DHA Modulates Immune Response and Mitochondrial Function of Atlantic Salmon Adipocytes after LPS Treatment
Abstract
1. Introduction
2. Results
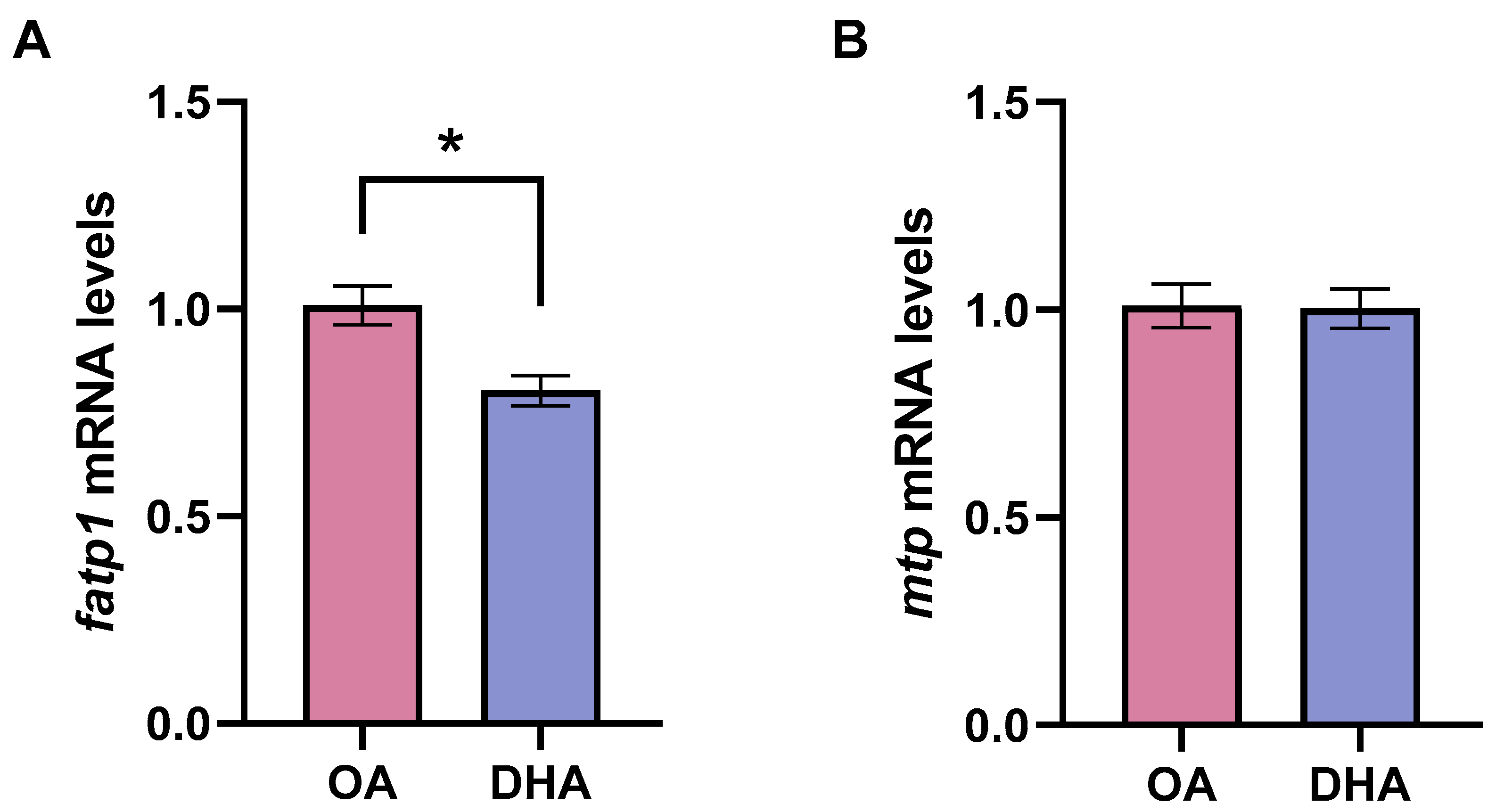
2.1. Characterization of Differentiated Experimental Adipocytes
2.2. Effect of FAs on Adipocytes’ Response to LPS Treatment
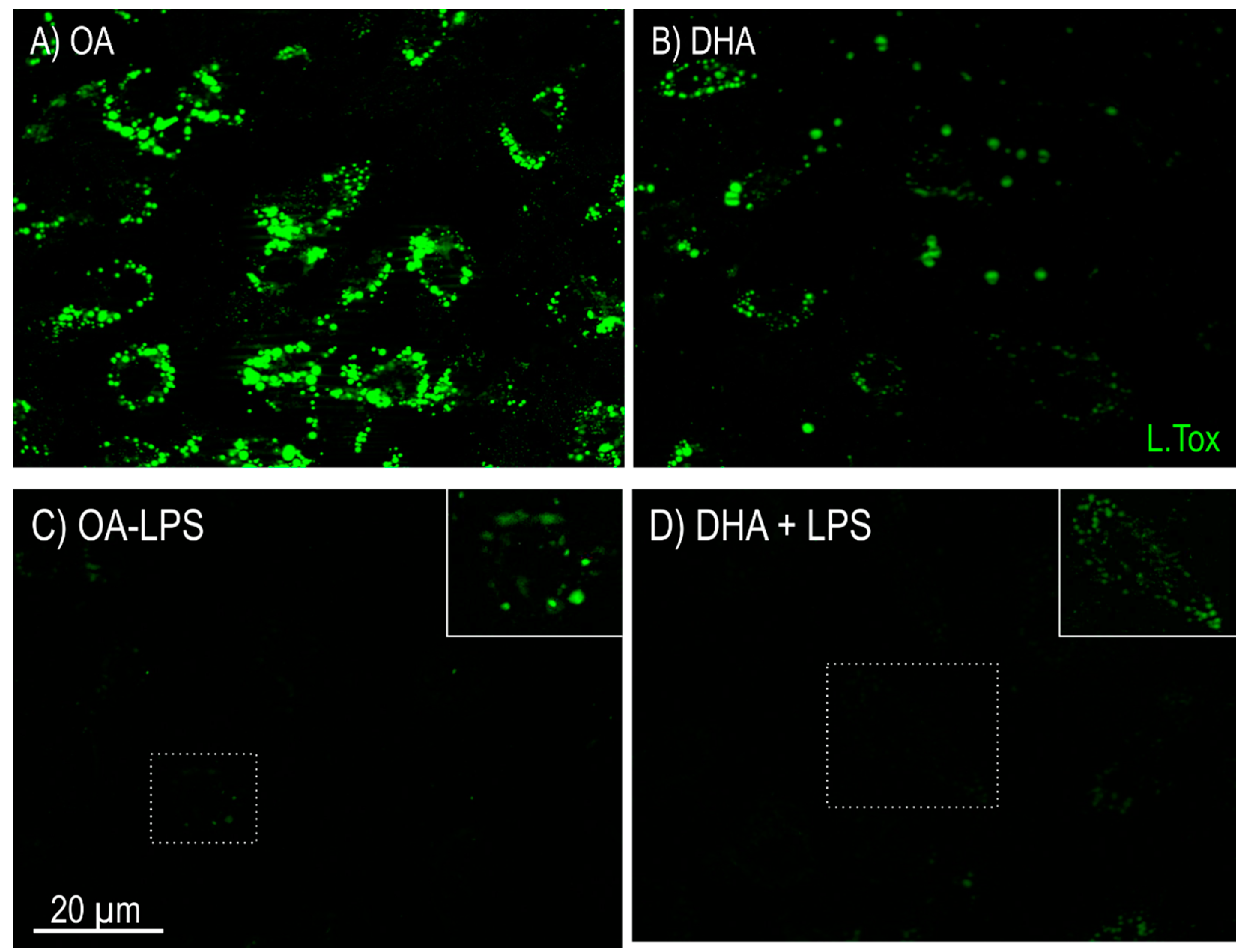
2.2.1. Lipid Mobilization
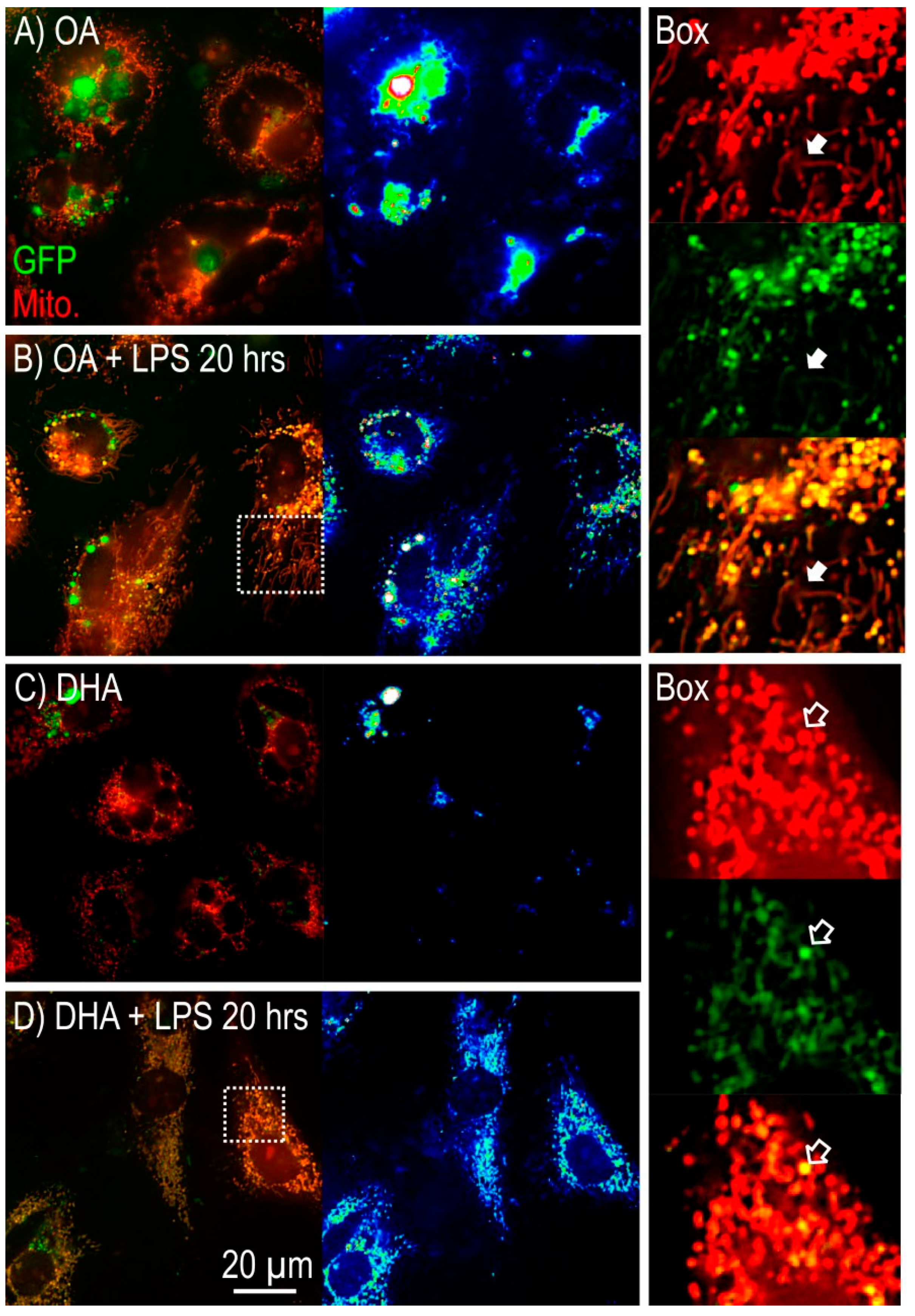
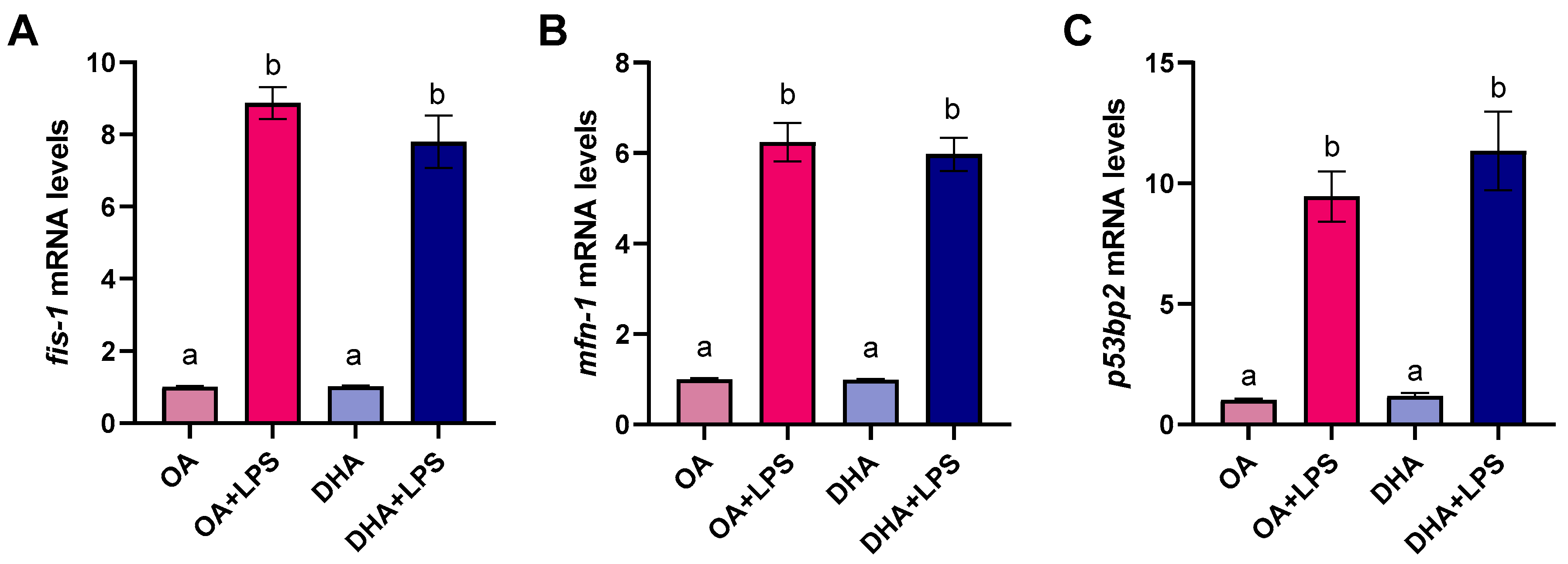
2.2.2. Mitochondrial Morphology
2.2.3. iNOS Activity
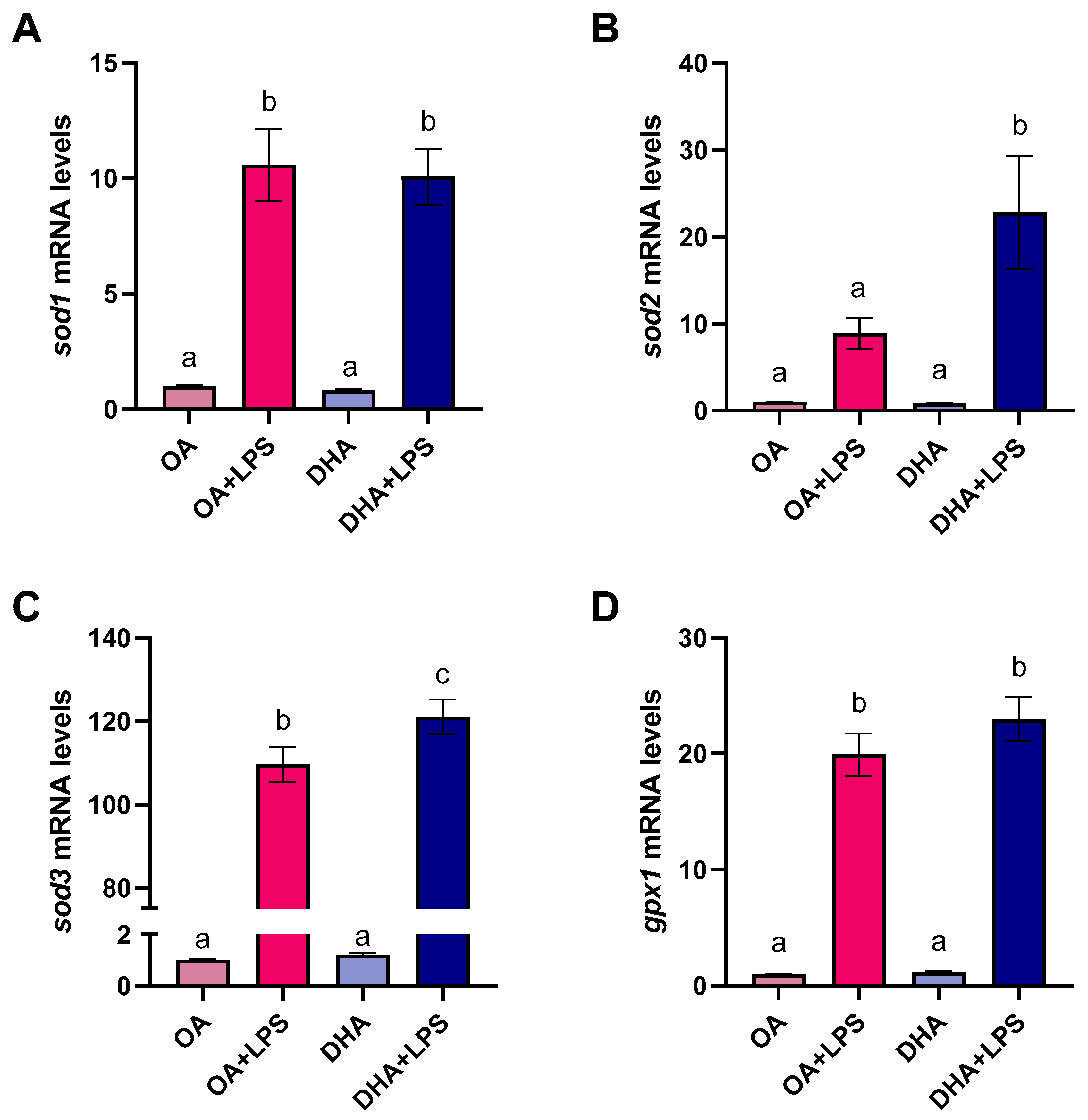
2.2.4. Antioxidant Enzyme Activity
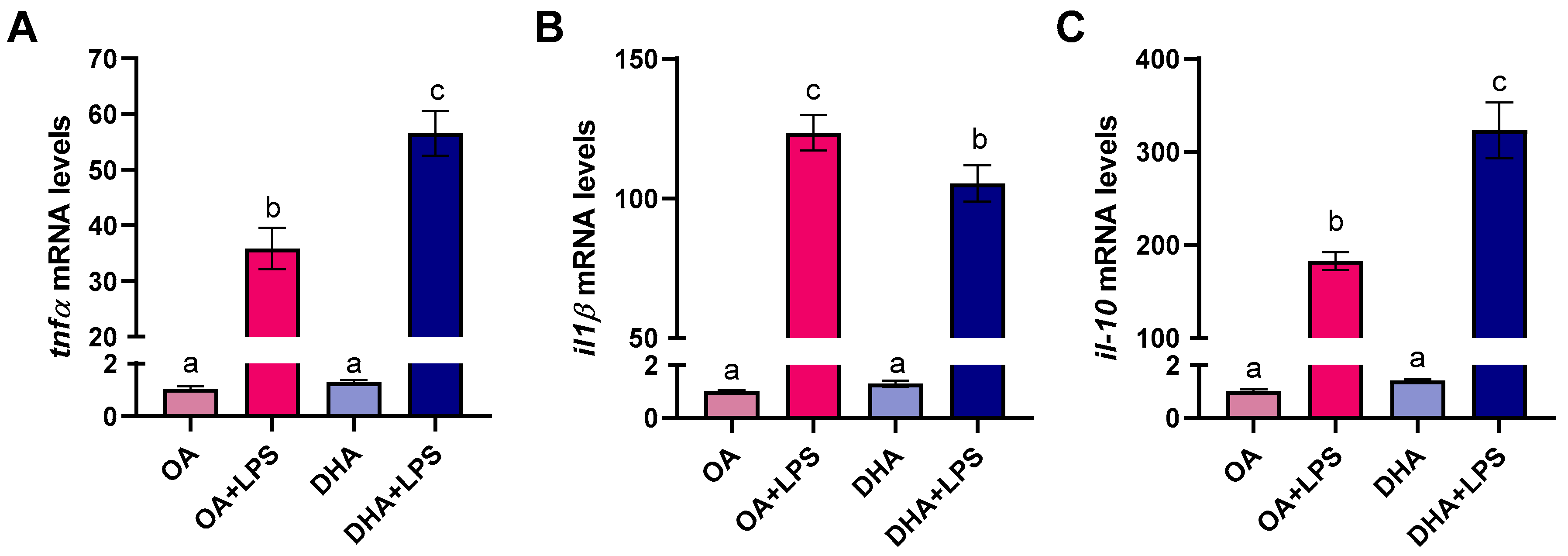
2.2.5. Transcriptional Responses to FAs and LPS Exposure
3. Discussion
4. Materials and Methods
4.1. Preadipocyte Isolation and Culture Conditions
4.2. Fatty Acid Composition
4.3. RNA Extraction and cDNA Synthesis
4.4. qPCR
4.5. SOD2-GFP Vector
4.6. Transfection
4.7. Immunofluorescence and Staining
4.8. Superoxide Dismutase (SOD) Assay
4.9. Catalase Assay
4.10. Statistics
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DHA | Docosahexaenoic acid |
| OA | Oleic acid |
| HUFAs | Highly unsaturated fatty acids |
| FAs | Fatty acids |
| LPS | Lipopolysaccharides |
References
- Aas, T.S.; Ytrestøyl, T.; Åsgård, T. Utilization of feed resources in the production of Atlantic salmon (Salmo salar) in Norway: An update for 2016. Aquac. Rep. 2019, 15, 100216. [Google Scholar] [CrossRef]
- Colombo, S.M.; Foroutani, M.B.; Parrish, C.C. Fats and Oils in Aquafeed Formulations. In Bailey’s Industrial Oil and Fat Products; Shahidi, F., Ed.; Wiley Online Library: Hoboken, NJ, USA, 2020; pp. 1–28. [Google Scholar]
- Todorčević, M.; Kjær, M.A.; Djaković, N.; Vegusdal, A.; Torstensen, B.E.; Ruyter, B. N-3 HUFAs affect fat deposition, susceptibility to oxidative stress, and apoptosis in Atlantic salmon visceral adipose tissue. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2009, 152, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Weil, C.; Lefèvre, F.; Bugeon, J. Characteristics and metabolism of different adipose tissues in fish. Rev. Fish Biol. Fish. 2013, 23, 157–173. [Google Scholar] [CrossRef]
- Bou, M.; Todorcevic, M.; Torgersen, J.; Skugor, S.; Navarro, I.; Ruyter, B. De novo lipogenesis in Atlantic salmon adipocytes. Biochim. Biophys. Acta 2016, 1860, 86–96. [Google Scholar] [CrossRef]
- Bou, M.; Berge, G.M.; Baeverfjord, G.; Sigholt, T.; Ostbye, T.K.; Ruyter, B. Low levels of very-long-chain n-3 PUFA in Atlantic salmon (Salmo salar) diet reduce fish robustness under challenging conditions in sea cages. J. Nutr. Sci. 2017, 6, e32. [Google Scholar] [CrossRef] [PubMed]
