARC: Lyon, France, 2015; Volume 109. [Google Scholar]
- Oberdorster, G.; Oberdorster, E.; Oberdorster, J. Nanotoxicology: An emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect 2005, 113, 823–839. [Google Scholar] [CrossRef] [PubMed]
- Gualtieri, M.; Mantecca, P.; Corvaja, V.; Longhin, E.; Perrone, M.G.; Bolzacchini, E.; Camatini, M. Winter fine particulate matter from Milan induces morphological and functional alterations in human pulmonary epithelial cells (A549). Toxicol. Lett. 2009, 188, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Longhin, E.; Capasso, L.; Battaglia, C.; Proverbio, M.C.; Cosentino, C.; Cifola, I.; Mangano, E.; Camatini, M.; Gualtieri, M. Integrative transcriptomic and protein analysis of human bronchial BEAS-2B exposed to seasonal urban particulate matter. Environ. Pollut. 2016, 209, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Longhin, E.; Holme, J.A.; Gutzkow, K.B.; Arlt, V.M.; Kucab, J.E.; Camatini, M.; Gualtieri, M. Cell cycle alterations induced by urban PM2.5 in bronchial epithelial cells: Characterization of the process and possible mechanisms involved. Part. Fibre Toxicol. 2013, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Gualtieri, M.; Longhin, E.; Mattioli, M.; Mantecca, P.; Tinaglia, V.; Mangano, E.; Proverbio, M.C.; Bestetti, G.; Camatini, M.; Battaglia, C. Gene expression profiling of A549 cells exposed to Milan PM2.5. Toxicol. Lett. 2012, 209, 136–145. [Google Scholar] [CrossRef]
- Deng, X.; Feng, N.; Zheng, M.; Ye, X.; Lin, H.; Yu, X.; Gan, Z.; Fang, Z.; Zhang, H.; Gao, M.; et al. PM2.5 exposure-induced autophagy is mediated by lncRNA loc146880 which also promotes the migration and invasion of lung cancer cells. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 112–125. [Google Scholar] [CrossRef]
- Reibman, J.; Hsu, Y.; Chen, L.C.; Kumar, A.; Su, W.C.; Choy, W.; Talbot, A.; Gordon, T. Size fractions of ambient particulate matter induce granulocyte macrophage colony-stimulating factor in human bronchial epithelial cells by mitogen-activated protein kinase pathways. Am. J. Respir. Cell Mol. Biol. 2002, 27, 455–462. [Google Scholar] [CrossRef]
- Jalava, P.I.; Salonen, R.O.; Halinen, A.I.; Penttinen, P.; Pennanen, A.S.; Sillanpaa, M.; Sandell, E.; Hillamo, R.; Hirvonen, M.R. In vitro inflammatory and cytotoxic effects of size-segregated particulate samples collected during long-range transport of wildfire smoke to Helsinki. Toxicol. Appl. Pharmacol. 2006, 215, 341–353. [Google Scholar] [CrossRef]
- Jalava, P.I.; Wang, Q.; Kuuspalo, K.; Ruusunen, J.; Hao, L.; Fang, D.; Väisänen, O.; Ruuskanen, A.; Sippula, O.; Happo, M.S.; et al. Day and night variation in chemical composition and toxicological responses of size segregated urban air PM samples in a high air pollution situation. Atmos. Environ. 2015, 120, 427–437. [Google Scholar] [CrossRef]
- Ramgolam, K.; Favez, O.; Cachier, H.; Gaudichet, A.; Marano, F.; Martinon, L.; Baeza-Squiban, A. Size-partitioning of an urban aerosol to identify particle determinants involved in the proinflammatory response induced in airway epithelial cells. Part. Fibre Toxicol. 2009, 6, 10. [Google Scholar] [CrossRef]
