Targeting KRAS Mutant Non-Small-Cell Lung Cancer: Past, Present and Future
Abstract
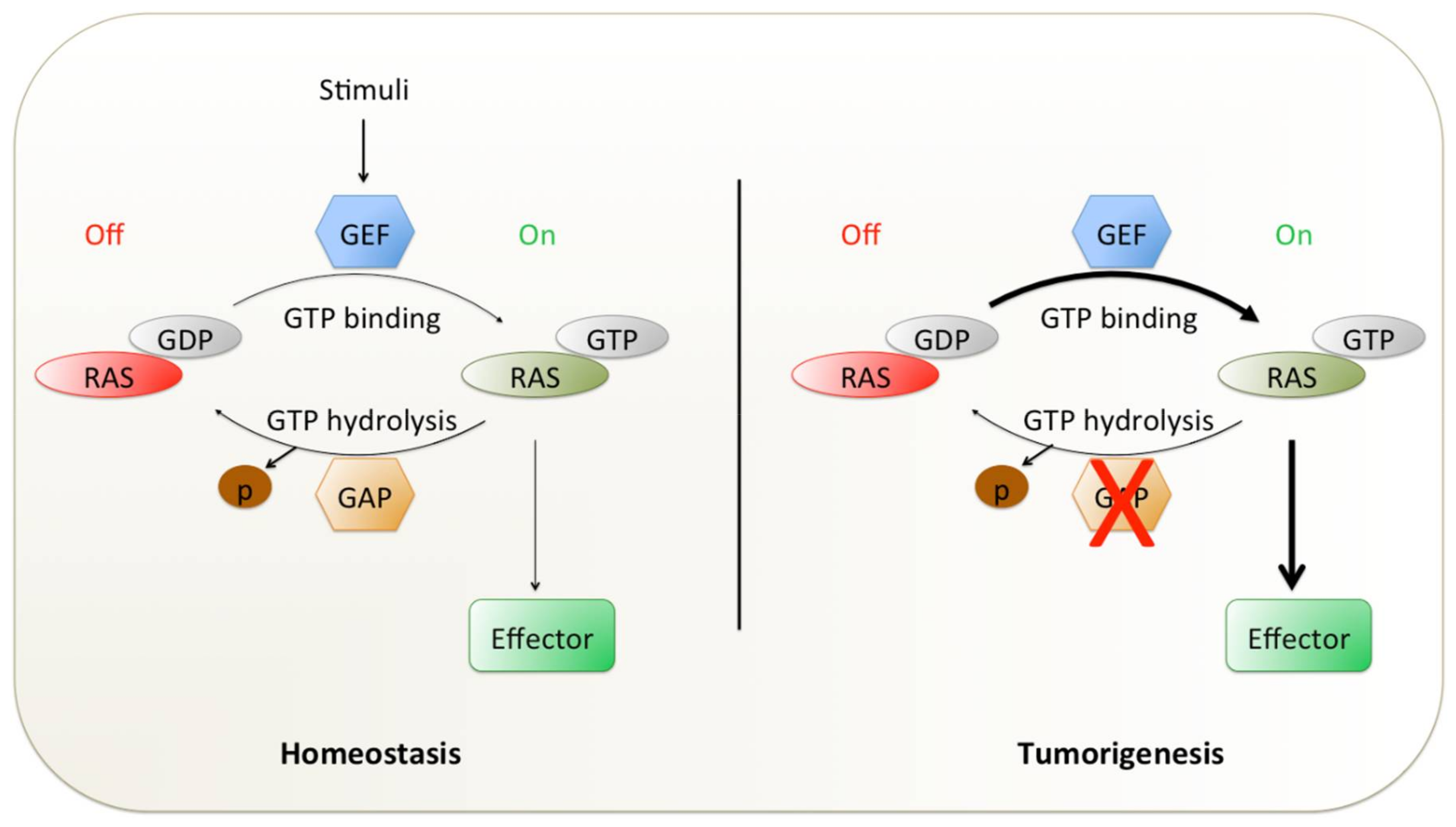
:1. Introduction
2. Molecular Diversity in KRAS Mutant NSCLC Influences Effective Targeting
3. Treatment Failures of the Past in Mutant KRAS NSCLC
3.1. Blocking KRAS Membrane Association
3.2. Synthetic Lethality Partners as Therapeutic Vulnerabilities
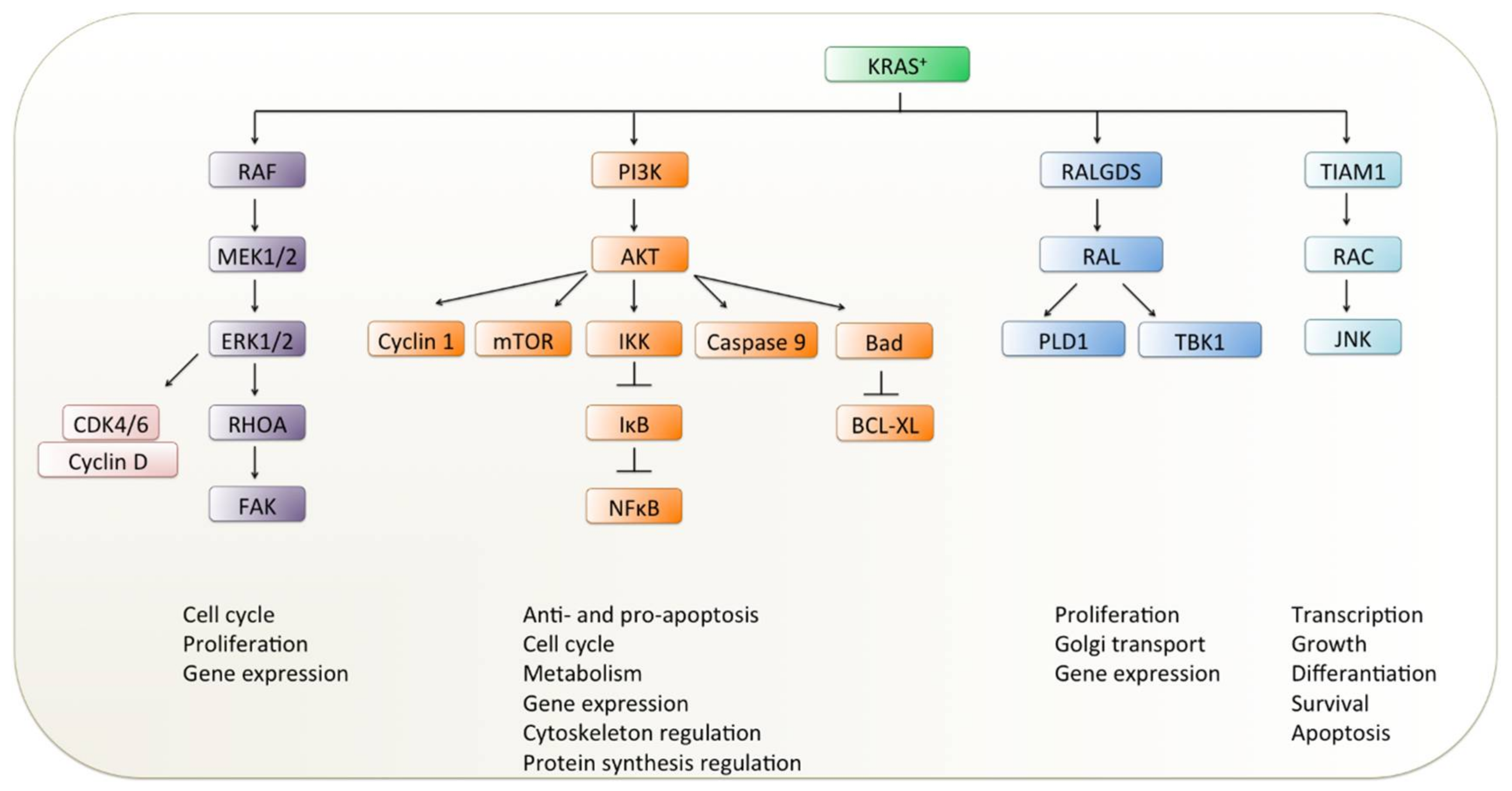
3.3. Targeting Downstream Effectors of KRAS Signaling
3.3.1. RAF Inhibitors
3.3.2. MEK Inhibitors
3.3.3. Other Downstream Inhibitors
4. New Optimism for Targeting KRAS Mutations
4.1. Covalent KRAS Inhibitors vs. KRAS Protein Degradation
4.2. Improved Combinatorial Downstream Strategies for KRAS Mutant Lung Tumors
4.3. Second Chance for EGFR-related Signaling in KRAS-driven NSCLC as Therapeutic Benefit
4.4. Metabolic Rewiring and Autophagy Inhibition
4.5. Immunotherapy
5. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| A-RAF | V-Raf murine sarcoma 3611 viral oncogene homolog 1 |
| AKT | Protein kinase B |
| ALK | Anaplastic lymphoma receptor tyrosine kinase |
| AMPK | AMP-activated protein kinase |
| APC/C | Anaphase-promoting complex/cyclosome |
| ASK1 | Apoptosis signal-regulating kinase 1 |
| ASO | Antisense-oligonucleotide |
| B-RAF | V-Raf murine sarcoma viral oncogene homolog B |
| Bad | BCL2-associated agonist of cell death |
| BCL-XL | B-cell lymphoma-extra large |
| BCL2 | B-cell lymphoma 2 |
| BET | Bromodomain and extraterminal domain |
| BRCA | Breast cancer gene |
| C-RAF | V-Raf-1 murine leukemia viral oncogene-like protein 1 |
| CDC25C | M-phase inducer phosphatase 3 |
| CDK | Cyclin-dependent kinase |
| CDKN2A/B | Cyclin-dependent kinase inhibitor 2A/B |
| Chk1 | Checkpoint kinase 1 |
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 |
| CSC | Cancer stem-like cell |
| CTLA-4 | Cytotoxic T-lymphocyte-associated protein 4 |
| DDR1 | Discoidin domain receptor 1 |
| EGFR | Epidermal growth factor receptor |
| ERK | Extracellular regulated kinase |
| FAK | Focal adhesion kinase |
| FDA | U.S. Food and Drug Administration |
| FGFR | Fibroblast growth factor receptor |
| FTI | Farnesyltransferase inhibitor |
| G12C | Glycine 12 to cysteine |
| G12D | Glycine 12 to aspartic acid |
| G12V | Glycine 12 to valine |
| GAP | GTPase-activating protein |
| GATA2 | GATA-binding protein 2 |
| GDP | Guanosine diphosphate |
| GEF | Guanine nucleotide exchange factor |
| GGTase I | Geranylgeranyltransferase I |
| GTP | Guanosine triphosphate |
| HER2–4/ERBB2–4 | Human epidermal growth factor receptors 2–4 |
| HLA | Human leukocyte antigen |
| HRAS | Harvey rat sarcoma viral oncogene homolog |
| HSP90 | Heat shock protein 90 |
| ICMT1 | Isoprenyl carboxyl methyltransferase |
| IGF1R | Insulin-like growth factor 1 receptor |
| IKK | IkappaB kinase |
| IκB | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor |
| JNK | c-Jun N-terminal kinase |
| KEAP1 | Kelch-like ECH-associated protein 1 |
| KRAS | Kirsten rat sarcoma viral oncogene homolog |
| LKB1 | Liver kinase B1 |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 |
| MAPK | Mitogen-activated protein kinase |
| MEK | Mitogen-activated protein kinase kinase |
| MHC II | Major histocompatibility complex class II |
| mTOR | Mechanistic target of rapamycin kinase |
| NF-κB | Nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells |
| NRAS | Neuroblastoma rat sarcoma viral oncogene homolog |
| NSCLC | Non-small-cell lung cancer |
| ORR | Objective response rate |
| p53 | Tumor protein p53 |
| p90RSK | p90 ribosomal S6 kinase |
| PARP | Poly(ADP-ribose)polymerase |
| PD-1 | Programmed cell death protein 1 |
| PD-L1 | Programmed cell death 1 ligand 1 |
| PDX | Patient-derived xenograft |
| pERK | Phosphorylated ERK |
| PFS | Progression-free survival |
| PI3K | Phosphoinositide 3-kinase |
| PLD1 | Phospholipase D1 |
| PLK1 | Polo-like kinase 1 |
| PROTAC | Proteolysis-targeting chimera |
| PTEN | Phosphatase and tensin homolog |
| PTPN11 | Protein tyrosine phosphatase non-receptor type 11 |
| RAC | Ras-related C3 botulinum toxin substrate 1 |
| RAF | Rat fibrosarcoma |
| RAL | Ras-like protein |
| RALGDS | Ral guanine nucleotide dissociation stimulator |
| RBD | RAS-binding domain |
| RCE1 | RAS-converting enzyme 1 |
| RET | RET proto-oncogene |
| RHOA | Ras homolog family member |
| RNAi | RNA interference |
| ROCK2 | Rho-associated coiled-coil containing protein kinase 2 |
| ROS | Proto-oncogene tyrosine-protein kinase ROS |
| RTK | Receptor tyrosine kinase |
| SNAIL2 | Snail family transcriptional repressor 2 |
| SRC | V-Src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| STK11 | Serine/threonine kinase 11 |
| STK3 | Serine/threonine-protein kinase 3 |
| TBK1 | TANK binding kinase 1 |
| TCA | Tricarboxylic acid cycle |
| TCR | T cell receptor |
| Th17 | T helper 17 |
| TIAM1 | T lymphoma invasion and metastasis-inducing protein 1 |
| Tim-3 | T cell immunoglobulin mucin-3 |
| TKI | Tyrosine kinase inhibitor |
| TPX2 | Targeting protein for Xklp2 |
| Treg | Regulatory T cells |
| ULK1 | Unc-51-like autophagy activating kinase 1 |
| VHL | von Hippel–Lindau disease tumor suppressor |
| WT1 | Wilms tumor 1 |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA. Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Socinski, M.A.; Evans, T.; Gettinger, S.; Hensing, T.A.; Van Dam Sequist, L.; Ireland, B.; Stinchcombe, T.E. Treatment of stage IV non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American college of chest physicians evidence-based clinical practice guidelines. Chest 2013, 143, e341S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polo, V.; Besse, B. Maintenance strategies in stage IV non-small-cell lung cancer (NSCLC): in which patients, with which drugs? Ann. Oncol. 2013, 25, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, M.A.T.; Gu, J.; Wu, X. Pharmacogenomics of platinum-based chemotherapy in NSCLC. Expert Opin. Drug Metab. Toxicol. 2009, 5, 745–755. [Google Scholar] [CrossRef] [Green Version]
- Ahmadzada, T.; Kao, S.; Reid, G.; Noyer, M.; Mahar, A.; Cooper, W.A. An Update on Predictive Biomarkers for Treatment Selection in Non-Small Cell Lung Cancer. J. Clin. Med. 2018, 7, 153. [Google Scholar] [CrossRef] [Green Version]
- Furugaki, K.; Mochizuki, M.; Kohno, M.; Shu, S.; Harada, N.; Yoshimura, Y. Expression of C-terminal ALK, RET, or ROS1 in lung cancer cells with or without fusion. BMC Cancer 2019, 19, 301. [Google Scholar] [CrossRef] [Green Version]
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 2014, 13, 828–851. [Google Scholar] [CrossRef] [Green Version]
- Kranenburg, O. The KRAS oncogene: past, present, and future. Biochim. Biophys. Acta. 2005, 1756, 81–82. [Google Scholar] [CrossRef]
