Abstract
The basic leucine zipper (bZIP) is a plant-specific transcription factor family that plays crucial roles in response to biotic and abiotic stresses. However, little is known about the function of bZIP genes in soybean. In this study, we isolated a bZIP gene, GmbZIP19, from soybean. A subcellular localization study of GmbZIP19 revealed its nucleus localization. We showed that GmbZIP19 expression was significantly induced by ABA (abscisic acid), JA (jasmonic acid) and SA (salicylic acid), but reduced under salt and drought stress conditions. Further, GmbZIP19 overexpression Arabidopsis lines showed increased resistance to S. sclerotiorum and Pseudomonas syringae associated with upregulated ABA-, JA-, ETH- (ethephon-)and SA-induced marker genes expression, but exhibited sensitivity to salt and drought stresses in association with destroyed stomatal closure and downregulated the salt and drought stresses marker genes’ expression. We generated a soybean transient GmbZIP19 overexpression line, performed a Chromatin immunoprecipitation assay and found that GmbZIP19 bound to promoters of ABA-, JA-, ETH-, and SA-induced marker genes in soybean. The yeast one-hybrid verified the combination. The current study suggested that GmbZIP19 is a positive regulator of pathogen resistance and a negative regulator of salt and drought stress tolerance.
1. Introduction
Soybean, an important economic crop and one of the main oil crops worldwide, is widely used as a source for plant oil and as a protein resource by humans. Extreme environmental conditions severely influence the productivity of soybean and weaken the crop productivity, ultimately affecting global food security [1]. Furthermore, various environmental conditions such as salinity, drought, temperature changes, nutritional deficiency and pathogen invasion significantly influence soybean growth and development, leading to reduced production. Evolutionarily, in order to overcome these stress conditions, plants have developed a series of response mechanisms at the physiological, morphological, cellular, biochemical and molecular levels [2,3,4]. Gene expression changes play an important role during stress response. Transcriptional factors can mediate some stress-related gene expressions and further regulate the responses to adverse stresses in plants [5,6]. One of these is the basic leucine zipper (bZIP) transcription factor.
The basic leucine zipper (bZIP) transcription factor family is one of the largest and the most diverse transcription factor (TF) families in plants. bZIP transcription factors own a highly conserved 40–80 amino acid bZIP domain, which consists of a conserved basic region and a leucine zipper [7,8]. The conserved basic region contains approximate 16 amino acid residues with a N-X7- R/K motif consists of a nuclear localization signal and DNA binding domain, whereas the leucine zipper is less conserved and forms a heptad repeat of leucine residues or other bulky hydrophobic amino acids (Ile, Val, Phe, or Met) [9,10]. It has been indicated that the plant bZIP proteins bind to DNA sequences with an ACGT core cis-element, especially ABRE (ABA-responsive element), G-box (CACGTG), C-box (GACGTC) and A-box (TACGTA) [11].
In plants, it has been indicated that bZIP transcription factors are involved in various biological processes, such as hormone signaling pathways and tolerance to stresses. Firstly, bZIP TFs (transcription factors) are involved in sugar and hormone signaling. For instance, a crucial repressor for ethylene biosynthesis in Arabidopsis, AtERF11, is modulated by the bZIP transcription factor HY5 [12]. In addition, previous studies have shown that bZIP transcription factors participate in the responses to biotic and abiotic stresses such as salinity, drought, cold and pathogen infections. TabZIP60, a novel wheat bZIP transcription factor, confers multiple abiotic stress tolerances in transgenic Arabidopsis [13]. bZIP transcription factors have been identified and studied in different kinds of plant species due to their high value in plant development and stress tolerance. 89 bZIPs were identified in Oryza sativa (rice) [14], 75 bZIP genes have been identified in Arabidopsis thaliana [8], 64 in cucumber [15], 125 in Zea mays (maize) [16] and 160 in Glycine max (soybean) [17].
Although there are several conserved and novel bZIP genes identified in different processes during soybean development, GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean [18], overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean [19] and GmbZIP2 confers drought and salt resistances in transgenic Arabidopsis and soybean [20]. In general, soybean bZIP genes were seldom studied in soybean, especially in stress responses. Previously, studies have shown that AtbZIP19 is essential for Arabidopsis adaptation to Zn deficiency in roots [21] and it can interact with AtbZIP23 to regulate the adaptation [22]. There are no studies that showed the stress responses of AtbZIP19. In the current study, GmbZIP19, a homologous gene of AtbZIP19 in soybean, was identified from a full-length soybean cDNA bank. Expression profile indicated that the expression of GmbZIP19 was induced by S. sclerotiorum, and exogenous hormones like JA (jasmonic acid), SA (salicylic acid), ETH (ethephon), ABA (abscisic acid) and BR (brassinolide) but reduced by salinity and drought. Overexpression of GmbZIP19 in Arabidopsis showed more resistance to S. sclerotiorum and Pseudomonas syringae, but more sensitivity to salinity, drought and different hormones, including ABA, JA and ETH, compared with wild-type (WT) plants. In summary, our results verified that GmbZIP19 plays an important role in multiple abiotic and biotic stress responses.