- Thomson, E.M.; Breznan, D.; Karthikeyan, S.; MacKinnon-Roy, C.; Charland, J.P.; Dabek-Zlotorzynska, E.; Celo, V.; Kumarathasan, P.; Brook, J.R.; Vincent, R. Cytotoxic and inflammatory potential of size-fractionated particulate matter collected repeatedly within a small urban area. Part. Fibre Toxicol. 2015, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Sharma, P.K.; Tiwari, R.; Rayavarapu, R.G.; Shankar, J.; Chauhan, L.K.S.; Pandey, A.K. Impaired lysosomal activity mediated autophagic flux disruption by graphite carbon nanofibers induce apoptosis in human lung epithelial cells through oxidative stress and energetic impairment. Part. Fibre Toxicol. 2017, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Stern, S.T.; Adiseshaiah, P.P.; Crist, R.M. Autophagy and lysosomal dysfunction as emerging mechanisms of nanomaterial toxicity. Part. Fibre Toxicol. 2012, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Long, J.M.; Liu, L.L.; He, T.; Jiang, L.Y.; Zhao, C.X.; Li, Z. A review of endoplasmic reticulum (ER) stress and nanoparticle (NP) exposure. Life Sci. 2017, 186, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef] [PubMed]
- Marvanova, S.; Kulich, P.; Skoupy, R.; Hubatka, F.; Ciganek, M.; Bendl, J.; Hovorka, J.; Machala, M. Size-segregated urban aerosol characterization by electron microscopy and dynamic light scattering and influence of sample preparation. Atmos. Environ. 2018, 178, 181–190. [Google Scholar] [CrossRef]
- Zhang, J.D.; Berntenis, N.; Roth, A.; Ebeling, M. Data mining reveals a network of early-response genes as a consensus signature of drug-induced in vitro and in vivo toxicity. Pharmacogen. J. 2014, 14, 208–216. [Google Scholar] [CrossRef]
- Prochazkova, J.; Strapacova, S.; Svrzkova, L.; Andrysik, Z.; Hyzd’alova, M.; Hruba, E.; Pencikova, K.; Libalova, H.; Topinka, J.; Klema, J.; et al. Adaptive changes in global gene expression profile of lung carcinoma A549 cells acutely exposed to distinct types of AhR ligands. Toxicol. Lett. 2018, 292, 162–174. [Google Scholar] [CrossRef]
- Chu, L.; Wang, T.; Hu, Y.; Gu, Y.; Su, Z.; Jiang, H. Activation of Egr-1 in human lung epithelial cells exposed to silica through MAPKs signaling pathways. PLoS ONE 2013, 8, e68943. [Google Scholar] [CrossRef][Green Version]
- Yan, F.; Wu, Y.; Liu, H.; Wu, Y.; Shen, H.; Li, W. ATF3 is positively involved in particulate matter-induced airway inflammation in vitro and in vivo. Toxicol. Lett. 2018, 287, 113–121. [Google Scholar] [CrossRef]
- Clark, B.J.; Bull, T.M.; Benson, A.B.; Stream, A.R.; Macht, M.; Gaydos, J.; Meadows, C.; Burnham, E.L.; Moss, M.; Investigators, A.N. Growth differentiation factor-15 and prognosis in acute respiratory distress syndrome: A retrospective cohort study. Crit. Care 2013, 17, R92. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Fang, F.; Tourtellotte, W.; Varga, J. Egr-1: New conductor for the tissue repair orchestra directs harmony (regeneration) or cacophony (fibrosis). J. Pathol. 2013, 229, 286–297. [Google Scholar] [CrossRef]