- Karnoub, A.E.; Weinberg, R.A. Ras oncogenes: Split personalities. Nat. Rev. Mol. Cell Biol. 2008, 9, 517–531. [Google Scholar] [CrossRef] [Green Version]
- Cox, A.D.; Der, C.J. Ras history: The saga continues. Small GTPases 2010, 1, 2–27. [Google Scholar] [CrossRef] [Green Version]
- Khosravi-Far, R.; Solski, P.A.; Clark, G.J.; Kinch, M.S.; Der, C.J. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol. Cell. Biol. 1995, 15, 6443–6453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leevers, S.J.; Paterson, H.F.; Marshall, C.J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature 1994, 369, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Ramjaun, A.R.; Haiko, P.; Wang, Y.; Warne, P.H.; Nicke, B.; Nye, E.; Stamp, G.; Alitalo, K.; Downward, J. Binding of Ras to Phosphoinositide 3-Kinase p110a Is Required for Ras-Driven Tumorigenesis in Mice. Cell 2007, 129, 957–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Viciana, P.; Warne, P.H.; Khwaj, A.A.; Marte, B.M.; Pappin, D.; Das, P.; Waterfield, M.D.; Ridley, A.; Downward, J. Role of Phosphoinositide 3-OH Kinase in Cell Transformation and Control of the Actin Cytoskeleton by Ras. Cell 1997, 89, 457–467. [Google Scholar] [CrossRef] [Green Version]
- Hofer, F.; Fields, S.; Schneider, C.; Martin, G.S. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc. Natl. Acad. Sci. USA 1994, 91, 11089–11093. [Google Scholar] [CrossRef] [Green Version]
- Bodemann, B.O.; White, M.A. Ral GTPases and cancer: Linchpin support of the tumorigenic platform. Nat. Rev. Cancer 2008, 8, 133–140. [Google Scholar] [CrossRef]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Medarde, A.; Santos, E. Ras in cancer and developmental diseases. Genes and Cancer 2011, 2, 344–358. [Google Scholar] [CrossRef] [Green Version]
- Dearden, S.; Stevens, J.; Wu, Y.-L.; Blowers, D. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann. Oncol. 2013, 24, 2371–2376. [Google Scholar] [CrossRef]
- Bar-Sagi, D.; Knelson, E.H.; Sequist, L.V. A bright future for KRAS inhibitors. Nat. Cancer 2020, 1, 25–27. [Google Scholar] [CrossRef] [Green Version]
- Skoulidis, F.; Byers, L.A.; Diao, L.; Papadimitrakopoulou, V.A.; Tong, P.; Izzo, J.; Behrens, C.; Kadara, H.; Parra, E.R.; Canales, J.R.; et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015, 5, 861–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riely, G.J.; Jordan, E.; Kim, H.R.; Yu, H.A.; Solit, D.B.; Kris, M.G.; Ni, A.; Arcila, M.E.; Ladanyi, M. Association of outcomes and co-occuring genomic alterations in patients with KRAS -mutant non-small cell lung cancer. J. Clin. Oncol. 2016, 34 15 SUPPL, 9019. [Google Scholar] [CrossRef]
- El Osta, B.E.; Behera, M.; Kim, S.; Berry, L.D.; Sica, G.; Pillai, R.N.; Owonikoko, T.K.; Kris, M.G.; Johnson, B.E.; Kwiatkowski, D.J.; et al. Characteristics and Outcomes of Patients With Metastatic KRAS-Mutant Lung Adenocarcinomas: The Lung Cancer Mutation Consortium Experience. J. Thorac. Oncol. 2019, 14, 876–889. [Google Scholar] [CrossRef] [PubMed]
- Dogan, S.; Shen, R.; Ang, D.C.; Johnson, M.L.; D’Angelo, S.P.; Paik, P.K.; Brozostowski, E.B.; Riely, G.J.; Kris, M.G.; Zakowski, M.F.; et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: Higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin. Cancer Res. 2012, 18, 6169–6177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Osta, B.E.; Behera, M.; Kim, S.; Berry, L.D.; Sica, G.; Pillai, R.N.; Owonikoko, T.K.; Kris, M.G.; Johnson, B.E.; Kwiatkowski, D.J.; et al. Characteristics and outcomes of patients (pts) with metastatic KRAS mutant lung adenocarcinomas: Lung Cancer Mutation Consortium (LCMC) database. J. Clin. Oncol. 2017, 35 15 SUPPL, 9021. [Google Scholar] [CrossRef]
- Redig, A.J.; Chambers, E.S.; Lydon, C.A.; Dahlberg, S.E.; Alden, R.S.; Janne, P.A. Genomic complexity in KRAS mutant non-small cell lung cancer (NSCLC) from never/light-smokers v smokers. J. Clin. Oncol. 2016, 34) 15 SUPPL, 9087. [Google Scholar] [CrossRef]
- Muñoz-Maldonado, C.; Zimmer, Y.; Medová, M. A comparative analysis of individual ras mutations in cancer biology. Front. Oncol. 2019, 9, 1088. [Google Scholar] [CrossRef] [Green Version]
- Ihle, N.T.; Byers, L.A.; Kim, E.S.; Sintigny, P.; Lee, J.J.; Blumenschein, G.R.; Tsao, A.; Liu, S.; Larsen, J.E.; Wang, J.; et al. Effect of KRAS oncogene substitutions on protein behavior: Implications for signaling and clinical outcome. J. Natl. Cancer Inst. 2012, 104, 228–239. [Google Scholar] [CrossRef]
- Nadal, E.; Chen, G.; Prensner, J.R.; Shiratsuchi, H.; Sam, C.; Zhao, L.; Kalemkerian, G.P.; Brenner, D.; Lin, J.; Reddy, R.M.; et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J. Thorac. Oncol. 2014, 9, 1513–1522. [Google Scholar] [CrossRef] [Green Version]
- Shepherd, F.A.; Domerg, C.; Hainaut, P.; Jänne, P.A.; Pignon, J.P.; Graziano, S.; Douillard, J.-Y.; Brambilla, E.; Chevalier, T.L.; Seymour, L.; et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non–small-cell lung cancer in four trials of adjuvant chemotherapy. J. Clin. Oncol. 2013, 31, 2173–2181. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.A.; Sima, C.S.; Shen, R.; Kass, S.; Gainor, J.; Shaw, A.; Hame, S.M.; Iam, S.W.; Aston, J.; Lovly, C.M.; et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J. Thorac. Oncol. 2015, 10, 431–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimmelman, A.C. Metabolic dependencies in RAS-driven cancers. Clin. Cancer Res. 2015, 21, 1828–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerr, E.M.; Gaude, E.; Turrell, F.K.; Frezza, C.; Martins, C.P. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 2016, 531, 110–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsay, C.R.; Jamal-Hanjani, M.; Forster, M.; Blackhall, F. KRAS: Reasons for optimism in lung cancer. Eur. J. Cancer 2018, 99, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Cox, A.D.; Der, C.J.; Philips, M.R. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin. Cancer Res. 2015, 21, 1819–1827. [Google Scholar] [CrossRef] [Green Version]
- Adjei, A.A.; Mauer, A.; Bruzek, L.; Marks, R.S.; Hillman, S.; Geyer, S.; Hanson, L.J.; Wright, J.J.; Erlichman, C.; Kaufmann, S.H.; et al. Phase II study of the farnesyl transferase inhibitor R115777 in patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 2003, 21, 1760–1766. [Google Scholar] [CrossRef]
- Kim, E.S.; Kies, M.S.; Fossella, F.V.; Glisson, B.S.; Zaknoen, S.; Statkevich, P.; Munden, R.F.; Summey, C.; Pisters, K.M.W.; Papadimitrakopoulou, V.; et al. Phase II study of the farnesyltransferase inhibitor lonafarnib with paclitaxel in patients with taxane-refractory/resistant nonsmall cell lung carcinoma. Cancer 2005, 104, 561–569. [Google Scholar] [CrossRef]
- Rowell, C.A.; Kowalczyk, J.J.; Lewis, M.D.; Garcia, A.M. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J. Biol. Chem. 1997, 272, 14093–14097. [Google Scholar] [CrossRef] [Green Version]