2. Results
2.1. Basic Bioinformatic Analyses of GmbZIP19
According to https://web.expasy.org, GmbZIP19 cDNA was predicted to be 1617 bp long, containing a 723-bp ORF (open reading frame), which encodes a polypeptide of 240 amino acids with a predicted molecular weight of 26.57 kDa and a theoretical pI of 6.59. Sequence alignment showed high levels of amino acid sequence similarity of GmbZIP19 with three other bZIP proteins from Arabidopsis and Oryza sativa (rice) (Figure S1). They shared a conserved bZIP DNA-binding domain, a basic DNA binding region and a leucine zipper dimerization motif. The basic DNA binding region is conserved and contains a 52-amino acid long basic region (N-x7-R/K-x9).
2.2. Subcellular Localization of GmbZIP19
To determine the subcellular localization of GmbZIP19 protein, the GmbZIP19 CDS was fused to the pGWB605-GFP vector. The recombinant vector (35S-GmbZIP19-GFP) and the 35S promoter-driven GFP control vector (35S-GFP) were transiently expressed in N. benthamiana leaves through Agrobacterium infection. As shown in Figure S2, the 35S-GFP control was observed in both the nucleus and cytoplasm membrane, whereas 35S-GmbZIP19-GFP was specifically localized in the nucleus.
2.3. Expression Profile of GmbZIP19 in Response to Biotic and Abiotic Stresses
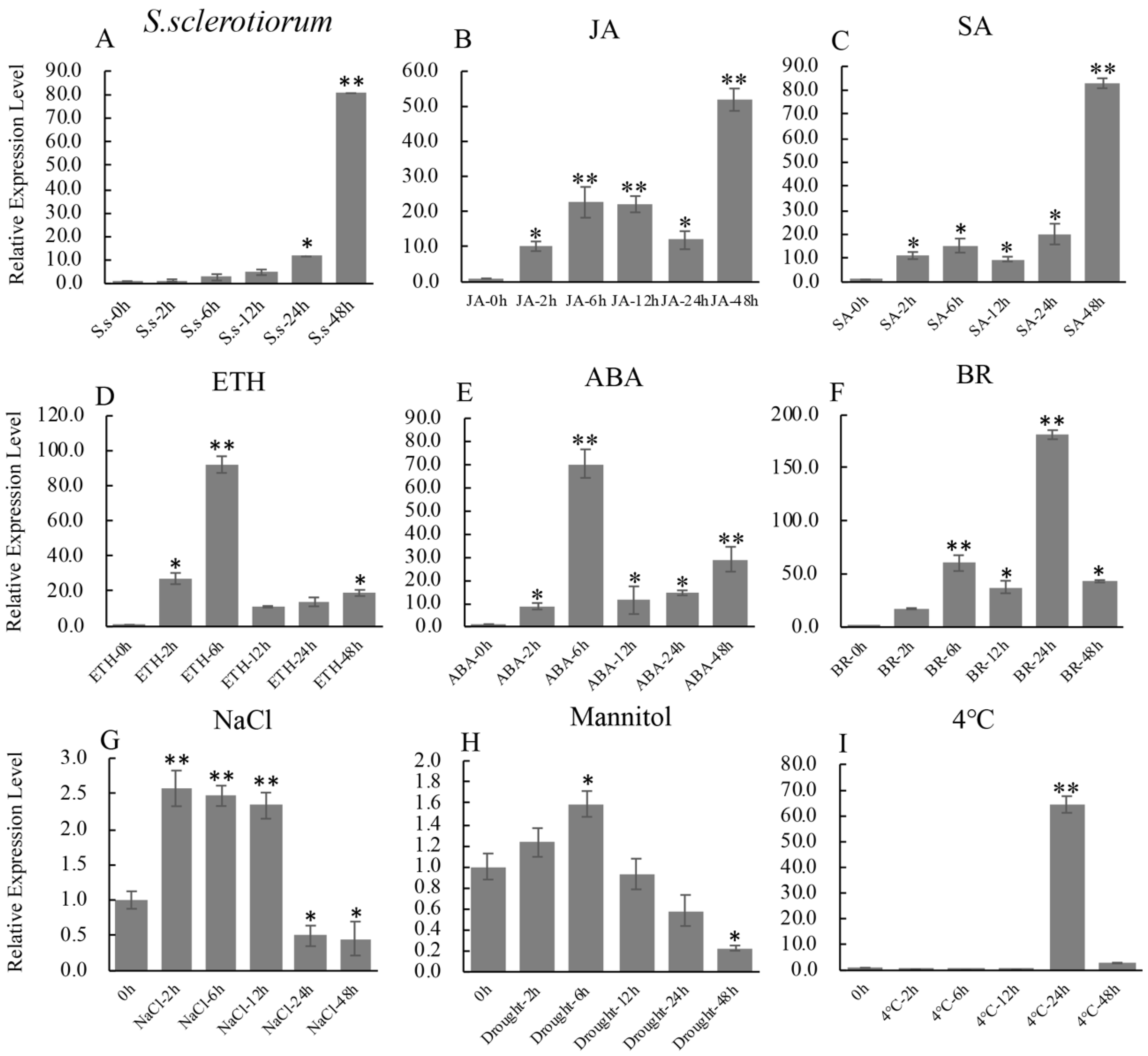
Bioinformatics analysis indicated that there are many predicted stress response-related cis-elements: G-box recognition site (1 hit), MYB recognition site (6 hits), CGTCA motif (1 hit) and TCA-element (3 hits) in the promoter of GmbZIP19 (Figure S3), suggesting that GmbZIP19 may be involved in biotic and abiotic stress responses. In order to further explore and predict the function of GmbZIP19, the expression changes of GmbZIP19 in soybean leaves under different biotic and abiotic treatments was evaluated by RT-qPCR. The expression of GmbZIP19 was obviously induced by S. sclerotiorum, a typical necrotrophic phytopathogen in soybean [23] (Figure 1A).