- Hai, T.; Wolford, C.C.; Chang, Y.S. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: Is modulation of inflammation a unifying component? Gene Expr. J. Liver Res. 2010, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rohini, M.; Haritha Menon, A.; Selvamurugan, N. Role of activating transcription factor 3 and its interacting proteins under physiological and pathological conditions. Int. J. Biol. Macromol. 2018, 120, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, K.K.; Moorthy, B.; Lingappan, K. Role of GDF15 (growth and differentiation factor 15) in pulmonary oxygen toxicity. Toxicol. Vitr. 2015, 29, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Wallentin, L.; Hijazi, Z.; Andersson, U.; Alexander, J.H.; De Caterina, R.; Hanna, M.; Horowitz, J.D.; Hylek, E.M.; Lopes, R.D.; Asberg, S.; et al. Growth differentiation factor 15, a marker of oxidative stress and inflammation, for risk assessment in patients with atrial fibrillation insights from the Apixaban for Reduction in Stroke and Other Thromboembolic Events in Atrial Fibrillation (ARISTOTLE) trial. Circulation 2014, 130, 1847–1858. [Google Scholar] [PubMed]
- Kannan, K.; Amariglio, N.; Rechavi, G.; Givol, D. Profile of gene expression regulated by induced p53: Connection to the TGF-beta family. FEBS Lett. 2000, 470, 77–82. [Google Scholar] [CrossRef]
- Rynning, I.; Neca, J.; Vrbova, K.; Libalova, H.; Rossner, P.; Holme, J.A.; Gutzkow, K.B.; Afanou, A.K.J.; Arnoldussen, Y.J.; Hruba, E.; et al. In vitro transformation of human bronchial epithelial cells by diesel exhaust particles: Gene expression profiling and early toxic responses. Toxicol. Sci. 2018, 166, 51–64. [Google Scholar] [CrossRef]
- Wang, D.Z.; Dubois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193. [Google Scholar] [CrossRef]
- Hruba, E.; Trilecova, L.; Marvanova, S.; Krcmar, P.; Vykopalova, L.; Milcova, A.; Libalova, H.; Topinka, J.; Starsichova, A.; Soucek, K.; et al. Genotoxic polycyclic aromatic hydrocarbons fail to induce the p53-dependent DNA damage response, apoptosis or cell-cycle arrest in human prostate carcinoma LNCaP cells. Toxicol. Lett. 2010, 197, 227–235. [Google Scholar] [CrossRef]
- Albino, A.P.; Huang, X.; Jorgensen, E.; Yang, J.; Gietl, D.; Traganos, F.; Darzynkiewicz, Z. Induction of H2AX phosphorylation in pulmonary cells by tobacco smoke: A new assay for carcinogens. Cell Cycle 2004, 3, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Zamarbide, M.; Martinez-Pinilla, E.; Ricobaraza, A.; Aragon, T.; Franco, R.; Perez-Mediavilla, A. Phenyl acyl acids attenuate the unfolded protein response in tunicamycin-treated neuroblastoma cells. PLoS ONE 2013, 8, e71082. [Google Scholar] [CrossRef] [PubMed]
- Simmons, S.O.; Fan, C.Y.; Ramabhadran, R. Cellular stress response pathway system as a sentinel ensemble in toxicological screening. Toxicol. Sci. 2009, 111, 202–225. [Google Scholar] [CrossRef] [PubMed]
- Etteieb, S.; Kawachi, A.; Han, J.; Elayni, F.; Tarhouni, J.; Isoda, H. Assessment of organic micropollutants occurrence in treated wastewater using heat shock protein 47 stress responses in Chinese hamster ovary cells and GC/MS-based non-target screening. Water Sci. Technol. 2016, 74, 2407–2416. [Google Scholar] [CrossRef] [PubMed]
- Wink, S.; Hiemstra, S.W.; Huppelschoten, S.; Klip, J.E.; van de Water, B. Dynamic imaging of adaptive stress response pathway activation for prediction of drug induced liver injury. Arch. Toxicol. 2018, 92, 1797–1814. [Google Scholar] [CrossRef]
- Zhao, D.Y.; Jacobs, K.M.; Hallahan, D.E.; Thotala, D. Silencing Egr1 attenuates radiation-induced apoptosis in normal tissues while killing cancer cells and delaying tumor growth. Mol. Cancer Ther. 2015, 14, 2343–2352. [Google Scholar] [CrossRef]