- Whyte, D.B.; Kirschmeier, P.; Hockenberry, T.N.; Nunew-Oliva, I.; James, L.; Catino, J.J.; Bishop, W.R.; Pai, J.K. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 1997, 272, 14459–14464. [Google Scholar] [CrossRef] [Green Version]
- Lobell, R.B.; Liu, D.; Buser, C.A.; Davide, J.P.; DePuy, E.; Hamilton, K.; Koblan, K.S.; Lee, Y.; Mosser, S.; Motzel, S.L.; et al. Preclinical and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol. Cancer Ther. 2002, 1, 747–758. [Google Scholar]
- Riely, G.J.; Johnson, M.L.; Medina, C.; Rizvi, N.A.; Miller, V.A.; Kris, M.G.; Pietanza, M.C.; Azzoli, C.G.; Krug, L.M.; Pao, W.; et al. A phase II trial of Salirasib in patients with lung adenocarcinomas with KRAS mutations. J. Thorac. Oncol. 2011, 6, 1435–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotblat, B.; Ehrlich, M.; Haklai, R.; Kloog, Y. The Ras inhibitor farnesylthiosalicylic acid (Salirasib) disrupts the spatiotemporal localization of active Ras: a potential treatment for cancer. Methods Enzymol. 2008, 439, 467–489. [Google Scholar] [PubMed]
- Zundelevich, A.; Elad-Sfadia, G.; Haklai, R.; Kloog, Y. Suppression of lung cancer tumor growth in a nude mouse model by the Ras inhibitor salirasib (farnesylthiosalicylic acid). Mol. Cancer Ther. 2007, 6, 1765–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahlstrom, A.M.; Cutts, B.A.; Karlsson, C.; Andersson, K.M.E.; Liu, M.; Sjogren, A.-K.M.; Swolin, B.; Young, S.G.; Bergo, M.O. Rce1 deficiency accelerates the development of K-RAS-induced myeloproliferative disease. Blood 2007, 109, 763–768. [Google Scholar] [CrossRef] [Green Version]
- Wahlstrom, A.M.; Cutts, B.A.; Liu, M.; Lindskog, A.; Karlsson, C.; Sjogren, A.-K.M.; Andersson, K.M.E.; Young, S.G.; Bergo, M.O. Inactivating Icmt ameliorates K-RAS-induced myeloproliferative disease. Blood 2008, 112, 1357–1365. [Google Scholar] [CrossRef] [Green Version]
- Court, H.; Amoyel, M.; Hackman, M.; Lee, K.E.; Xu, R.; Miller, G.; Bar-Sagi, D.; Bach, E.A.; Bergö, M.O.; Philips, M.R. Isoprenylcysteine carboxylmethyltransferase deficiency exacerbates KRAS-driven pancreatic neoplasia via Notch suppression. J. Clin. Invest. 2013, 123, 4681–4694. [Google Scholar] [CrossRef] [Green Version]
- Majmudar, J.D.; Hodges-Loaiza, H.B.; Hahne, K.; Donelson, J.L.; Song, J.; Shrestha, L.; Harrison, M.L.; Hrycyna, C.A.; Gibbs, R.A. Amide-modified prenylcysteine based Icmt inhibitors: Structure-activity relationships, kinetic analysis and cellular characterization. Bioorg. Med. Chem. 2012, 20, 283–295. [Google Scholar] [CrossRef] [Green Version]
- Manandhar, S.P.; Hildebrandt, E.R.; Schmidt, W.K. Small-molecule inhibitors of the Rce1p CaaX protease. J. Biomol. Screen 2007, 12, 983–993. [Google Scholar] [CrossRef] [Green Version]
- Winter-Vann, A.M.; Baron, R.A.; Wong, W.; Dela Cruz, J.; York, J.D.; Gooden, D.M.; Bergo, M.O.; Young, S.G.; Toone, E.J.; Casey, P.J. A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc. Natl. Acad. Sci. USA 2005, 102, 4336–4341. [Google Scholar] [CrossRef] [Green Version]
- Bridges, C.B. The Origin of Variations in Sexual and Sex-Limited Characters. Am. Nat. 1922, 56, 51–63. [Google Scholar] [CrossRef]
- Dobzhansky, T. Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. Genetics 1946, 31, 269–290. [Google Scholar] [PubMed]
- Farmer, H.; McCabe, H.; Lord, C.J.; Tutt, A.N.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Balmaña, J.; Domchek, S.M.; Tutt, A.; Garber, J.E. Stumbling blocks on the path to personalized medicine in breast cancer: The case of PARP inhibitors for BRCA1/2 -associated cancers. Cancer Discov. 2011, 1, 29–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.A.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S.; Hancock, D.C.; Molina-Arcas, M.; Steckel, M.; East, P.; Diefenbacher, M.; Armenteros-Monterroso, E.; Lassailly, F.; Matthews, N.; Nye, E.; et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell 2012, 149, 642–655. [Google Scholar] [CrossRef] [Green Version]
- Steckel, M.; Molina-Arcas, M.; Weigelt, B.; Marani, M.; Warne, P.H.; Kuznetsov, H.; Kelly, G.; Saunders, B.; Howell, M.; Downward, J.; et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 2012, 22, 1227–1245. [Google Scholar] [CrossRef] [Green Version]
- Ferrer, I.; Zugazagoitia, J.; Herbertz, S.; John, W.; Paz-Ares, L.; Schmid-Bindert, G. KRAS-Mutant non-small cell lung cancer: From biology to therapy. Lung Cancer 2018, 124, 53–64. [Google Scholar] [CrossRef] [Green Version]
- Downward, J. RAS Synthetic Lethal Screens Revisited: Still Seeking the Elusive Prize? Clin. Cancer Res. 2015, 21, 1802–1809. [Google Scholar] [CrossRef] [Green Version]
- Scholl, C.; Fröhling, S.; Dunn, I.F.; Schinzel, A.C.; Barbie, D.A.; Kim, S.Y.; Silver, S.J.; Tamayo, P.; Wadlow, R.C.; Ramaswamy, S.; et al. Synthetic Lethal Interaction between Oncogenic KRAS Dependency and STK33 Suppression in Human Cancer Cells. Cell 2009, 137, 821–834. [Google Scholar] [CrossRef] [Green Version]
- Luo, T.; Masson, K.; Jaffe, J.D.; Silkworth, W.; Ross, N.T.; Scherer, C.A.; Scholl, C.; Fröhling, S.; Carr, S.A.; Stern, A.M.; et al. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proc. Natl. Acad. Sci. USA 2012, 109, 2860–2865. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.; Mao, C.-Q.; Yang, X.-Z.; Du, X.-J.; Liu, Y.; Zhu, Y.-H.; Wang, J. Cationic lipid-assisted polymeric nanoparticle mediated GATA2 siRNA delivery for synthetic lethal therapy of KRAS mutant non-small-cell lung carcinoma. Mol. Pharm. 2014, 11, 2612–2622. [Google Scholar] [CrossRef] [PubMed]
- Tessema, M.; Yingling, C.M.; Snider, A.M.; Do, K.; Juri, D.E.; Picchi, M.A.; Zhang, X.; Liu, Y.; Leng, S.; Tellez, C.S.; et al. GATA2 is epigenetically repressed in human and mouse lung tumors and is not requisite for survival of KRAS mutant lung cancer. J. Thorac. Oncol. 2014, 9, 784–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litvak, A.M.; Drilon, A.E.; Rekhtman, N.; Pietanza, M.C.; Chaft, J.E.; Woo, K.; Paik, P.K.; Kris, M.G.; Riely, G.J. Phase II trial of bortezomib in KRAS G12D mutant lung cancers. J. Clin. Oncol. 2015, 33 15 SUPPL, e19002. [Google Scholar] [CrossRef]
- Drilon, A.; Schoenfeld, A.J.; Arbour, K.C.; Litvak, A.; Ni, A.; Montecalvi, J.; Yu, H.A.; Panora, E.; Ahn, L.; Kennedy, M.; et al. Exceptional responders with invasive mucinous adenocarcinomas: a phase 2 trial of bortezomib in patients with KRAS G12D-mutant lung cancers. Cold Spring Harb. Mol. case Stud. 2019, 5, a003665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corcoran, R.B.; Cheng, K.A.; Hata, A.N.; Faber, A.C.; Ebi, H.; Coffee, E.M.; Greninger, P.; Brown, R.D.; Godfrey, J.T.; Cohoon, T.J.; et al. Synthetic Lethal Interaction of Combined BCL-XL and MEK Inhibition Promotes Tumor Regressions in KRAS Mutant Cancer Models. Cancer Cell 2013, 23, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Ambrogio, C.; Gómez-López, G.; Falcone, M.; Vidal, A.; Nadal, E.; Crosetto, N.; Blasco, R.B.; Fernández-Marcos, P.J.; Sánchez-Céspedes, M.; Ren, X.; et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat. Med. 2016, 22, 270–277. [Google Scholar] [CrossRef]