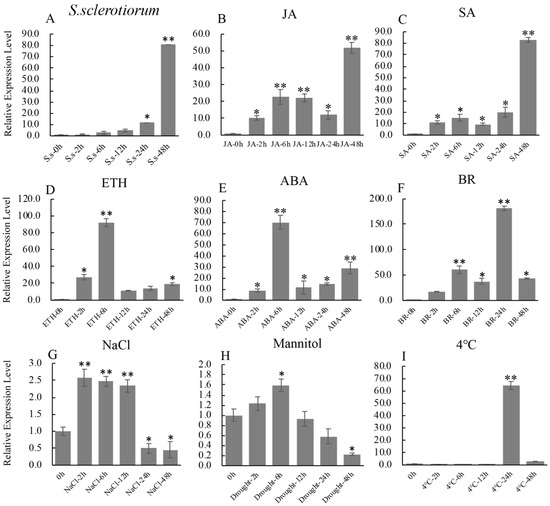
Figure 1.
The expression profile of GmbZIP19 in response to biotic and abiotic stresses. (A–I) The expression profile of GmbZIP19 in response to S. sclerotiorum, 100 μM JA (jasmonic acid), 1 mM SA (salicylic acid), 100 μM ABA (abscisic acid), 1 mM ETH (ethephon), 100 μM BR (brassinolide), 100 mM NaCl for salt condition, 250 mM Mannitol for drought condition and 4 °C for low temperature. Three technical replicates were performed for each of the three independent biological replicates. The error bars were obtained from multiple replicates of the RT-qPCR and indicate ±SD (n = 3 replicates). Asterisks indicate significant differences for the indicated comparisons based on a Student’s t-test (** p < 0.01; * p < 0.05).
Previous studies have shown that plant defense to pathogen was associated with the mediation of various plant hormones, including SA, JA, ABA and ETH. Therefore, we examined the expression of GmbZIP19 under different hormones. The result showed that GmbZIP19 expression was induced rapidly by JA, SA, ETH, ABA and BR within 2 h after treatment (Figure 1B–F). GmbZIP19 maintained gradually increased expression under JA and SA treatments (Figure 1B,C) while the expression level of GmbZIP19 in response to ETH and ABA peaked at 6 h and decreased rapidly (Figure 1D,E). These results indicated that the expression of GmbZIP19 may be related to the defense responses mediated by JA, SA, ETH and ABA in different manners.
In addition, the expression of GmbZIP19 was significantly induced by NaCl at 2, 6 and 12 h, but significantly reduced at 24 and 48 h (Figure 1G). The expression of GmbZIP19 was also induced by mannitol (drought) treatment and reached to the peak at 6 h, and then gradually decreased and reached to the lowest level at 48 h after treatment (Figure 1H), while cold (4 °C) induced the expression of GmbZIP19 at 24 h (Figure 1I).
