Hypoxia-Regulated miRNAs in Human Mesenchymal Stem Cells: Exploring the Regulatory Effects in Ischemic Disorders
Abstract
1. Introduction
2. Results
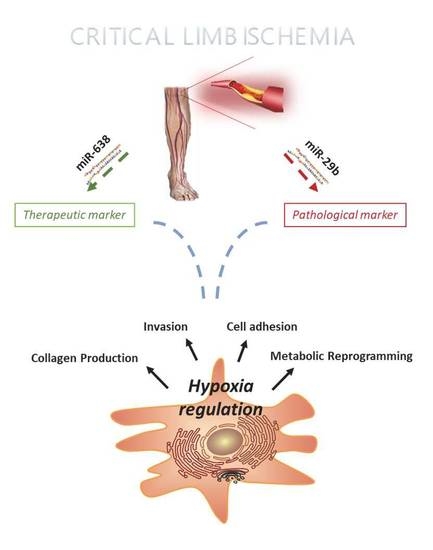
2.1. miRNA Profiling of hMSC Derived from CLI Patients
2.2. microRNA 638 (miR-638) as a Potential Therapeutic Target
3. Discussion
4. Methods
4.1. Isolation of Human BMSCs (hBMSCs)
4.2. Human Bone Marrow Stem Cell Harvesting
4.3. RNA Isolation and miRNA Expression Analysis
4.4. Real-Time PCR
4.5. miRNA Microarray Profiling and Data Analysis
4.6. Computational Prediction of miRNA Target Genes
4.7. Gene-Enrichment and Functional Annotation Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human mesenchymal stem cells—Current trends and future prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, J.; Sensebe, L. Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities. Cell Stem Cell 2018, 22, 824–833. [Google Scholar] [CrossRef]
- Abdul Wahid, S.F.; Ismail, N.A.; Wan Jamaludin, W.F.; Muhamad, N.A.; Abdul Hamid, M.K.A.; Harunarashid, H.; Lai, N.M. Autologous cells derived from different sources and administered using different regimens for ‘no-option’ critical lower limb ischaemia patients. Cochrane Database Syst. Rev. 2018, 8, CD010747. [Google Scholar] [CrossRef]
- Qadura, M.; Terenzi, D.C.; Verma, S.; Al-Omran, M.; Hess, D.A. Concise Review: Cell Therapy for Critical Limb Ischemia: An Integrated Review of Preclinical and Clinical Studies. Stem Cells 2018, 36, 161–171. [Google Scholar] [CrossRef]
- Yu, H.; Lu, K.; Zhu, J.; Wang, J. Stem cell therapy for ischemic heart diseases. Br. Med. Bull. 2017, 121, 135–154. [Google Scholar] [CrossRef]
- Peeters Weem, S.M.; Teraa, M.; de Borst, G.J.; Verhaar, M.C.; Moll, F.L. Bone Marrow derived Cell Therapy in Critical Limb Ischemia: A Meta-analysis of Randomized Placebo Controlled Trials. Eur. J. Vasc. Endovasc. Surg. 2015, 50, 775–783. [Google Scholar] [CrossRef]
- Von Bahr, L.; Batsis, I.; Moll, G.; Hagg, M.; Szakos, A.; Sundberg, B.; Uzunel, M.; Ringden, O.; Le Blanc, K. Analysis of tissues following mesenchymal stromal cell therapy in humans indicates limited long-term engraftment and no ectopic tissue formation. Stem Cells 2012, 30, 1575–1578. [Google Scholar] [CrossRef] [PubMed]
- Forster, R.; Liew, A.; Bhattacharya, V.; Shaw, J.; Stansby, G. Gene therapy for peripheral arterial disease. Cochrane Database Syst. Rev. 2018, 10, CD012058. [Google Scholar] [CrossRef] [PubMed]
- Pugh, C.W.; Ratcliffe, P.J. Regulation of angiogenesis by hypoxia: Role of the HIF system. Nat. Med. 2003, 9, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.R.; Teixeira, G.Q.; Santos, S.G.; Barbosa, M.A.; Almeida-Porada, G.; Goncalves, R.M. Mesenchymal Stromal Cell Secretome: Influencing Therapeutic Potential by Cellular Pre-conditioning. Front. Immunol. 2018, 9, 2837. [Google Scholar] [CrossRef] [PubMed]
- De Becker, A.; Riet, I.V. Homing and migration of mesenchymal stromal cells: How to improve the efficacy of cell therapy? World J. Stem Cells 2016, 8, 73–87. [Google Scholar] [CrossRef]
- Nitzsche, F.; Muller, C.; Lukomska, B.; Jolkkonen, J.; Deten, A.; Boltze, J. Concise Review: MSC Adhesion Cascade-Insights into Homing and Transendothelial Migration. Stem Cells 2017, 35, 1446–1460. [Google Scholar] [CrossRef] [PubMed]