- Puyol, M.; Martín, A.; Dubus, P.; Mulero, F.; Pizcueta, P.; Khan, G.; Guerra, C.; Santamaría, D.; Barbacid, M. A Synthetic Lethal Interaction between K-Ras Oncogenes and Cdk4 Unveils a Therapeutic Strategy for Non-small Cell Lung Carcinoma. Cancer Cell 2010, 18, 63–73. [Google Scholar] [CrossRef]
- Patnaik, A.; Rosen, L.S.; Tolaney, S.M.; Tolcher, A.W.; Goldman, J.W.; Gandhi, L.; Papadopoulos, K.P.; Beeram, M.; Rasco, D.W.; Hilton, J.F.; et al. Efficacy and safety of Abemaciclib, an inhibitor of CDK4 and CDK6, for patients with breast cancer, non–small cell lung cancer, and other solid tumors. Cancer Discov. 2016, 6, 740–753. [Google Scholar] [CrossRef] [Green Version]
- Goldman, J.W.; Shi, P.; Reck, M.; Paz-Ares, L.; Koustenis, A.; Hurt, K.C. Treatment Rationale and Study Design for the JUNIPER Study: A Randomized Phase III Study of Abemaciclib With Best Supportive Care Versus Erlotinib With Best Supportive Care in Patients With Stage IV Non-Small-Cell Lung Cancer With a Detectable KRAS Mutation Whose Disease Has Progressed After Platinum-Based Chemotherapy. Clin. Lung Cancer 2016, 17, 80–84. [Google Scholar]
- Dietlein, F.; Kalb, B.; Jokic, M.; Noll, E.M.; Strong, A.; Tharun, L.; Ozretić, L.; Künstlinger, H.; Kambartel, K.; Randerath, W.J.; et al. A Synergistic Interaction between Chk1- and MK2 Inhibitors in KRAS-Mutant Cancer. Cell 2015, 162, 146–159. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Liang, S.-Q.; Schmid, R.A.; Peng, R.-W. New horizons in KRAS-mutant lung cancer: Dawn after darkness. Front. Oncol. 2019, 9, 953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drosten, M.; Barbacid, M. Targeting the MAPK Pathway in KRAS-Driven Tumors. Cancer Cell 2020, 37, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Blumenschein, G.R., Jr.; Saintigny, P.; Liu, S.; Kim, E.S.; Tsao, A.S.; Herbst, R.S.; Alden, C.; Lee, J.J.; Tang, X.; Stewart, D.J.; et al. Comprehensive biomarker analysis and final efficacy results of sorafenib in the BATTLE (Biomarker-Integrated Approaches of Targeted Therapy for Lung Cancer Elimination) trial. Clin. Cancer Res. 2013, 19, 6967–6975. [Google Scholar]
- Dingemans, A.-M.C.; Mellema, W.W.; Groen, H.J.M.; Van Wijk, A.; Burgers, S.A.; Kunst, P.W.A.; Thunnissen, E.; Heideman, D.A.M.; Smit, E.F. A phase II study of sorafenib in patients with platinum-pretreated, advanced (Stage IIIb or IV) non-small cell lung cancer with a KRAS mutation. Clin. Cancer Res. 2013, 19, 743–751. [Google Scholar] [CrossRef] [Green Version]
- Papadimitrakopoulou, V.; Lee, J.J.; Wistuba, I.I.; Tsao, A.S.; Fossella, F.V.; Kalhor, N.; Gupta, S.; Byers, L.A.; Izzo, J.G.; Gettinger, S.N.; et al. The BATTLE-2 study: A biomarker-integrated targeted therapy study in previously treated patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 2016, 34, 3638–3647. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Hirsh, V.; Zhang, L.; De Marinis, F.; Yang, J.C.-H.; Wakelee, H.A.; Seto, T.; Wu, Y.L.; Novello, S.; Juhász, E.; et al. Monotherapy Administration of Sorafenib in Patients with Non-Small Cell Lung Cancer (MISSION) Trial: A Phase III, Multicenter, Placebo-Controlled Trial of Sorafenib in Patients with Relapsed or Refractory Predominantly Nonsquamous Non-Small-Cell Lung Cancer after 2 or 3 Previous Treatment Regimens. J. Thorac. Oncol. 2015, 10, 1745–1753. [Google Scholar]
- Blasco, R.B.; Francoz, S.; Santamaría, D.; Cañamero, M.; Dubus, P.; Charron, J.; Baccarini, M.; Barbacid, M. C-Raf, but Not B-Raf, Is Essential for Development of K-Ras Oncogene-Driven Non-Small Cell Lung Carcinoma. Cancer Cell 2011, 19, 652–663. [Google Scholar] [CrossRef] [Green Version]
- Karreth, F.A.; Frese, K.K.; DeNicola, G.M.; Baccarini, M.; Tuveson, D.A. C-Raf is required for the initiation of lung cancer by K-Ras G12D. Cancer Discov. 2011, 1, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Sanclemente, M.; Francoz, S.; Esteban-Burgos, L.; Bousquet-Mur, E.; Djurec, M.; Lopez-Casas, P.P.; Hidalgo, M.; Guerra, C.; Drosten, M.; Musteanu, M.; et al. c-RAF Ablation Induces Regression of Advanced Kras/Trp53 Mutant Lung Adenocarcinomas by a Mechanism Independent of MAPK Signaling. Cancer Cell 2018, 33, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Cui, D.; Chen, X.; Xiong, X.; Zhao, Y. PROTACs: An Emerging Targeting Technique for Protein Degradation in Drug Discovery. Bioessays 2018, 40, e1700247. [Google Scholar] [CrossRef]
- Yaeger, R.; Corcoran, R.B. Targeting Alterations in the RAF–MEK Pathway. Cancer Discov. 2019, 9, 329–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lito, P.; Saborowski, A.; Yue, J.; Solomon, M.; Joseph, E.; Gadal, S.; Saborowski, M.; Kastenhuber, E.; Fellmann, C.; Ohara, K.; et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 2014, 25, 697–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, C.A.; Rajan, A.; Keen, C.; Szabo, E.; Khozin, S.; Thomas, A.; Brzezniak, C.; Guha, U.; Doyle, L.A.; Steinberg, S.M.; et al. Selumetinib with and without erlotinib in KRAS mutant and KRAS wild-type advanced nonsmall-cell lung cancer. Ann. Oncol. 2016, 27, 693–699. [Google Scholar] [CrossRef]
- Hainsworth, J.D.; Cebotaru, C.L.; Kanarev, V.; Ciuleanu, T.E.; Damyanov, D.; Stella, P.; Ganchev, H.; Pover, G.; Morris, C.; Tzekova, V. A phase II, open-label, randomized study to assess the efficacy and safety of AZD6244 (ARRY-142886) versus pemetrexed in patients with non-small cell lung cancer who have failed one or two prior chemotherapeutic regimens. J. Thorac. Oncol. 2010, 5, 1630–1636. [Google Scholar] [CrossRef] [Green Version]
- Jänne, P.A.; Shaw, A.T.; Pereira, J.R.; Jeannin, G.; Vansteenkist, J.; Barrios, C.; Franke, F.A.; Grinsted, L.; Zazulina, V.; Smith, P.; et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: A randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013, 14, 38–47. [Google Scholar]
- Jänne, P.A.; Van Den Heuvel, M.M.; Barlesi, F.; Cobo, M.; Mazieres, J.; Crinò, L.; Orlov, S.; Blackhall, F.; Wolf, J.; Garrido, P.; et al. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: The SELECT-1 randomized clinical trial. J. Am. Med. Assoc. 2017, 317, 1844–1853. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Cheng, K.; Walton, Z.; Wang, Y.; Ebi, H.; Shimamura, T.; Liu, Y.; Tupper, T.; Ouyang, J.; Li, J.; et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012, 483, 613–617. [Google Scholar] [CrossRef]