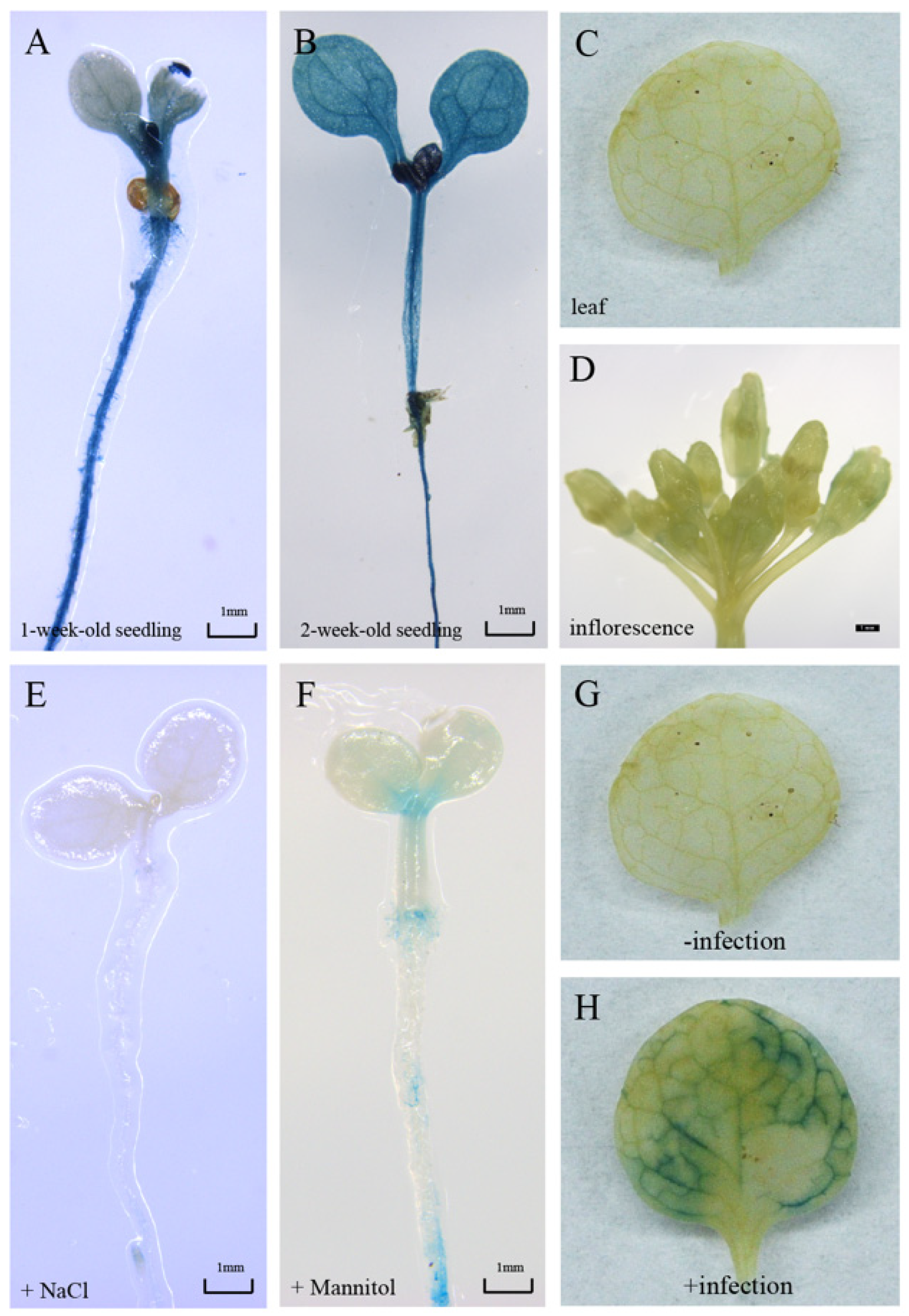
In order to verify the expression pattern of GmbZIP19, we generated pGmbZIP19-GUS fusion transgenic Arabidopsis plants (pGmbZIP19-GUS). GUS staining was detected in the shoot apical meristem (SAM), hypocotyl and root of one-week-old pGmbZIP19-GUS seedlings (Figure 2A). A similar GUS expression pattern was also examined in two-week-old pGmbZIP19-GUS seedlings (Figure 2B) but no GUS signal was found in mature pGmbZIP19-GUS plants, including in the leaves and inflorescence (Figure 2C,D). Interestingly, the pGmbZIP19-GUS signal was significantly reduced when one-week-old pGmbZIP19-GUS seedlings were treated with 150 mM NaCl or 300 mM mannitol for 7 days (Figure 2E,F), suggesting that the expression of GmbZIP19 was inhibited by salt and drought. These results were consistent with the reduced expression of GmbZIP19 evaluated by RT-qPCR after salt and drought (mimic using mannitol) treatment after 24 h (Figure 1), which indicated that GmbZIP19 may be involved in regulating different abiotic stress tolerance.

Figure 2.
The responses of the GmbZIP19 promoter to salt, drought and S. sclerotiorum. (A) GUS staining result of 1-week-old pGmbZIP19-GUS Arabidopsis seedlings. (B) GUS staining result of two-week-old pGmbZIP19-GUS Arabidopsis seedlings. Bar = 1 mm. (C) GUS staining result of 4-week-old pGmbZIP19-GUS Arabidopsis leaf. (D) GUS staining result of 6-week-old pGmbZIP19-GUS Arabidopsis inflorescence. Bar = 1 mm. (E) GUS staining result of 1-week-old pGmbZIP19-GUS Arabidopsis seedlings under 150 mM NaCl for 6 days. Bar = 1 mm. (F) GUS staining result of 1-week-old pGmbZIP19-GUS Arabidopsis seedlings under 300 mM mannitol for 6 days. Bar = 1 mm. (G) GUS staining result of pGmbZIP19-GUS Arabidopsis leaf under 0 h S. scleroterium infection. (H) GUS staining result of pGmbZIP19-GUS Arabidopsis leaf under 6 h S. scleroterium infection.