- Todorcevic, M.; Skugor, S.; Krasnov, A.; Ruyter, B. Gene expression profiles in Atlantic salmon adipose-derived stromo-vascular fraction during differentiation into adipocytes. BMC Genom. 2010, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Ytrestøyl, T.; Aas, T.S.; Åsgård, T. Utilisation of feed resources in production of Atlantic salmon (Salmo salar) in Norway. Aquaculture 2015, 448, 365–374. [Google Scholar] [CrossRef]
- Montero, D.; Mathlouthi, F.; Tort, L.; Afonso, J.; Torrecillas, S.; Fernández-Vaquero, A.; Negrin, D.; Izquierdo, M. Replacement of dietary fish oil by vegetable oils affects humoral immunity and expression of pro-inflammatory cytokines genes in gilthead sea bream Sparus aurata. Fish Shellfish Immunol. 2010, 29, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Todorčević, M.; Hodson, L. The Effect of Marine Derived n-3 Fatty Acids on Adipose Tissue Metabolism and Function. J. Clin. Med. 2015, 5, 3. [Google Scholar] [CrossRef]
- Bou, M.; Berge, G.M.; Baeverfjord, G.; Sigholt, T.; Østbye, T.-K.; Romarheim, O.H.; Hatlen, B.; Leeuwis, R.; Venegas, C.; Ruyter, B.J.B.J.O.N. Requirements of n-3 very long-chain PUFA in Atlantic salmon (Salmo salar L): Effects of different dietary levels of EPA and DHA on fish performance and tissue composition and integrity. Br. J. Nutr. 2017, 117, 30–47. [Google Scholar] [CrossRef]
- Todorčević, M.; Vegusdal, A.; Gjoen, T.; Sundvold, H.; Torstensen, B.E.; Kjaer, M.A.; Ruyter, B. Changes in fatty acids metabolism during differentiation of Atlantic salmon preadipocytes; effects of n-3 and n-9 fatty acids. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2008, 1781, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Bou, M.; Wang, X.; Todorčević, M.; Østbye, T.-K.K.; Torgersen, J.; Ruyter, B. Lipid Deposition and Mobilisation in Atlantic Salmon Adipocytes. Int. J. Mol. Sci. 2020, 21, 2332. [Google Scholar] [CrossRef] [PubMed]
- Hafidi, M.E.; Buelna-Chontal, M.; Sanchez-Munoz, F.; Carbo, R. Adipogenesis: A Necessary but Harmful Strategy. Int. J. Mol. Sci. 2019, 20, 3657. [Google Scholar] [CrossRef] [PubMed]
- Vishvanath, L.; Gupta, R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J. Clin. Investig. 2019, 129, 4022–4031. [Google Scholar] [CrossRef] [PubMed]
- Burhans, M.S.; Hagman, D.K.; Kuzma, J.N.; Schmidt, K.A.; Kratz, M. Contribution of Adipose Tissue Inflammation to the Development of Type 2 Diabetes Mellitus. Compr. Physiol. 2018, 9, 1–58. [Google Scholar]
- Szabova, M.; Jahnova, E.; Horvathova, M.; Ilavska, S.; Pruzincova, V.; Nemessanyi, T.; Tulinska, J.; Wsolova, L.; Volkovova, K. Changes in immunologic parameters of humoral immunity and adipocytokines in obese persons are gender dependent. Hum. Immunol. 2012, 73, 486–492. [Google Scholar] [CrossRef]