- Blumenschein, G.R., Jr.; Smit, E.F.; Planchard, D.; Kim, D.-W.; Cadranel, J.; De Pas, T.; Dunphy, F.; Udud, K.; Ahn, M.-J.; Hanna, N.H.; et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC). Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 894–901. [Google Scholar] [CrossRef]
- Gadgeel, S.M.; Miao, J.; Riess, J.W.; Mack, P.C.; Gerstner, G.J.; Burns, T.F.; Taj, A.; Akerley, W.L.; Dragnev, K.H.; Moon, J.; et al. S1507: Phase II study of docetaxel and trametinib in patients with G12C or non-G12C KRAS mutation positive (+) recurrent non-small cell lung cancer (NSCLC). J. Clin. Oncol. 2019, 37 15 SUPPL, 9021. [Google Scholar] [CrossRef]
- Castellano, E.; Downward, J. Ras interaction with PI3K: More than just another effector pathway. Genes and Cancer 2011, 2, 261–274. [Google Scholar] [CrossRef] [Green Version]
- Salt, M.B.; Bandyopadhyay, S.; McCormick, F. Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov. 2014, 4, 186–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misale, S.; Fatherree, J.P.; Cortez, E.; Li, C.; Bilton, S.; Timonina, D.; Myers, D.T.; Lee, D.; Gomez-Caraballo, M.; Greenberg, M.; et al. KRAS G12C NSCLC models are sensitive to direct targeting of KRAS in combination with PI3K inhibition. Clin. Cancer Res. 2019, 25, 796–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riely, G.J.; Brahmer, J.R.; Planchard, D.; Crinò, L.; Doebele, R.C.; Mas Lopez, L.A.; Gettinger, S.N.; Schumann, C.; Li, X.; Atkins, B.M.; et al. A randomized discontinuation phase II trial of ridaforolimus in non-small cell lung cancer (NSCLC) patients with KRAS mutations. J. Clin. Oncol. 2012, 30 15 SUPPL, 7531. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Patnaik, A.; Papadopoulos, K.P.; Rasco, D.W.; Becerra, C.R.; Allred, A.J.; Orford, K.; Aktan, G.; Ferron-Brady, G.; Ibrahim, N.; et al. Phase I study of the MEK inhibitor trametinib in combination with the AKT inhibitor afuresertib in patients with solid tumors and multiple myeloma. Cancer Chemother. Pharmacol. 2015, 75, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Mita, M.; Fu, S.; Piha-Paul, S.A.; Janku, F.; Mita, A.; Natale, R.; Guo, W.; Zhao, C.; Kurzrock, R.; Naing, A. Phase I trial of MEK 1/2 inhibitor pimasertib combined with mTOR inhibitor temsirolimus in patients with advanced solid tumors. Invest. New Drugs 2017, 35, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Schram, A.M.; Gandhi, L.; Mita, M.M.; Damstrup, L.; Campana, F.; Hidalgo, M.; Grande, E.; Hyman, D.M.; Heist, R.S. A phase Ib dose-escalation and expansion study of the oral MEK inhibitor pimasertib and PI3K/MTOR inhibitor voxtalisib in patients with advanced solid tumours. Br. J. Cancer 2018, 119, 1471–1476. [Google Scholar] [CrossRef]
- Konstantinidou, G.; Ramadori, G.; Torti, F.; Kangasniemi, K.; Ramirez, R.E.; Cai, Y.; Behrens, C.; Dellinger, M.T.; Brekken, R.A.; Wistuba, I.I.; et al. RHOA-FAK is a required signaling axis for the maintenance of KRAS-driven lung adenocarcinomas. Cancer Discov. 2013, 3, 444–457. [Google Scholar] [CrossRef] [Green Version]
- Gerber, D.E.; Ross Camidge, D.; Morgensztern, D.; Cetnar, J.; Kelly, R.J.; Ramalingam, S.S.; Spigel, D.R.; Jeong, W.; Scaglioni, P.P.; Zhang, S.; et al. Phase 2 study of the focal adhesion kinase inhibitor defactinib (VS-6063) in previously treated advanced KRAS mutant non-small cell lung cancer. Lung Cancer 2020, 139, 60–67. [Google Scholar] [CrossRef]
- Mak, G.; Soria, J.-C.; Blagden, S.P.; Plummer, R.; Fleming, R.A.; Nebot, N.; Zhang, J.; Mazumdar, J.; Rogan, D.; Gazzah, A.; et al. A phase Ib dose-finding, pharmacokinetic study of the focal adhesion kinase inhibitor GSK2256098 and trametinib in patients with advanced solid tumours. Br. J. Cancer. 2019, 120, 975–981. [Google Scholar] [CrossRef]
- Park, K.-S.; Yang, H.; Choi, J.; Seo, S.; Kim, D.; Lee, C.H.; Jeon, H.; Kim, S.-W.; Lee, D.H. The HSP90 inhibitor, NVP-AUY922, attenuates intrinsic PI3K inhibitor resistance in KRAS-mutant non-small cell lung cancer. Cancer Lett. 2017, 406, 47–53. [Google Scholar] [CrossRef]
- Park, K.-S.; Oh, B.; Lee, M.H.; Jin, H.R.; Nam, K.-Y.; Yang, H.; Choi, Y.; Kim, S.-W.; Lee, D.H. The HSP90 inhibitor, NVP-AUY922, sensitizes KRAS-mutant non-small cell lung cancer with intrinsic resistance to MEK inhibitor, trametinib. Cancer Lett. 2016, 372, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Le, H.T.; Nguyen, H.T.; Min, H.-Y.; Hyun, S.Y.; Kwon, S.; Lee, Y.; Van Le, T.H.; Lee, J.; Park, J.H.; Lee, H.-Y. Panaxynol, a natural Hsp90 inhibitor, effectively targets both lung cancer stem and non-stem cells. Cancer Lett. 2018, 412, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Huang, E.H.-B.; Christie, I.; Kurland, B.F.; Burns, T.F. Acquired resistance to the Hsp90 inhibitor, ganetespib, in KRAS-Mutant NSCLC is mediated via reactivation of the ERK-p90RSK-mTOR signaling network. Mol. Cancer Ther. 2017, 16, 793–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, S.; Huang, E.H.-B.; Christie, I.; Burns, T.F. Reactivation of the p90RSK–CDC25C pathway leads to bypass of the ganetespib-induced G2–M arrest and mediates acquired resistance to ganetespib in KRAS-mutant NSCLC. Mol. Cancer Ther. 2017, 16, 1658–1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrem, J.M.L.; Shokat, K.M. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat. Rev. Drug Discov. 2016, 15, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.M.; Westover, K.D.; Ficarro, S.B.; Harrison, R.A.; Choi, H.G.; Pacold, M.E.; Carrasco, M.; Hunter, J.; Kim, N.D.; Xie, T.; et al. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew. Chem. Int. Ed. Engl. 2014, 53, 199–204. [Google Scholar] [CrossRef]
- Ostrem, J.M.; Peters, U.; Sos, M.L.; Wells, J.A.; Shokat, K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503, 548–551. [Google Scholar] [CrossRef] [Green Version]
- Lito, P.; Solomon, M.; Li, L.S.; Hansen, R.; Rosen, N. Cancer therapeutics: Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 2016, 351, 604–608. [Google Scholar] [CrossRef] [Green Version]
- Patricelli, M.P.; Janes, M.R.; Li, L.-S.; Hansen, R.; Peters, U.; Kessler, L.V.; Chen, Y.; Kucharski, J.M.; Feng, J.; Ely, T.; et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016, 6, 316–329. [Google Scholar] [CrossRef] [Green Version]
- Janes, M.R.; Zhang, J.; Li, L.-S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Hallin, J.; Engstrom, L.D.; Hargi, L.; Calinisan, A.; Aranda, R.; Briere, D.M.; Sudhakar, N.; Bowcut, V.; Baer, B.R.; Ballard, J.A.; et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020, 10, 54–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seton-Rogers, S. KRAS-G12C in the crosshairs. Nat. Rev. Cancer 2020, 20, 3. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.B.; Fece de la Cruz, F.; Phat, S.; Myers, S.T.; Wong, W.; Shahzade, H.A.; Hong, C.B.; Corcoran, R.B. Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS G12C Inhibition. Clin. Cancer Res. 2020, 26, 1633–1643. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.Y.; Zhao, Y.; Aronowitz, J.; Mai, T.T.; Vides, A.; Qeriqi, B.; Kim, D.; Li, C.; De Stanchina, E.; Mazutis, L.; et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 2020, 577, 421–425. [Google Scholar] [CrossRef]