In addition, we compared the GUS expression in the leaf of mature pGmbZIP19-GUS plants before and after inoculation of S. sclerotiorum. In line with the induced expression of GmbZIP19 in plants after S. sclerotiorum inoculation (Figure 1A), an increased GUS signal was detected in pGmbZIP19-GUS plants after inoculation of S. sclerotiorum (Figure 2G,H). These results indicated that GmbZIP19 was induced by this pathogen and it may be involved in the pathogen defense process.
2.4. Overexpression of GmbZIP19 in Arabidopsis Increases the Tolerance to Pathogen
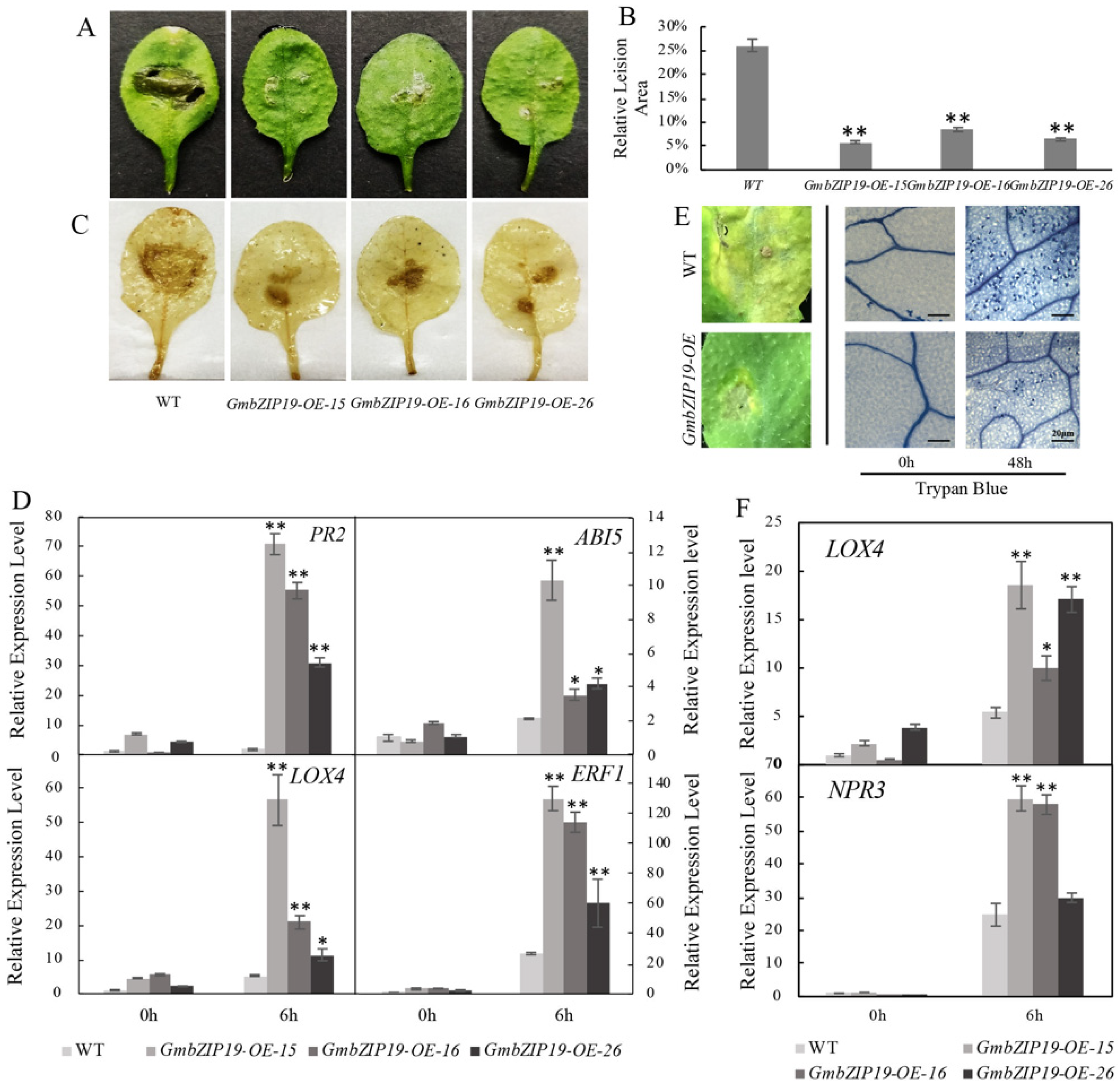
In order to determine the function of GmbZIP19 in response to pathogen, we generated 35S promoter-driven overexpression of GmbZIP19 Arabidopsis plants (GmbZIP19-OE) and inoculated the same size of S.sclerotiorum, a typical pathogen that can cause considerable damage and severe production loss in soybean [23], on the surface of leaves of WT and GmbZIP19-OE plants. The result showed that the relative lesion areas of three different GmbZIP19-OE lines were much smaller than WT at 12 h post-inoculation (hpi) (Figure 3A,B), suggesting that GmbZIP19-OE plants exhibited stronger resistance to S. sclerotiorum than WT. DAB staining showed that brown precipitates, representing accumulation of H2O2, in leaves of GmbZIP19-OE lines after infection, was lighter than that in WT (Figure 3C). The result indicates that overexpression of GmbZIP19 in Arabidopsis significantly improved plant tolerance to S. sclerotiorum.