- Asghar, A.; Sheikh, N. Role of immune cells in obesity induced low grade inflammation and insulin resistance. Cell. Immunol. 2017, 315, 18–26. [Google Scholar] [CrossRef]
- Skugor, S.; Skugor, A.; Todorcevic, M.; Torgersen, J.; Ruyter, B.; Krasnov, A. Exposure to lipopolysaccharide induces immune genes in cultured preadipocytes of Atlantic salmon. Fish Shellfish Immunol. 2010, 29, 817–824. [Google Scholar] [CrossRef]
- Prostek, A.; Gajewska, M.; Kamola, D.; Balasinska, B. The influence of EPA and DHA on markers of inflammation in 3T3-L1 cells at different stages of cellular maturation. Lipids Health Dis. 2014, 13, 3. [Google Scholar] [CrossRef]
- Bjørndal, B.; Burri, L.; Staalesen, V.; Skorve, J.; Berge, R.K. Different adipose depots: Their role in the development of metabolic syndrome and mitochondrial response to hypolipidemic agents. J. Obes. 2011, 2011, 490650. [Google Scholar] [CrossRef]
- Jelenik, T.; Roden, M. Mitochondrial plasticity in obesity and diabetes mellitus. Antioxid. Redox Signal. 2013, 19, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Bournat, J.C.; Brown, C.W. Mitochondrial dysfunction in obesity. Curr. Opin. Endocrinol. Diabetes Obes. 2010, 17, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Busiello, R.A.; Savarese, S.; Lombardi, A. Mitochondrial uncoupling proteins and energy metabolism. Front. Physiol. 2015, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Lustig, M.E.; Boyle, K.E.; Woodlief, T.L.; Kane, D.A.; Lin, C.T.; Price, J.W., 3rd; Kang, L.; Rabinovitch, P.S.; Szeto, H.H.; et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J. Clin. Investig. 2009, 119, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Bondia-Pons, I.; Ryan, L.; Martinez, J.A. Oxidative stress and inflammation interactions in human obesity. J. Physiol. Biochem. 2012, 68, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Kjaer, M.; Vegusdal, A.; Gjøen, T.; Rustan, A.; Todorčević, M.; Ruyter, B. Effect of rapeseed oil and dietary n-3 fatty acids on triacylglycerol synthesis and secretion in Atlantic salmon hepatocytes. Biochim. Biophys. Acta 2008, 1781, 112–122. [Google Scholar] [CrossRef]
- Sethi, J.K.; Vidal-Puig, A.J. Thematic review series: Adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J. Lipid Res. 2007, 48, 1253–1262. [Google Scholar] [CrossRef]
- Blüher, M. Adipose tissue inflammation: A cause or consequence of obesity-related insulin resistance? Clin. Sci. (Lond.) 2016, 130, 1603–1614. [Google Scholar] [CrossRef]