- Karp, J.M.; Leng Teo, G.S. Mesenchymal stem cell homing: The devil is in the details. Cell Stem Cell 2009, 4, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Poissonnier, L.; Villain, G.; Soncin, F.; Mattot, V. miR126-5p repression of ALCAM and SetD5 in endothelial cells regulates leucocyte adhesion and transmigration. Cardiovasc. Res. 2014, 102, 436–447. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Hernando, C.; Suarez, Y. MicroRNAs in endothelial cell homeostasis and vascular disease. Curr. Opin. Hematol. 2018, 25, 227–236. [Google Scholar] [CrossRef]
- Nicoli, S.; Knyphausen, C.P.; Zhu, L.J.; Lakshmanan, A.; Lawson, N.D. miR-221 is required for endothelial tip cell behaviors during vascular development. Dev. Cell 2012, 22, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Landskroner-Eiger, S.; Qiu, C.; Perrotta, P.; Siragusa, M.; Lee, M.Y.; Ulrich, V.; Luciano, A.K.; Zhuang, Z.W.; Corti, F.; Simons, M.; et al. Endothelial miR-17 approximately 92 cluster negatively regulates arteriogenesis via miRNA-19 repression of WNT signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 12812–12817. [Google Scholar] [CrossRef]
- Caporali, A.; Emanueli, C. MicroRNA-503 and the extended microRNA-16 family in angiogenesis. Trends Cardiovasc. Med. 2011, 21, 162–166. [Google Scholar] [CrossRef]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef]
- Dewdney, B.; Trollope, A.; Moxon, J.; Thomas Manapurathe, D.; Biros, E.; Golledge, J. Circulating MicroRNAs as Biomarkers for Acute Ischemic Stroke: A Systematic Review. J. Stroke Cerebrovasc. Dis. 2018, 27, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Pogribny, I.P. MicroRNAs as biomarkers for clinical studies. Exp. Biol. Med. 2018, 243, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wu, R.; Shehadeh, L.A.; Zhou, Q.; Jiang, C.; Huang, X.; Zhang, L.; Gao, F.; Liu, X.; Yu, H.; et al. Severe hypoxia exerts parallel and cell-specific regulation of gene expression and alternative splicing in human mesenchymal stem cells. BMC Genom. 2014, 15, 303. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ouyang, R.; Wang, Z.; Zhou, W.; Chen, H.; Jiang, Y.; Zhang, Y.; Li, H.; Liao, M.; Wang, W.; et al. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci. Rep. 2016, 6, 39001. [Google Scholar] [CrossRef]
- Lin, T.P.; Chang, Y.T.; Lee, S.Y.; Campbell, M.; Wang, T.C.; Shen, S.H.; Chung, H.J.; Chang, Y.H.; Chiu, A.W.; Pan, C.C.; et al. REST reduction is essential for hypoxia-induced neuroendocrine differentiation of prostate cancer cells by activating autophagy signaling. Oncotarget 2016, 7, 26137–26151. [Google Scholar] [CrossRef]
- Arsenault, P.R.; Song, D.; Bergkamp, M.; Ravaschiere, A.M.; Navalsky, B.E.; Lieberman, P.M.; Lee, F.S. Identification of Small-Molecule PHD2 Zinc Finger Inhibitors that Activate Hypoxia Inducible Factor. ChemBioChem 2016, 17, 2316–2323. [Google Scholar] [CrossRef]
- Celic, T.; Metzinger-Le Meuth, V.; Six, I.; Massy, Z.A.; Metzinger, L. The mir-221/222 Cluster is a Key Player in Vascular Biology via the Fine-Tuning of Endothelial Cell Physiology. Curr. Vasc. Pharmacol. 2017, 15, 40–46. [Google Scholar] [CrossRef]