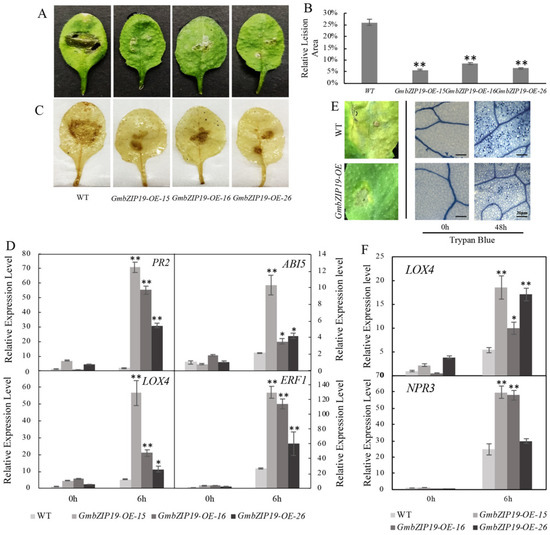
Figure 3.
Overexpression of GmbZIP19 confers improved disease resistance against S. sclerotiorum and Pseudomonas syringae. (A) The phenotype of GmbZIP19-OE lines and WT after S. scleroterium infection. (B) The relative lesion area of GmbZIP19-OE lines and WT under S. scleroterium infection. Error bars indicate ± SD (n = 5 leaves). (C) The DAB staining result (accumulation of H2O2 in leaves) of GmbZIP19-OE lines and WT after S. scleroterium infection. (D) qPCR analysis of transcription levels in GmbZIP19-OE transgenic and WT plants after S. scleroterium infection. (E) The phenotype and trypan blue staining result of GmbZIP19-OE and WT plants under Pseudomonas syringae infection. Bar = 20 μm. (F) qPCR analysis of transcription levels in GmbZIP19-OE transgenic and WT plants after pst. DC3000 infection. The error bars were obtained from multiple replicates of the RT-qPCR and indicate ±SD (n = 3 replicates). Asterisks indicate significant differences for the indicated comparisons based on a Students’ t-test (** p < 0.01; * p < 0.05).
To evaluate the possible role of plant hormones in the resistance to S. sclerotiorum conferred by GmbZIP19 in Arabidopsis, the relative expression levels of stress-responsive genes AtPR2, AtABI5, AtLOX4 and AtERF1 in association with SA, ABA, JA and ETH respectively, were examined by RT-qPCR. The expression of these four marker genes in GmbZIP19-OE plants has almost no significant difference with WT when there is no treatment. In contrast, at 6 hpi to S. sclerotiorum, the relative expression levels of these four genes in three GmbZIP19-OE transgenic lines significantly increased compared to WT (Figure 3D). Induced expression of SA-, ABA-, JA- and ETH-dependent genes in GmbZIP19-OE upon S. sclerotiorum inoculation suggested the involvement of these plant hormones in the regulation of GmbZIP19 to S. sclerotiorum resistance.
Furthermore, we investigated whether GmbZIP19-OE in Arabidopsis can also enhance plant tolerance to Pseudomonas syringae (pst DC3000), a (hemi) biotrophic bacterial pathogen that can cause bacterial leaf spot disease [24]. After 48 h of Pseudomonas syringae infection, WT plants exhibited extremely severe chlorosis symptoms showing large disease spots and a strong trypan blue staining signal, representing the large number of dead cells, in the whole leaf. Nevertheless, the leaves of GmbZIP19-OE lines had smaller disease spots and a lighter trypan blue staining signal after Pseudomonas syringae inoculation (Figure 3E). The result demonstrated that GmbZIP19 promotes plant tolerance to Pseudomonas syringae as well. Moreover, the increased transcription levels of JA-related gene, AtLOX4, and SA-related gene, AtNPR3, confirmed the phenotype and suggested that JA- and SA-dependent genes are involved in the regulation of GmbZIP19 to pst. DC3000 (Figure 3F).