- Huang, C.-W.; Chien, Y.-S.; Chen, Y.-J.; Ajuwon, K.M.; Mersmann, H.M.; Ding, S.-T. Role of n-3 Polyunsaturated Fatty Acids in Ameliorating the Obesity-Induced Metabolic Syndrome in Animal Models and Humans. Int. J. Mol. Sci. 2016, 17, 1689. [Google Scholar] [CrossRef]
- Todorcevic, M.; Skugor, S.; Ruyter, B. Alterations in oxidative stress status modulate terminal differentiation in Atlantic salmon adipocytes cultivated in media rich in n-3 fatty acids. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010, 156, 309–318. [Google Scholar] [CrossRef]
- Kim, H.-K.; Della-Fera, M.; Lin, J.; Baile, C.A. Docosahexaenoic acid inhibits adipocyte differentiation and induces apoptosis in 3T3-L1 preadipocytes. J. Nutr. 2006, 136, 2965–2969. [Google Scholar] [CrossRef] [PubMed]
- Barber, E.; Sinclair, A.J.; Cameron-Smith, D. Comparative actions of omega-3 fatty acids on in-vitro lipid droplet formation. Prostaglandins Leukot. Essent. Fatty Acids 2013, 89, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Manickam, E.; Sinclair, A.J.; Cameron-Smith, D. Suppressive actions of eicosapentaenoic acid on lipid droplet formation in 3T3-L1 adipocytes. Lipids Health Dis. 2010, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Belzung, F.; Raclot, T.; Groscolas, R. Fish oil n-3 fatty acids selectively limit the hypertrophy of abdominal fat depots in growing rats fed high-fat diets. Am. J. Physiol. 1993, 264, R1111–R1118. [Google Scholar] [CrossRef] [PubMed]
- Hainault, I.; Carlotti, M.; Hajduch, E.; Guichard, C.; Lavau, M. Fish oil in a high lard diet prevents obesity, hyperlipemia, and adipocyte insulin resistance in rats. Ann. N. Y. Acad. Sci. 1993, 683, 98. [Google Scholar] [CrossRef] [PubMed]
- Ruzickova, J.; Rossmeisl, M.; Prazak, T.; Flachs, P.; Sponarova, J.; Vecka, M.; Tvrzicka, E.; Bryhn, M.; Kopecky, J. Omega-3 PUFA of marine origin limit diet-induced obesity in mice by reducing cellularity of adipose tissue. Lipids 2004, 39, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Flachs, P.; Rossmeisl, M.; Kuda, O.; Kopecky, J. Stimulation of mitochondrial oxidative capacity in white fat independent of UCP1: A key to lean phenotype. Biochim. Biophys. Acta 2013, 1831, 986–1003. [Google Scholar] [CrossRef] [PubMed]
- Adamcova, K.; Horakova, O.; Bardova, K.; Janovska, P.; Brezinova, M.; Kuda, O.; Rossmeisl, M.; Kopecky, J. Reduced Number of Adipose Lineage and Endothelial Cells in Epididymal fat in Response to Omega-3 PUFA in Mice Fed High-Fat Diet. Mar. Drugs 2018, 16, 515. [Google Scholar] [CrossRef]