- Chang, Y.; Lee, H.H.; Chen, Y.T.; Lu, J.; Wu, S.Y.; Chen, C.W.; Takada, K.; Tsai, C.H. Induction of the early growth response 1 gene by Epstein-Barr virus lytic transactivator Zta. J. Virol. 2006, 80, 7748–7755. [Google Scholar] [CrossRef]
- Martinez, J.M.; Baek, S.J.; Mays, D.M.; Tithof, P.K.; Eling, T.E.; Walker, N.J. EGR1 is a novel target for AhR agonists in human lung epithelial cells. Toxicol. Sci. 2004, 82, 429–435. [Google Scholar] [CrossRef]
- Brauze, D.; Zawierucha, P.; Kiwerska, K.; Bednarek, K.; Oleszak, M.; Rydzanicz, M.; Jarmuz-Szymczak, M. Induction of expression of aryl hydrocarbon receptor-dependent genes in human HepaRG cell line modified by shRNA and treated with beta-naphthoflavone. Mol. Cell Biochem. 2017, 425, 59–75. [Google Scholar] [CrossRef]
- Reynolds, P.R.; Cosio, M.G.; Hoidal, J.R. Cigarette smoke-induced Egr-1 upregulates proinflammatory cytokines in pulmonary epithelial cells. Am. J. Respir. Cell Mol. Biol. 2006, 35, 314–319. [Google Scholar] [CrossRef]
- Shen, N.; Gong, T.; Wang, J.D.; Meng, F.L.; Qiao, L.; Yang, R.L.; Xue, B.; Pan, F.Y.; Zhou, X.J.; Chen, H.Q.; et al. Cigarette smoke-induced pulmonary inflammatory responses are mediated by EGR-1/GGPPS/MAPK signaling. Am. J. Pathol. 2011, 178, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; Ning, W.; Matthay, M.A.; Feghali-Bostwick, C.A.; Choi, A.M. MAPK pathway mediates EGR-1-HSP70-dependent cigarette smoke-induced chemokine production. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 292, L1297–L1303. [Google Scholar] [CrossRef] [PubMed]
- Sarill, M.; Zago, M.; Sheridan, J.A.; Nair, P.; Matthews, J.; Gomez, A.; Roussel, L.; Rousseau, S.; Hamid, Q.; Eidelman, D.H.; et al. The aryl hydrocarbon receptor suppresses cigarette-smoke-induced oxidative stress in association with dioxin response element (DRE)-independent regulation of sulfiredoxin 1. Free Radic. Biol. Med. 2015, 89, 342–357. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Liu, C.T.; Zhang, W.D. IL-17a and GDF15 are able to induce epithelial-mesenchymal transition of lung epithelial cells in response to cigarette smoke. Exp. Ther. Med. 2018, 16, 12–20. [Google Scholar] [CrossRef]
- Menendez, D.; Inga, A.; Resnick, M.A. The expanding universe of p53 targets. Nat. Rev. Cancer 2009, 9, 724–737. [Google Scholar] [CrossRef]
- Li, Z.D.; Wang, K.; Yang, X.W.; Zhuang, Z.G.; Wang, J.J.; Tong, X.W. Expression of aryl hydrocarbon receptor in relation to p53 status and clinicopathological parameters in breast cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 7931–7937. [Google Scholar]
- Kochhar, A.; Kopelovich, L.; Sue, E.; Guttenplan, J.B.; Herbert, B.S.; Dannenberg, A.J.; Subbaramaiah, K. P53 modulates Hsp90 ATPase activity and regulates aryl hydrocarbon receptor signaling. Cancer Prev. Res. (Phila) 2014, 7, 596–606. [Google Scholar] [CrossRef]
- Li, X.B.; Zhou, X.X.; Li, Y.W.; Zu, L.L.; Pan, H.L.; Liu, B.N.; Shen, W.; Fan, Y.G.; Zhou, Q.H. Activating transcription factor 3 promotes malignance of lung cancer cells in vitro. Thorac. Cancer 2017, 8, 181–191. [Google Scholar] [CrossRef]