2.5. Overexpression GmbZIP19 Arabidopsis Exhibits Sensitivity to Salinity
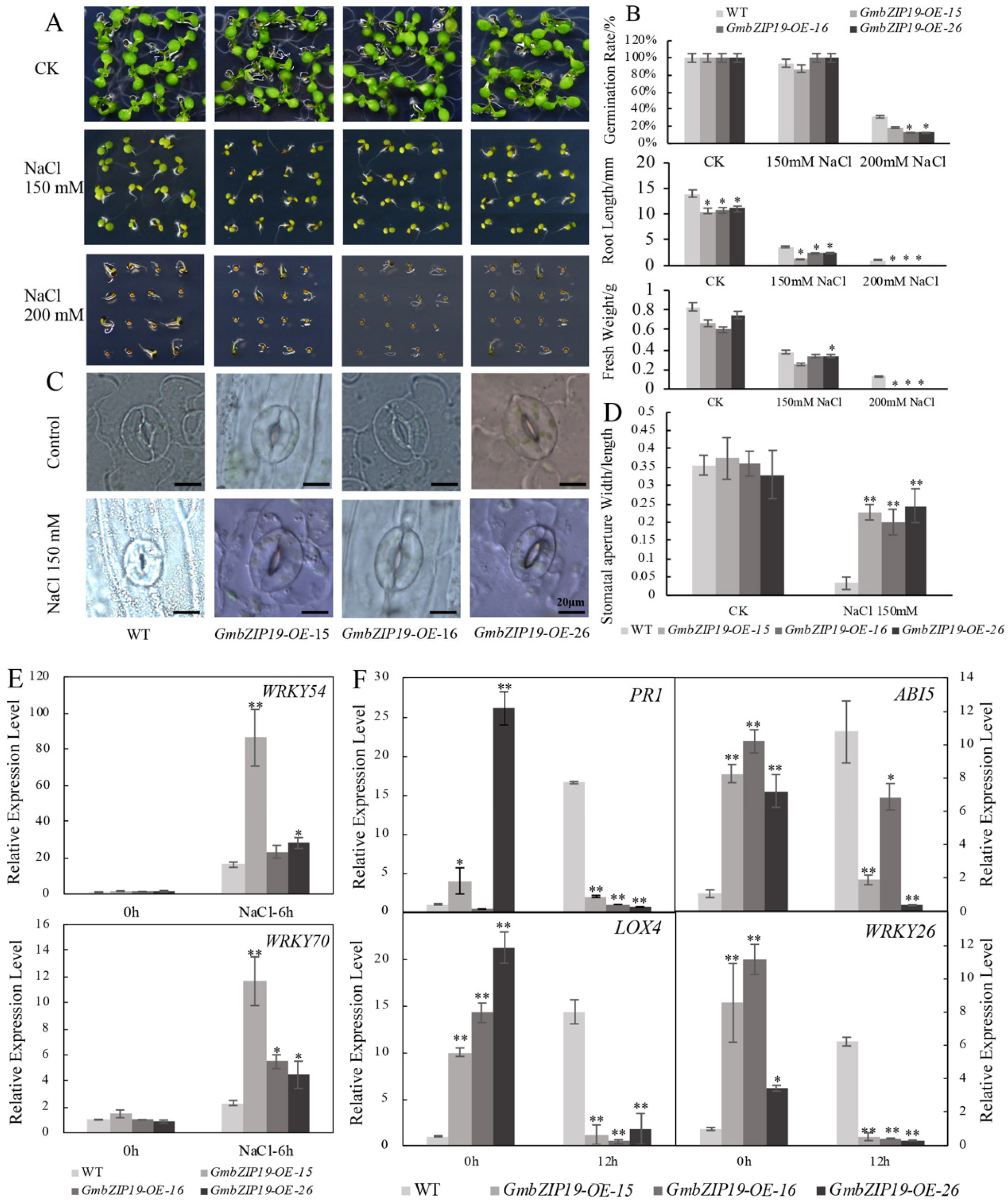
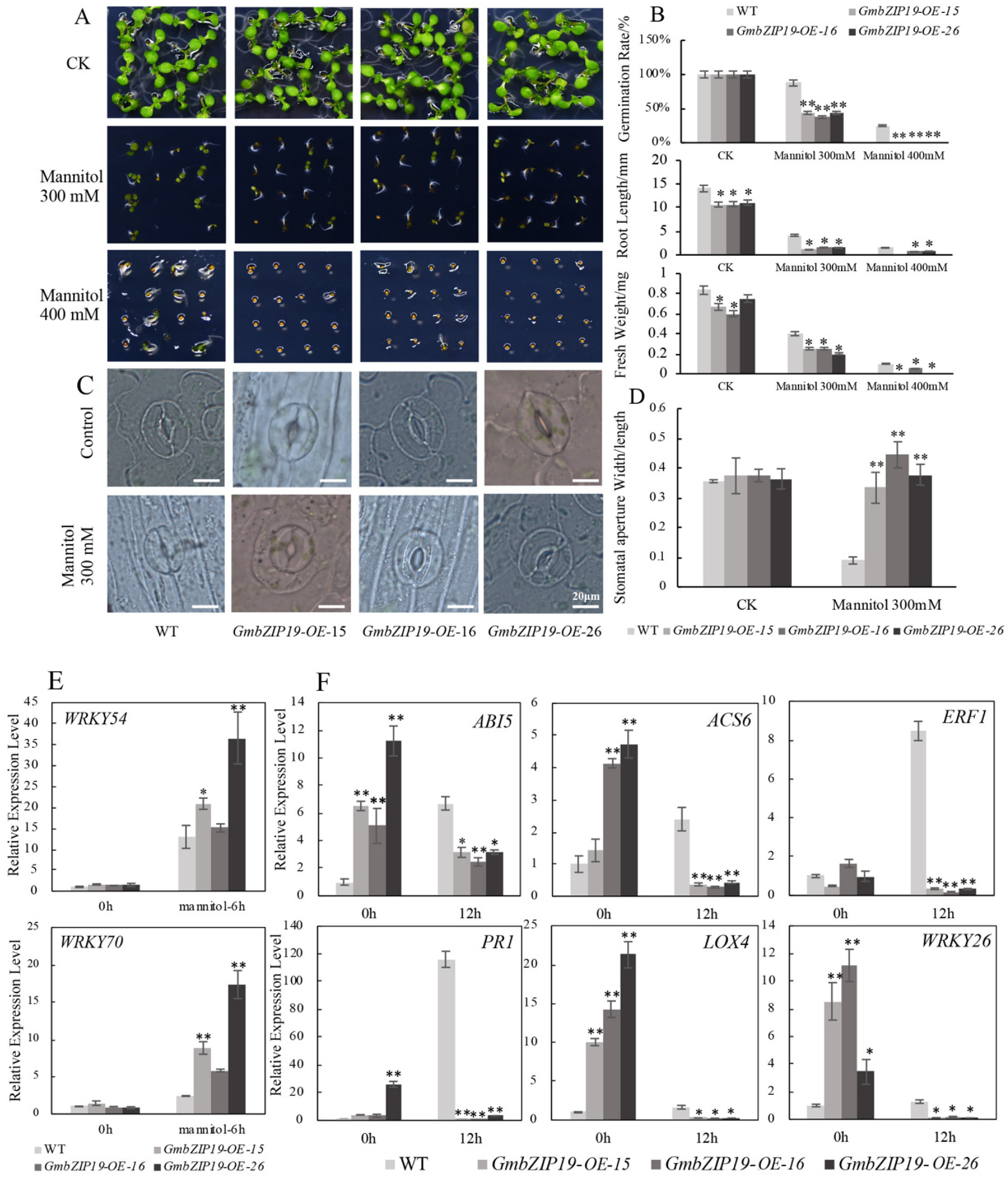
In order to determine the function of GmbZIP19 in response to salinity in plants, seeds of GmbZIP19-OE lines and WT were planted in 1/2 MS plates containing 0, 150 and 200 mM NaCl, respectively. As is shown in Figure 4A, under normal condition (no treatment), there was no obvious difference in growth between WT and GmbZIP19-OE plants. However, when exposed to 150 mM and 200 mM NaCl, a continuous decline of root length and fresh weight in both WT and GmbZIP19-OE plants was observed with the increasing of salt concentration, but the seed germination only dropped significantly under 150 mM NaCl treatment (Figure 4A,B). Although seed germination showed no significant difference between the WT and GmbZIP19-OE lines under control (non-salinity) and 150 mM NaCl, the seed germination rate was declined in GmbZIP19-OE lines