- Bjursell, M.; Xu, X.; Admyre, T.; Böttcher, G.; Lundin, S.; Nilsson, R.; Stone, V.M.; Morgan, N.G.; Lam, Y.Y.; Storlien, L.H.; et al. The Beneficial Effects of n-3 Polyunsaturated Fatty Acids on Diet Induced Obesity and Impaired Glucose Control Do Not Require Gpr120. PLoS ONE 2014, 9, e114942. [Google Scholar] [CrossRef]
- Martinez-Fernandez, L.; Laiglesia, L.M.; Huerta, A.E.; Martinez, J.A.; Moreno-Aliaga, M.J. Omega-3 fatty acids and adipose tissue function in obesity and metabolic syndrome. Prostaglandins Other Lipid Mediat. 2015, 121, 24–41. [Google Scholar] [CrossRef]
- Murumalla, R.K.; Gunasekaran, M.K.; Padhan, J.K.; Bencharif, K.; Gence, L.; Festy, F.; Cesari, M.; Roche, R.; Hoareau, L. Fatty acids do not pay the toll: Effect of SFA and PUFA on human adipose tissue and mature adipocytes inflammation. Lipids Health Dis. 2012, 11, 175. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.C.; Ding, S.T. N-3 polyunsaturated fatty acids regulate lipid metabolism through several inflammation mediators: Mechanisms and implications for obesity prevention. J. Nutr. Biochem. 2010, 21, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Kratz, M.; Kuzma, J.N.; Hagman, D.K.; van Yserloo, B.; Matthys, C.C.; Callahan, H.S.; Weigle, D.S. n3 PUFAs do not affect adipose tissue inflammation in overweight to moderately obese men and women. J. Nutr. 2013, 143, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Vachharajani, V.; Granger, D.N. Adipose tissue: A motor for the inflammation associated with obesity. IUBMB Life 2009, 61, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Investig. 2004, 114, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Taube, A.; Schlich, R.; Sell, H.; Eckardt, K.; Eckel, J. Inflammation and metabolic dysfunction: Links to cardiovascular diseases. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H2148–H2165. [Google Scholar] [CrossRef]
- Lasker, S.; Rahman, M.M.; Parvez, F.; Zamila, M.; Miah, P.; Nahar, K.; Kabir, F.; Sharmin, S.B.; Subhan, N.; Ahsan, G.U.; et al. High-fat diet-induced metabolic syndrome and oxidative stress in obese rats are ameliorated by yogurt supplementation. Sci. Rep. 2019, 9, 20026. [Google Scholar] [CrossRef]
- Vona, R.; Gambardella, L.; Cittadini, C.; Straface, E.; Pietraforte, D. Biomarkers of Oxidative Stress in Metabolic Syndrome and Associated Diseases. Oxid. Med. Cell. Longev. 2019, 2019, 8267234. [Google Scholar] [CrossRef]
- Hernández-Aguilera, A.; Rull, A.; Rodríguez-Gallego, E.; Riera-Borrull, M.; Luciano-Mateo, F.; Camps, J.; Menéndez, J.A.; Joven, J. Mitochondrial Dysfunction: A Basic Mechanism in Inflammation-Related Non-Communicable Diseases and Therapeutic Opportunities. Mediat. Inflamm. 2013, 2013, 1–13. [Google Scholar] [CrossRef]