- Fan, F.; Jin, S.; Amundson, S.A.; Tong, T.; Fan, W.; Zhao, H.; Zhu, X.; Mazzacurati, L.; Li, X.; Petrik, K.L.; et al. ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene 2002, 21, 7488–7496. [Google Scholar] [CrossRef]
- Harizi, H.; Corcuff, J.B.; Gualde, N. Arachidonic-acid-derived eicosanoids: Roles in biology and immunopathology. Trends Mol. Med. 2008, 14, 461–469. [Google Scholar] [CrossRef]
- Maayah, Z.H.; El-Kadi, A.O.S. The role of mid-chain hydroxyeicosatetraenoic acids in the pathogenesis of hypertension and cardiac hypertrophy. Arch. Toxicol. 2016, 90, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Lawal, A.; Ricks, J.; Fox, J.R.; Larson, T.; Navab, M.; Fogelman, A.M.; Rosenfeld, M.E.; Araujo, J.A. Diesel exhaust induces systemic lipid peroxidation and development of dysfunctional pro-oxidant and pro-inflammatory high-density lipoprotein. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Cathcart, M.C.; Reynolds, J.V.; O’Byrne, K.J.; Pidgeon, G.P. The role of prostacyclin synthase and thromboxane synthase signaling in the development and progression of cancer. Biochim. Biophys. Acta 2010, 1805, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Umeno, A.; Shichiri, M. Lipid peroxidation biomarkers for evaluating oxidative stress and assessing antioxidant capacity in vivo. J. Clin. Biochem. Nutr. 2013, 52, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, K.; Tran, L.; Jimenez, L.A.; Duffin, R.; Newby, D.E.; Mills, N.; MacNee, W.; Stone, V. Combustion-derived nanoparticles: A review of their toxicology following inhalation exposure. Part. Fibre Toxicol. 2005, 2, 10. [Google Scholar] [CrossRef]
- Billet, S.; Abbas, I.; Le Goff, J.; Verdin, A.; Andre, V.; Lafargue, P.E.; Hachimi, A.; Cazier, F.; Sichel, F.; Shirali, P.; et al. Genotoxic potential of polycyclic aromatic hydrocarbons-coated onto airborne particulate matter (PM 2.5) in human lung epithelial A549 cells. Cancer Lett. 2008, 270, 144–155. [Google Scholar] [CrossRef]
- Libalova, H.; Krckova, S.; Uhlirova, K.; Klema, J.; Ciganek, M.; Rossner, P., Jr.; Sram, R.J.; Vondracek, J.; Machala, M.; Topinka, J. Analysis of gene expression changes in A549 cells induced by organic compounds from respirable air particles. Mutat. Res. 2014, 770, 94–105. [Google Scholar] [CrossRef]
- Abbas, I.; Badran, G.; Verdin, A.; Ledoux, F.; Roumie, M.; Lo Guidice, J.M.; Courcot, D.; Garcon, G. In vitro evaluation of organic extractable matter from ambient PM2.5 using human bronchial epithelial BEAS-2B cells: Cytotoxicity, oxidative stress, pro-inflammatory response, genotoxicity, and cell cycle deregulation. Environ. Res. 2019, 171, 510–522. [Google Scholar] [CrossRef]
- Souček, K.; Malenovská, A.; Kahounová, Z.; Remšík, J.; Holubcová, Z.; Soukup, T.; Kurfürstová, D.; Bouchal, J.; Suchánková, T.; Slabáková, E.; et al. Presence of growth/differentiation factor-15 cytokine in human follicular fluid, granulosa cells, and oocytes. J. Assist. Reprod. Genet. 2018, 35, 1407–1417. [Google Scholar] [CrossRef]
- Van Schadewijk, A.; van’t Wout, E.F.; Stolk, J.; Hiemstra, P.S. A quantitative method for detection of spliced x-box binding protein-1 (XBP1) mrna as a measure of endoplasmic reticulum (ER) stress. Cell Stress Chaperon. 2012, 17, 275–279. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Ho Sui, S.J.; Mortimer, J.R.; Arenillas, D.J.; Brumm, J.; Walsh, C.J.; Kennedy, B.P.; Wasserman, W.W. Opossum: Identification of over-represented transcription factor binding sites in co-expressed genes. Nucl. Acids Res. 2005, 33, 3154–3164. [Google Scholar] [CrossRef] [PubMed]