- Ross, S.J.; Revenko, A.S.; Hanson, L.L.; Ellston, R.; Staniszewska, A.; Whally, N.; Pandey, S.K.; Revill, M.; Rooney, C.; Buckett, L.K.; et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl. Med. 2017, 9, eaal5253. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.; Xiong, Y.; Safaee, N.; Nowal, R.P.; Donovan, K.A.; Yuan, C.J.; Gero, T.W.; Feru, F.; Li, L. Exploring Targeted Degradation Strategy for Oncogenic KRASG12C. Cell Chem. Biol. 2020, 27, 19–31. [Google Scholar] [CrossRef]
- Bond, M.J.; Chu, L.; Nalawansha, D.A.; Li, K.; Crews, C. Targeted Degradation of Oncogenic KRASG12C by VHL-recruiting PROTACs. ChemRxiv 2020. [Google Scholar]
- Merlet, J.; Burger, J.; Gomes, J.-E.; Pintard, L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell. Mol. Life Sci. 2009, 66, 1924–1938. [Google Scholar] [CrossRef]
- Engelman, J.A.; Chen, L.; Tan, X.; Crosby, K.; Guimaraes, A.R.; Upadhyay, R.; Maira, M.; McNamara, K.; Perera, S.A.; Song, Y.; et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008, 14, 1351–1356. [Google Scholar] [CrossRef] [Green Version]
- Tolcher, A.W.; Bendell, J.C.; Papadopoulus, K.P.; Burris, H.A., 3rd; Patnaik, A.; Jones, S.F.; Rasco, D.; Cox, D.S.; Durante, M.; Bellew, K.M.; et al. A phase IB trial of the oral MEK inhibitor trametinib (GSK1120212) in combination with everolimus in patients with advanced solid tumors. Ann. Oncol. 2015, 26, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Broutin, S.; Stewart, A.; Thavasu, P.; Paci, A.; Bidart, J.-M.; Banerji, U. Insights into significance of combined inhibition of MEK and m-TOR signalling output in KRAS mutant non-small-cell lung cancer. Br. J. Cancer 2016, 115, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.-Q.; Bührer, E.D.; Berezowska, S.; Marti, T.M.; Xu, D.; Froment, L.; Yang, H.; Hall, S.R.R.; Vassella, E.; Yang, Z.; et al. mTOR mediates a mechanism of resistance to chemotherapy and defines a rational combination strategy to treat KRAS-mutant lung cancer. Oncogene 2019, 38, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Khan, K.; Ong, M.; Banerji, U.; Papadimitrakopoulou, V.; Gandara, D.R.; Patnaik, A.; Baird, R.D.; Olmos, D.; Garrett, C.R.; et al. Antitumor activity in ras-driven tumors by blocking akt and mek. Clin. Cancer Res. 2015, 21, 739–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerra, S.L.; Maertens, O.; Kuzmickas, R.; De Raedt, T.; Adeyemi, R.O.; Guild, C.J.; Guillemette, S.; Redig, A.J.; Chambers, E.S.; Xu, M.; et al. A Deregulated HOX Gene Axis Confers an Epigenetic Vulnerability in KRAS-Mutant Lung Cancers. Cancer Cell 2020, 37, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Manchado, E.; Weissmueller, S.; Morris, J.P., 4th; Chen, C.-C.; Wullenkord, R.; Lujambio, A.; De Stanchia, E.; Poirier, J.T.; Gainor, J.F.; Corcoran, R.B.; et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 2016, 534, 647–651. [Google Scholar] [CrossRef] [Green Version]
- Molina-Arcas, M.; Moore, C.; Rana, S.; Van Maldegem, F.; Mugarza, E.; Romero-Clavijo, P.; Herbert, E.; Horswell, S.; Li, L.-S.; Janes, M.R.; et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci. Transl. Med. 2019, 11, eaaw7999. [Google Scholar] [CrossRef]
- Chenard-Poirier, M.; Kaiser, M.; Boyd, K.; Sriskandarajah, P.; Constantinidou, A.; Harris, S.J.; Fandos, S.S.; Ryan, A.; Witt, K.; Dawes, J.C.; et al. Results from the biomarker-driven basket trial of RO5126766 (CH5127566), a potent RAF/MEK inhibitor, in RAS- or RAF-mutated malignancies including multiple myeloma. J. Clin. Oncol. 2017, 35 15 SUPPL, 2506. [Google Scholar] [CrossRef]
- Athuluri-Divakar, S.K.; Vasquez-Del Carpio, R.; Dutta, K.; Baker, S.J.; Cosenza, S.C.; Basu, I.; Gupta, Y.K.; Reddy, M.V.R.; Ueno, L.; Hart, J.R.; et al. A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling. Cell 2016, 165, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Citri, A.; Yarden, Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 2006, 7, 505–516. [Google Scholar] [CrossRef]
- Massarelli, E.; Varella-Garcia, M.; Tang, X.; Xavier, A.C.; Ozburn, N.C.; Liu, D.D.; Bekele, B.N.; Herbst, R.S.; Wistuba, I.I. KRAS Mutation Is an Important Predictor of Resistance to Therapy With Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancer. Clin. Cancer Res. 2007, 13, 2890–2896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cadranel, J.; Mauguen, A.; Faller, M.; Zalcman, G.; Buisine, M.-P.; Westeel, V.; Longchampt, E.; Wislez, M.; Coudert, B.; Daniel, C.; et al. Impact of systematic EGFR and KRAS mutation evaluation on progression-free survival and overall survival in patients with advanced non-small-cell lung cancer treated by erlotinib in a French prospective cohort (ERMETIC project--part 2). J. Thorac. Oncol. 2012, 7, 1490–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rulli, E.; Marabese, M.; Torri, V.; Farina, G.; Veronese, S.; Bettini, A.; Longo, F.; Moscetti, L.; Ganzinelli, M.; Lauricella, C.; et al. Value of KRAS as prognostic or predictive marker in NSCLC: results from the TAILOR trial. Ann. Oncol. 2015, 26, 2079–2084. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Qiu, L.-X.; Liao, R.-Y.; Du, F.-B.; Ding, H.; Yang, W.-C.; Li, J.; Chen, Q. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: A meta-analysis of 22 studies. Lung Cancer 2010, 69, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Linardou, H.; Dahabreh, I.J.; Kanaloupiti, D.; Siannis, F.; Bafaloukos, D.; Kosmidis, P.; Papadimitriou, C.A.; Murray, S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet. Oncol. 2008, 9, 962–972. [Google Scholar] [CrossRef]
- Molina-Arcas, M.; Hancock, D.C.; Sheridan, C.; Kumar, M.S.; Downward, J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov. 2013, 3, 548–563. [Google Scholar] [CrossRef] [Green Version]
- Navas, C.; Hernández-Porras, I.; Schuhmacher, A.J.; Sibilia, M.; Guerra, C.; Barbacid, M. EGF Receptor Signaling Is Essential for K-Ras Oncogene-Driven Pancreatic Ductal Adenocarcinoma. Cancer Cell 2012, 22, 318–330. [Google Scholar] [CrossRef] [Green Version]
- Ardito, C.M.; Grüner, B.M.; Takeuchi, K.K.; Lubeseder-Martellato, C.; Teichmann, N.; Mazur, P.K.; Delgiorno, K.E.; Carpenter, E.S.; Halbrook, C.J.; Hall, J.C.; et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 2012, 22, 304–317. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Hobor, S.; Bertotti, A.; Zecchin, D.; Huang, S.; Galimi, F.; Cottino, F.; Prahallad, A.; Grernrum, W.; Tzani, A.; et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014, 7, 86–93. [Google Scholar] [CrossRef] [Green Version]