- De Mello, A.H.; Costa, A.B.; Engel, J.D.G.; Rezin, G.T. Mitochondrial dysfunction in obesity. Life Sci. 2018, 192, 26–32. [Google Scholar] [CrossRef]
- Perreault, M.; Marette, A. Targeted disruption of inducible nitric oxide synthase protects against obesity-linked insulin resistance in muscle. Nat. Med. 2001, 7, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chomyn, A.; Chan, D.C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005, 280, 26185–26192. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-H.; Lin, C.-C.; Yang, M.-C.; Wei, C.-C.; Liao, H.-D.; Lin, R.-C.; Tu, W.-Y.; Kao, T.-C.; Hsu, C.-M.; Cheng, J.-T. GSK3beta-mediated Drp1 phosphorylation induced elongated mitochondrial morphology against oxidative stress. PLoS ONE 2012, 7, e49112. [Google Scholar] [CrossRef] [PubMed]
- Barbour, J.A.; Turner, N. Mitochondrial stress signaling promotes cellular adaptations. Int. J. Cell Biol. 2014, 2014, 156020. [Google Scholar] [CrossRef]
- Katyare, S.S.; Mali, A.V. Omega-3 Fatty Acids and Mitochondrial Functions. In Omega-3 Fatty Acids: Keys to Nutritional Health; Hegde, M.V., Zanwar, A.A., Adekar, S.P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 229–233. [Google Scholar]
- Dabkowski, E.R.; O’Connell, K.A.; Xu, W.; Ribeiro, R.F., Jr.; Hecker, P.A.; Shekar, K.C.; Daneault, C.; Des Rosiers, C.; Stanley, W.C. Docosahexaenoic acid supplementation alters key properties of cardiac mitochondria and modestly attenuates development of left ventricular dysfunction in pressure overload-induced heart failure. Cardiovasc. Drugs Ther. 2013, 27, 499–510. [Google Scholar] [CrossRef]
- Khairallah, R.J.; Sparagna, G.C.; Khanna, N.; O’Shea, K.M.; Hecker, P.A.; Kristian, T.; Fiskum, G.; Des Rosiers, C.; Polster, B.M.; Stanley, W.C. Dietary supplementation with docosahexaenoic acid, but not eicosapentaenoic acid, dramatically alters cardiac mitochondrial phospholipid fatty acid composition and prevents permeability transition. Biochim. Biophys. Acta 2010, 1797, 1555–1562. [Google Scholar] [CrossRef]
- Ovide-Bordeaux, S.; Grynberg, A. Docosahexaenoic acid affects insulin deficiency- and insulin resistance-induced alterations in cardiac mitochondria. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004, 286, R519–R527. [Google Scholar] [CrossRef][Green Version]
- Vegusdal, A.; Sundvold, H.; Gjoen, T.; Ruyter, B. An in vitro method for studying the proliferation and differentiation of Atlantic salmon preadipocytes. Lipids 2003, 38, 289–296. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
- Mason, M.E.; Waller, G.R. Dimethoxypropane induced transesterification of fats and oils in preparation of methyl esters for gas chromatographic analysis. Anal. Chem. 1964, 36, 583–586. [Google Scholar] [CrossRef]