- Pencikova, K.; Svrzkova, L.; Strapacova, S.; Neca, J.; Bartonkova, I.; Dvorak, Z.; Hyzdalova, M.; Pivnicka, J.; Palkova, L.; Lehmler, H.J.; et al. In vitro profiling of toxic effects of prominent environmental lower-chlorinated PCB congeners linked with endocrine disruption and tumor promotion. Environ. Pollut. 2018, 237, 473–486. [Google Scholar] [CrossRef] [PubMed]





| PGE2 | 13,14-Dihydro-15-Keto-PGE2 | PGD2 | 6-Keto-PGF1α | TxB2 | 8-Iso-PGF2α | |
|---|---|---|---|---|---|---|
| Ctrl | 9.1 ± 3.6 | 10.0 ± 3.5 | 15.6 ± 3.1 | 2.6 ± 1.1 | 74.4 ± 6.7 | 16.7 ± 4.2 |
| PM0.5 | 9.4 ± 2.3 | 5.3 ± 1.8 * | 23.7 ± 5.1 * | 4.0 ± 1.1 | 65.2 ± 4.8 | 13.9 ± 5.6 |
| TF Matrices Family | hEGR1 | hGDF15 |
|---|---|---|
| V$AHRR | 2.12 | 2.04 |
| V$P53F | – | 4.24 |
| V$EGRF | 6.42 | 8.32 |
| Gene Symbol/RefSeq Code | Sequences | |
|---|---|---|
| CDKN1A (p21) | NM_00389.4 | F: 5′-CCGAAGTCAGTTCCTTGTGG-3′ R: 5′-CATGGGTTCTGACGGACAT-3′ P: #82 |
| DDIT3 | NM_001195056.1 NM_001195054.1 NM_001195053.1 NM_001195055.1 NM_004083.5 NM_001195057.1 | F: 5′-AAGGCACTGAGCGTATCATGT-3′ R: 5′-TGAAGATACACTTCCTTCTTGAACA-3′ P: #21 |
| HSPA5 | NM_005347.4 | F: 5′-AGCCTGGCGACAAGAGTG-3′ R: 5′-TCCTTGGGCAGTATTGGATT-3′ P: #39 |
| ATF3 | NM_001206486.2 NM_001206488.2 NM_001040619.2 NM_001030287.3 NM_001674.3 NM_001206484.2 | F: 5′-TTTGCCATCCAGAACAAGC-3′ R: 5′-CATCTTCTTCAGGGGCTACCT-3′ P: #53 |
| EGR1 | NM_001964.2 | F: 5′-AGCCCTACGAGCACCTGAC-3′ R: 5′-GGTTTGGCTGGGGTAACTG-3′ P: #22 |
| NOXA (PMAIP1) | NM_021127.2 | F: 5′-GGAGATGCCTGGGAAGAAG -3′ R: 5′-CCTGAGTTGAGTAGCACACTCG -3′ P: #67 |
| PUMA (BBC3) | NM_014417.3 NM_001127240.1 | F: 5′-GACCTCAACGCACAGTACGA -3′ R: 5′-GAGATTGTACAGGACCCTCCA -3′ P: #68 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šimečková, P.; Marvanová, S.; Kulich, P.; Králiková, L.; Neča, J.; Procházková, J.; Machala, M. Screening of Cellular Stress Responses Induced by Ambient Aerosol Ultrafine Particle Fraction PM0.5 in A549 Cells. Int. J. Mol. Sci. 2019, 20, 6310. https://doi.org/10.3390/ijms20246310
Šimečková P, Marvanová S, Kulich P, Králiková L, Neča J, Procházková J, Machala M. Screening of Cellular Stress Responses Induced by Ambient Aerosol Ultrafine Particle Fraction PM0.5 in A549 Cells. International Journal of Molecular Sciences. 2019; 20(24):6310. https://doi.org/10.3390/ijms20246310
Chicago/Turabian StyleŠimečková, Pavlína, Soňa Marvanová, Pavel Kulich, Lucie Králiková, Jiří Neča, Jiřina Procházková, and Miroslav Machala. 2019. "Screening of Cellular Stress Responses Induced by Ambient Aerosol Ultrafine Particle Fraction PM0.5 in A549 Cells" International Journal of Molecular Sciences 20, no. 24: 6310. https://doi.org/10.3390/ijms20246310
APA StyleŠimečková, P., Marvanová, S., Kulich, P., Králiková, L., Neča, J., Procházková, J., & Machala, M. (2019). Screening of Cellular Stress Responses Induced by Ambient Aerosol Ultrafine Particle Fraction PM0.5 in A549 Cells. International Journal of Molecular Sciences, 20(24), 6310. https://doi.org/10.3390/ijms20246310