- Moll, H.P.; Pranz, K.; Musteanu, M.; Grabner, B.; Hruschka, N.; Mohrherr, J.; Aigner, P.; Stiedl, P.; Brcic, L.; Laszlo, V.; et al. Afatinib restrains K-RAS–driven lung tumorigenesis. Sci. Transl. Med. 2018, 10, eaao2301. [Google Scholar] [CrossRef] [Green Version]
- Kruspig, B.; Monteverde, T.; Neidler, S.; Hock, A.; Kerr, E.; Nixon, C.; Clark, W.; Hedley, A.; Laing, S.; Coffelt, S.B.; et al. The ERBB network facilitates KRAS-driven lung tumorigenesis. Sci. Transl. Med. 2018, 10, eaao2565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Lu, W.; Chen, G.; Wang, P.; Chen, Z.; Zhou, Y.; Ogasawara, M.; Trachootham, D.; Feng, L.; Pelicano, H.; et al. K-ras(G12V) transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 2012, 22, 399–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Kerr, E.M.; Martins, C.P. Metabolic rewiring in mutant Kras lung cancer. FEBS J. 2018, 285, 28–41. [Google Scholar] [CrossRef]
- Pylayeva-Gupta, Y.; Grabocka, E.; Bar-Sagi, D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer 2011, 11, 761–774. [Google Scholar] [CrossRef] [Green Version]
- Bryant, K.L.; Mancias, J.D.; Kimmelman, A.C.; Der, C.J. KRAS: feeding pancreatic cancer proliferation. Trends Biochem. Sci. 2014, 39, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Osugi, J.; Yamaura, T.; Muto, S.; Okabe, N.; Matsumura, Y.; Hoshino, M.; Higuchi, M.; Suzuki, H.; Gotoh, M. Prognostic impact of the combination of glucose transporter 1 and ATP citrate lyase in node-negative patients with non-small lung cancer. Lung Cancer 2015, 88, 310–318. [Google Scholar] [CrossRef]
- Chen, G.; Gharib, T.G.; Wang, H.; Huang, C.-C.; Kucik, R.; Thomas, D.G.; Shedden, K.A.; Misek, D.E.; Taylor, J.M.G.; Giordano, T.; et al. Protein profiles associated with survival in lung adenocarcinoma. Proc. Natl. Acad. Sci. USA 2003, 100, 13537–13542. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Jin, C.C.; Choi, J.E.; Hong, M.J.; Jung, D.K.; Do, S.K.; Baek, S.A.; Kang, H.J.; Kang, H.-G.; Choi, S.H.; et al. Genetic polymorphisms in glycolytic pathway are associated with the prognosis of patients with early stage non-small cell lung cancer. Sci. Rep. 2016, 6, 35603. [Google Scholar] [CrossRef]
- Puzone, R.; Savarino, G.; Salvi, S.; Dal Bello, M.G.; Barletto, G.; Genova, C.; Rijavec, E.; Sini, C.; Esposito, A.I.; Ratto, G.B.; et al. Glyceraldehyde-3-phosphate dehydrogenase gene over expression correlates with poor prognosis in non small cell lung cancer patients. Mol. Cancer 2013, 12, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Moothart, D.R.; Lowy, D.R.; Qian, X. The expression of glyceraldehyde-3-phosphate dehydrogenase associated cell cycle (GACC) genes correlates with cancer stage and poor survival in patients with solid tumors. PLoS One 2013, 8, e61262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patra, K.C.; Wang, Q.; Bhaskar, P.T.; Miller, L.; Wang, Z.; Wheaton, W.; Chandel, N.; Laakso, M.; Muller, W.J.; Allen, E.L.; et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 2013, 24, 213–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.; Zhang, Y.; Chen, T.; Wang, Y.; Xue, J.; Zhang, Y.; X, W.; Mo, X.; Lu, Y. Efficacy of RNAi targeting of pyruvate kinase M2 combined with cisplatin in a lung cancer model. J. Cancer Res. Clin. Oncol. 2011, 137, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Li, D.; Zhang, J.; Wang, Y.-S.; Yang, L.; Zhang, H.-L.; Wang, X.-H.; Mu, B.; Wang, W.; Ma, Y.; et al. Silencing of pkm2 increases the efficacy of docetaxel in human lung cancer xenografts in mice. Cancer Sci. 2010, 101, 1447–1453. [Google Scholar] [CrossRef]
- Xie, H.; Hanai, J.-I.; Ren, J.-G.; Kats, L.; Burgess, K.; Bhargava, P.; Signoretti, S.; Billiard, J.; Duffy, K.J.; Grant, A.; et al. Targeting lactate dehydrogenase-A inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014, 19, 795–809. [Google Scholar] [CrossRef] [Green Version]
- Altman, J.K.; Szilard, A.; Goussetis, D.J.; Sassano, A.; Colamonici, M.; Gounaris, W.; Frankfurt, O.; Giles, F.J.; Eklund, E.A.; Beauchamp, E.M.; et al. Autophagy is a survival mechanism of acute myelogenous leukemia precursors during dual mTORC2/mTORC1 targeting. Clin. Cancer Res. 2014, 20, 2400–2409. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, K.; Guo, H.; Tang, H.; Yuan, Y.; Jun, Z.; Lingxia, Z.; Kun, H.; Xin, Z. Gastrin enhances autophagy and promotes gastric carcinoma proliferation via inducing AMPKα. Oncol. Res. 2017, 25, 1399–1407. [Google Scholar]
- Masui, A.; Hamada, M.; Kameyama, H.; Wakabayashi, K.; Takasu, A.; Imai, T.; Iwai, S.; Yura, Y. Autophagy as a survival mechanism for squamous cell carcinoma cells in endonuclease g-mediated apoptosis. PLoS One 2016, 11, e0162786. [Google Scholar] [CrossRef] [Green Version]
- Tan, Q.; Wang, M.; Yu, M.; Zhang, J.; Bristow, R.G.; Hill, R.P.; Tannock, I.F. Role of Autophagy as a Survival Mechanism for Hypoxic Cells in Tumors. Neoplasia 2016, 18, 347–355. [Google Scholar] [CrossRef]
- Fitzwalter, B.E.; Thorburn, A. Recent insights into cell death and autophagy. FEBS J. 2015, 282, 4279–4288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.Y.; Chen, H.Y.; Mathew, R.; Fan, J.; Strohecker, A.M.; Karsli-Uzunbas, G.; Kamphorst, J.J.; Chen, G.; Lemons, J.M.S.; Karantza, V.; et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011, 25, 460–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Wang, X.; Contino, G.; Liesa, M.; Sahin, E.; Ying, H.; Bause, A.; Li, Y.; Stommel, J.M.; Dell’antonio, G.; et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011, 25, 717–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eng, C.H.; Wang, Z.; Tkac, H.D.; Toral-Barza, L.; Ugwonali, S.; Liu, S.; Fitzgerald, S.L.; George, E.; Frias, E.; Cochran, N.; et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc. Natl. Acad. Sci. USA 2016, 113, 182–187. [Google Scholar] [CrossRef] [Green Version]
- Bryant, K.L.; Stalnecker, C.A.; Zeitouni, D.; Klomp, J.E.; Peng, S.; Tikunov, A.P.; Gunda, V.; Pierobon, M.; Waters, A.M.; George, S.D.; et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 2019, 25, 628–640. [Google Scholar] [CrossRef]
- Kinsey, C.G.; Camolotto, S.A.; Boespflug, A.M.; Guillen, K.P.; Foth, M.; Truong, A.; Schuman, S.S.; Shea, J.E.; Seipp, M.T.; Yap, J.T.; et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 2019, 25, 620–627. [Google Scholar] [CrossRef]
- Lee, C.-S.; Lee, L.C.; Yuan, T.L.; Chakka, S.; Fellmann, C.; Lowe, S.W.; Caplen, N.J.; McCormick, F.; Luo, J. MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc. Natl. Acad. Sci. USA 2019, 116, 4508–4517. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.Y.; Karsli-Uzunbas, G.; Mathew, R.; Aisner, S.C.; Kamphorst, J.J.; Strohecker, A.M.; Chen, G.; Price, S.; Lu, W.; Teng, X.; et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013, 27, 1447–1461. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, F.; Hamanaka, R.; Wheaton, W.W.; Weinberg, S.; Joseph, J.; Lopez, M.; Kalyanaraman, B.; Mutlu, G.M.; Budinger, G.R.S.; Chandel, N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA 2010, 107, 8788–8793. [Google Scholar] [CrossRef] [Green Version]