- Hoshi, M.; Williams, M.; Kishimoto, Y. Esterification of fatty acids at room temperature by chloroform-methanolic HCl–cupric acetate. J. Lipid Res. 1973, 14, 599–601. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35 (Suppl. 2), W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Gurmaches, J.; Ostbye, T.K.; Navarro, I.; Torgersen, J.; Hevroy, E.M.; Ruyter, B.; Torstensen, B.E. In vivo and in vitro insulin and fasting control of the transmembrane fatty acid transport proteins in Atlantic salmon (Salmo salar). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, R947–R957. [Google Scholar] [CrossRef]
- Ebbesson, L.O.; Tipsmark, C.K.; Holmqvist, B.; Nilsen, T.; Andersson, E.; Stefansson, S.O.; Madsen, S.S. Nitric oxide synthase in the gill of Atlantic salmon: Colocalization with and inhibition of Na+,K+-ATPase. J. Exp. Biol. 2005, 208, 1011–1017. [Google Scholar] [CrossRef]
- Baudhuin, P.; Beaufay, H.; Rahman-Li, Y.; Sellinger, O.Z.; Wattiaux, R.; Jacques, P.; De Duve, C. Tissue fractionation studies. 17. Intracellular distribution of monoamine oxidase, aspartate aminotransferase, alanine aminotransferase, D-amino acid oxidase and catalase in rat-liver tissue. Biochem. J. 1964, 92, 179–184. [Google Scholar] [CrossRef]



| OA | DHA | p | |
|---|---|---|---|
| 14:0 | 1.0 ± 0.3 | 1.1 ± 0.3 | 0.803 |
| 16:0 | 10.2 ± 0.8 | 11.3 ± 0.2 | 0.231 |
| 18:0 | 7.4 ± 0.2 a | 8.2 ± 0.2 b | 0.032 |
| ƩSFA 1 | 20.2 ± 0.9 | 21.4 ± 0.6 | 0.318 |
| 16:1n-7 | 1.2 ± 0.5 | 1.7 ± 0.3 | 0.457 |
| 16:1n-9 | 0.4 ± 0.2 | 0.6 ± 0.1 | 0.338 |
| 18:1n-7 | 2.3 ± 0.1 | 2.5 ± 0.1 | 0.208 |
| 18:1n-9 | 34.8 ± 1.1 a | 19.2 ± 0.6 b | 0.0003 |
| 18:1n-11 | 0.8 ± 0.1 | 0.7 ± 0.1 | 0.728 |
| 20:1n-9 | 2.5 ± 0.3 | 2.2 ± 0.1 | 0.5523 |
| 22:1n-11 | 1.0 ± 0.2 | 1.2 ± 0.1 | 0.349 |
| ƩMUFA 2 | 45.4 ± 1.4 a | 30.7 ± 0.7 b | 0.0008 |
| 18:2n-6 | 3.9 ± 0.1 | 4.6 ± 0.2 | 0.035 |
| 20:2n-6 | 0.6 ± 0.0 | 0.4 ± 0.2 | 0.670 |
| 20:4n-6 | 5.3 ± 0.1 | 4.9 ± 0.4 | 0.412 |
| Ʃn-6 3 | 11.1 ± 0.6 | 11.1 ± 0.7 | 0.961 |
| 18:3n-3 | 1.3 ± 0.2 | 1.5 ± 0.3 | 0.680 |
| 20:5n-3 | 4.2 ± 0.3 | 4.3 ± 0.6 | 0.923 |
| 22:5n-3 | 2.9 ± 0.1 | 2.8 ± 0.2 | 0.576 |
| 22:6n-3 | 9.0 ± 0.6 a | 23.2 ± 1.0 b | 0.0003 |
| Ʃn-3 4 | 18.0 ± 0.7 a | 32.2 ± 1.1 b | 0.0003 |
| PUFA 5 | 29.9 ± 0.7 a | 44.8 ± 1.2 b | 0.0005 |
| Gene | Accession No. | Direction | Primer Sequence 5′→3′ |
|---|---|---|---|
| ef1α | AF321836 | Forward | CACCACCGGCCATCTGATCTACAA |
| Reverse | TCAGCAGCCTCCTTCTGAACTTC | ||
| etif3 | DW542195 | Forward | CAGGATGTTGTTGCTGGATGGG |
| Reverse | ACCCAACTGGGCAGGTCAAGA | ||