- Tan, X.; Fu, Y.; Chen, L.; Lee, W.; Lai, Y.; Rezaei, K.; Tabbara, S.; Latham, P.; Teal, C.B.; Man, Y.G.; et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget 2016, 7, 293–307. [Google Scholar] [CrossRef]
- Li, P.; Liu, Y.; Yi, B.; Wang, G.; You, X.; Zhao, X.; Summer, R.; Qin, Y.; Sun, J. MicroRNA-638 is highly expressed in human vascular smooth muscle cells and inhibits PDGF-BB-induced cell proliferation and migration through targeting orphan nuclear receptor NOR1. Cardiovasc. Res. 2013, 99, 185–193. [Google Scholar] [CrossRef]
- Krueger, T.E.G.; Thorek, D.L.J.; Denmeade, S.R.; Isaacs, J.T.; Brennen, W.N. Concise Review: Mesenchymal Stem Cell-Based Drug Delivery: The Good, the Bad, the Ugly, and the Promise. Stem Cells Transl. Med. 2018, 7, 651–663. [Google Scholar] [CrossRef]
- Singh, A.; Singh, A.; Sen, D. Mesenchymal stem cells in cardiac regeneration: A detailed progress report of the last 6 years (2010–2015). Stem Cell Res. Ther. 2016, 7, 82. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Botti, C.; Caiafa, I.; Coppola, A.; Cuomo, F.; Miceli, M.; Altucci, L.; Cobellis, G. SIRT1 inhibition affects angiogenic properties of human MSCs. Biomed. Res. Int. 2014, 2014, 783459. [Google Scholar] [CrossRef] [PubMed]
- Maione, C.; Botti, C.; Coppola, C.A.; Silvestroni, C.; Lillo, S.; Schiavone, V.; Sica, G.; Sica, V.; Kumar, V.; Cobellis, G. Effect of autologous transplantation of bone marrow cells concentrated with the MarrowXpress system in patients with critical limb ischemia. Transplant. Proc. 2013, 45, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Benoit, E.; O’Donnell, T.F.; Patel, A.N. Safety and efficacy of autologous cell therapy in critical limb ischemia: A systematic review. Cell Transplant. 2013, 22, 545–562. [Google Scholar] [CrossRef] [PubMed]
- Wahid, F.S.A.; Ismail, N.A.; Wan Jamaludin, W.F.; Muhamad, N.A.; Mohamad Idris, M.A.; Lai, N.M. Efficacy and Safety of Autologous Cell-based Therapy in Patients with No-option Critical Limb Ischaemia: A Meta-Analysis. Curr. Stem Cell Res. Ther. 2018, 13, 265–283. [Google Scholar] [CrossRef] [PubMed]
- Dengler, V.L.; Galbraith, M.; Espinosa, J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cuomo, F.; Coppola, A.; Botti, C.; Maione, C.; Forte, A.; Scisciola, L.; Liguori, G.; Caiafa, I.; Ursini, M.V.; Galderisi, U.; et al. Pro-inflammatory cytokines activate hypoxia-inducible factor 3alpha via epigenetic changes in mesenchymal stromal/stem cells. Sci. Rep. 2018, 8, 5842. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cai, B.; Shen, L.; Dong, Y.; Lu, Q.; Sun, S.; Liu, S.; Ma, S.; Ma, P.X.; Chen, J. MiRNA-29b suppresses tumor growth through simultaneously inhibiting angiogenesis and tumorigenesis by targeting Akt3. Cancer Lett. 2017, 397, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Khalil, R.A. Matrix Metalloproteinases, Vascular Remodeling, and Vascular Disease. Adv. Pharmacol. 2018, 81, 241–330. [Google Scholar] [CrossRef]
- Wang, T.; Hou, J.; Jian, S.; Luo, Q.; Wei, J.; Li, Z.; Wang, X.; Bai, P.; Duan, B.; Xing, J.; et al. miR-29b negatively regulates MMP2 to impact gastric cancer development by suppress gastric cancer cell migration and tumor growth. J. Cancer 2018, 9, 3776–3786. [Google Scholar] [CrossRef]