- Shackelford, D.B.; Abt, E.; Gerken, L.; Vasquez, D.S.; Seki, A.; Leblanc, M.; Wei, L.; Fishbein, M.C.; Czernin, J.; Mischel, P.S.; et al. LKB1 Inactivation Dictates Therapeutic Response of Non-Small Cell Lung Cancer to the Metabolism Drug Phenformin. Cancer Cell 2013, 23, 143–158. [Google Scholar] [CrossRef] [Green Version]
- Ho, P.-C.; Liu, P.-S. Metabolic communication in tumors: A new layer of immunoregulation for immune evasion. J. Immunother. Cancer 2016, 4, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfriend, E.; Schwarz, S.; Rother, G.; Hoves, S.; et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef] [PubMed]
- Pollizzi, K.N.; Sun, I.-H.; Patel, C.H.; Lo, Y.-C.; Oh, M.-H.; Waickman, A.T.; Tam, A.J.; Blosser, R.L.; Wen, J.; Delgoffe, G.M.; et al. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8+ T cell differentiation. Nat. Immunol. 2016, 17, 704–711. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Pavlides, S.; Howell, A.; Pestell, R.G.; Tanowitz, H.B.; Sotgia, F.; Lisanti, M.P. Stromal-epithelial metabolic coupling in cancer: Integrating autophagy and metabolism in the tumor microenvironment. Int. J. Biochem. Cell Biol. 2011, 43, 1045–1051. [Google Scholar] [CrossRef] [Green Version]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [Green Version]
- Rittmeyer, A.; Barlesi, F.; Waterkamp, D.; Park, K.; Ciardiello, F.; Von Pawel, J.; Gadgeel, S.M.; Hida, T.; Kowalski, D.M.; Dols, M.C.; et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017, 389, 255–265. [Google Scholar] [CrossRef]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Garcia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [Green Version]
- Zugazagoitia, J.; Molina-Pinel, S.; Lopez-Rios, F.; Paz-Ares, L. Biological therapies in nonsmall cell lung cancer. Eur. Respir. J. 2017, 49, 1601520. [Google Scholar] [CrossRef] [Green Version]
- Adderley, H.; Blackhall, F.H.; Lindsay, C.R. KRAS-mutant non-small cell lung cancer: Converging small molecules and immune checkpoint inhibition. EBioMedicine 2019, 41, 711–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.K.; Man, J.; Lord, S.; Cooper, W.; Links, M.; Gebski, V.; Herbst, R.S.; Gralla, R.J.; Mok, T.; Yang, J.C.-H. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non-small cell lung carcinoma: A systematic review and meta-analysis. JAMA Oncol. 2018, 4, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, A.J.; Rizvi, H.; Bandlamudi, C.; Sauter, J.L.; Travis, W.D.; Rekhtman, N.; Plodkowski, A.J.; Perez-Johnston, R.; Sawan, P.; Beras, A.; et al. Clinical and molecular correlates of PD-L1 expression in patients with lung adenocarcinomas. Ann. Oncol. 2020, 31, 599–608. [Google Scholar] [CrossRef]
- Gianoncelli, L.; Spitaleri, G.; Passaro, A.; Radice, D.; Fumagalli, C.; Del Signore, W.; Sati, V.; Catania, C.M.; Guerini-Rocco, E.; Barberis, M.; et al. Efficacy of Anti-PD1/PD-L1 Therapy (IO) in KRAS Mutant Non-small Cell Lung Cancer Patients: A Retrospective Analysis. Anticancer Res. 2020, 40, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Kauffmann-Guerrero, D.; Tufman, A.; Kahnert, K.; Bollmann, B.A.; Reu, S.; Syunyaeva, Z.; Schneider, C.; Manapov, F.; Huber, R.M.; Golpon, H. Response to Checkpoint Inhibition in Non-Small Cell Lung Cancer with Molecular Driver Alterations. Oncol. Res. Treat. 2020, 43, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Heymach, J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Goldberg, M.E.; Greenawalt, D.M.; Hellmann, M.D.; Awad, M.M.; Gainor, J.F.; Schrock, A.B.; Hartmaier, R.J.; Trabucco, S.E. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018, 8, 822–835. [Google Scholar] [CrossRef] [Green Version]
- Hellmann, M.D.; Nathanson, T.; Rizvi, H.; Creelan, B.C.; Sanchez-Vega, F.; Ahuja, A.; Ni, A.; Novik, J.B.; Mangarin, L.M.B.; Abu-Akeel, M.; et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018, 33, 843–852. [Google Scholar] [CrossRef] [Green Version]
- Koyama, S.; Akbay, E.A.; Li, Y.Y.; Aref, A.R.; Skoulidis, F.; Herter-Sprie, G.S.; Buczkowski, K.A.; Liu, Y.; Awad, M.M.; Denning, W.L.; et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res. 2016, 76, 999–1008. [Google Scholar] [CrossRef] [Green Version]
- Arbour, K.C.; Jordan, E.; Kim, H.R.; Dienstag, J.; Yu, H.A.; Sanchez-Vega, F.; Lito, P.; Berger, M.; Solit, D.B.; Hellmann, M.; et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin. Cancer Res. 2018, 24, 334–340. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.-Y.; Zhong, W.-Z.; Zhang, X.-C.; Su, J.; Xie, Z.; Liu, S.-Y.; Tu, H.-Y.; Chen, H.-J.; Sun, Y.-L.; Zhou, Q.; et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 2017, 23, 3012–3024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuwelt, A.J.; Kimball, A.K.; Johnson, A.M.; Arnold, B.W.; Bullock, B.L.; Kaspar, R.E.; Kleczko, E.K.; Kwak, J.W.; Wu, M.-H.; Heasley, L.E.; et al. Cancer cell-intrinsic expression of MHC II in lung cancer cell lines is actively restricted by MEK/ERK signaling and epigenetic mechanisms. J. Immunother. Cancer 2020, 8, e000441. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Deng, J.; Li, S.; Sali, T.; Dong, L.; Brea, E.J.; Houghton, S.; Redmond, D.; Zhong, H.; Boiarsky, J.; et al. Pulsatile MEK Inhibition Improves Anti-tumor Immunity and T Cell Function in Murine Kras Mutant Lung Cancer. Cell Rep. 2019, 27, 806–819.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, E.; Robbins, P.F.; Lu, Y.-C.; Prickett, T.D.; Gartner, J.J.; Jia, L.; Pasetto, A.; Zheng, Z.; Ray, S.; Groh, E.M.; et al. T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 2016, 375, 2255–2262. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uras, I.Z.; Moll, H.P.; Casanova, E. Targeting KRAS Mutant Non-Small-Cell Lung Cancer: Past, Present and Future. Int. J. Mol. Sci. 2020, 21, 4325. https://doi.org/10.3390/ijms21124325
Uras IZ, Moll HP, Casanova E. Targeting KRAS Mutant Non-Small-Cell Lung Cancer: Past, Present and Future. International Journal of Molecular Sciences. 2020; 21(12):4325. https://doi.org/10.3390/ijms21124325
Chicago/Turabian StyleUras, Iris Z., Herwig P. Moll, and Emilio Casanova. 2020. "Targeting KRAS Mutant Non-Small-Cell Lung Cancer: Past, Present and Future" International Journal of Molecular Sciences 21, no. 12: 4325. https://doi.org/10.3390/ijms21124325
APA StyleUras, I. Z., Moll, H. P., & Casanova, E. (2020). Targeting KRAS Mutant Non-Small-Cell Lung Cancer: Past, Present and Future. International Journal of Molecular Sciences, 21(12), 4325. https://doi.org/10.3390/ijms21124325