| β-actin | AF012125 | Forward | ACATCAAGGAGAAGCTGTGC |
| Reverse | GACAACGGAACCTCTCGTTA | ||
| fatp1 | CA373015 | Forward | TGGGAGCTTGTGGGTTCAA |
| Reverse | ACTTTCATGAGGCGGATTGG | ||
| mtp | CA042356.1 | Forward | CAAAGACCAGCGTCAACAACAA |
| Reverse | CGCCTCTGTCTCAAAGCTCACT | ||
| sod1 | BT057716 | Forward | TTCTGTTGTACGCTGTCCCAAAAGC |
| Reverse | GCAGCTTGGTACGCAAAGTGAACA | ||
| sod2 | BT060259 | Forward | TTCCGAGTGTGCCCAGTCTTGT |
| Reverse | ACAGGGAGGTGAAAGTGCATGGT | ||
| sod3 | BT046917 | Forward | TCATCGACAGCAGAGAAGAAGGGGA |
| Reverse | CTGCGATGAAAGGTGGTGAGCG | ||
| gpx1 | CA345853 | Forward | CCTTCCAGTACCTGGAGTTGAATGC |
| Reverse | CTCATGATTGTCTCCTGGCTCCTGT | ||
| tnfα | NM_001123589 | Forward | AGGTTGGCTATGGAGGCTGT |
| Reverse | TCTGCTTCAATGTATGGTGGG | ||
| il1β | CA377361 | Forward | GTATCCCATCACCCCATCAC |
| Reverse | TTGAGCAGGTCCTTGTCCTT | ||
| il10 | EF165028 | Forward | ATGAGGCTAATGACGAGCTGGAGA |
| Reverse | GGTGTAGAATGCCTTCGTCCAACA | ||
| mfn1 | BT072406 | Forward | AGTGTGTCCAGTCTTCCGCACA |
| Reverse | ACAGGCTACAGCACCCAACCTT | ||
| fis1 | BT072691 | Forward | CCCCAGGGGGCATCCTGTCTTA |
| Reverse | TTGCAGCTGGCCGATCTAGCG | ||
| p53bp2 | XM014195885.1 | Forward | TTTTCCAGGCATCACCAATGAC |
| Reverse | CCAGATCAGCCATGATAGCGT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bou, M.; Torgersen, J.S.; Østbye, T.-K.K.; Ruyter, B.; Wang, X.; Škugor, S.; Kristiansen, I.Ø.; Todorčević, M. DHA Modulates Immune Response and Mitochondrial Function of Atlantic Salmon Adipocytes after LPS Treatment. Int. J. Mol. Sci. 2020, 21, 4101. https://doi.org/10.3390/ijms21114101
Bou M, Torgersen JS, Østbye T-KK, Ruyter B, Wang X, Škugor S, Kristiansen IØ, Todorčević M. DHA Modulates Immune Response and Mitochondrial Function of Atlantic Salmon Adipocytes after LPS Treatment. International Journal of Molecular Sciences. 2020; 21(11):4101. https://doi.org/10.3390/ijms21114101
Chicago/Turabian StyleBou, Marta, Jacob Seilø Torgersen, Tone-Kari Knutsdatter Østbye, Bente Ruyter, Xinxia Wang, Stanko Škugor, Inger Øien Kristiansen, and Marijana Todorčević. 2020. "DHA Modulates Immune Response and Mitochondrial Function of Atlantic Salmon Adipocytes after LPS Treatment" International Journal of Molecular Sciences 21, no. 11: 4101. https://doi.org/10.3390/ijms21114101
APA StyleBou, M., Torgersen, J. S., Østbye, T.-K. K., Ruyter, B., Wang, X., Škugor, S., Kristiansen, I. Ø., & Todorčević, M. (2020). DHA Modulates Immune Response and Mitochondrial Function of Atlantic Salmon Adipocytes after LPS Treatment. International Journal of Molecular Sciences, 21(11), 4101. https://doi.org/10.3390/ijms21114101