- Chen, Q.; Jin, M.; Yang, F.; Zhu, J.; Xiao, Q.; Zhang, L. Matrix metalloproteinases: Inflammatory regulators of cell behaviors in vascular formation and remodeling. Mediat. Inflamm. 2013, 2013, 928315. [Google Scholar] [CrossRef] [PubMed]
- Belo, V.A.; Guimaraes, D.A.; Castro, M.M. Matrix Metalloproteinase 2 as a Potential Mediator of Vascular Smooth Muscle Cell Migration and Chronic Vascular Remodeling in Hypertension. J. Vasc. Res. 2015, 52, 221–231. [Google Scholar] [CrossRef]
- Bellayr, I.H.; Kumar, A.; Puri, R.K. MicroRNA expression in bone marrow-derived human multipotent Stromal cells. BMC Genom. 2017, 18, 605. [Google Scholar] [CrossRef]
- Yan, S.; Dang, G.; Zhang, X.; Jin, C.; Qin, L.; Wang, Y.; Shi, M.; Huang, H.; Duan, Q. Downregulation of circulating exosomal miR-638 predicts poor prognosis in colon cancer patients. Oncotarget 2017, 8, 72220–72226. [Google Scholar] [CrossRef]
- Li, M.; Wang, J.; Liu, H. Downregulation of miR-638 promotes progression of breast cancer and is associated with prognosis of breast cancer patients. OncoTargets Ther. 2018, 11, 6871–6877. [Google Scholar] [CrossRef]
- Cobellis, G.; Silvestroni, A.; Lillo, S.; Sica, G.; Botti, C.; Maione, C.; Schiavone, V.; Rocco, S.; Brando, G.; Sica, V. Long-term effects of repeated autologous transplantation of bone marrow cells in patients affected by peripheral arterial disease. Bone Marrow Transplant. 2008, 42, 667–672. [Google Scholar] [CrossRef]
- Dell’Aversana, C.; Giorgio, C.; D’Amato, L.; Lania, G.; Matarese, F.; Saeed, S.; Di Costanzo, A.; Belsito Petrizzi, V.; Ingenito, C.; Martens, J.H.A.; et al. miR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia 2018, 32, 573. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dell’Aversana, C.; Cuomo, F.; Botti, C.; Maione, C.; Carissimo, A.; Casamassimi, A.; Altucci, L.; Cobellis, G. Hypoxia-Regulated miRNAs in Human Mesenchymal Stem Cells: Exploring the Regulatory Effects in Ischemic Disorders. Int. J. Mol. Sci. 2019, 20, 1340. https://doi.org/10.3390/ijms20061340
Dell’Aversana C, Cuomo F, Botti C, Maione C, Carissimo A, Casamassimi A, Altucci L, Cobellis G. Hypoxia-Regulated miRNAs in Human Mesenchymal Stem Cells: Exploring the Regulatory Effects in Ischemic Disorders. International Journal of Molecular Sciences. 2019; 20(6):1340. https://doi.org/10.3390/ijms20061340
Chicago/Turabian StyleDell’Aversana, Carmela, Francesca Cuomo, Chiara Botti, Ciro Maione, Annamaria Carissimo, Amelia Casamassimi, Lucia Altucci, and Gilda Cobellis. 2019. "Hypoxia-Regulated miRNAs in Human Mesenchymal Stem Cells: Exploring the Regulatory Effects in Ischemic Disorders" International Journal of Molecular Sciences 20, no. 6: 1340. https://doi.org/10.3390/ijms20061340
APA StyleDell’Aversana, C., Cuomo, F., Botti, C., Maione, C., Carissimo, A., Casamassimi, A., Altucci, L., & Cobellis, G. (2019). Hypoxia-Regulated miRNAs in Human Mesenchymal Stem Cells: Exploring the Regulatory Effects in Ischemic Disorders. International Journal of Molecular Sciences, 20(6), 1340. https://doi.org/10.3390/ijms20061340