compared to that in WT when subjected to 200 mM NaCl treatment (Figure 4B). The root length and fresh weight were obviously decreased in GmbZIP19-OE lines compared with WT treated with increasing NaCl concentrates (Figure 4A,B). These results indicated that overexpression of GmbZIP19 in Arabidopsis resulted in plant sensitivity to salinity.
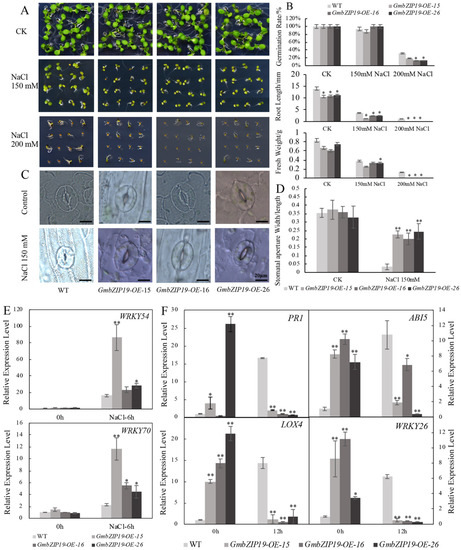
Figure 4.
GmbZIP19-overexpressed plants display sensitivity to salinity. (A) The phenotype of GmbZIP19-OE lines and the WT under 150 mM and 200 mM NaCl. (B) Quantification of the germination rate, root length and fresh weight of GmbZIP19-OE lines and the WT under 150 mM and 200 mM NaCl. The error bars indicate ±SD (n > 10 seedlings). (C) The stomatal aperture phenotype of GmbZIP19-OE lines and WT under 150 mM NaCl. Bar = 20 μm. (D) The quantitative analysis of stomatal aperture of GmbZIP19-OE lines and WT under 150 mM NaCl. (E) The expression levels of stomata movement marker genes in GmbZIP19-OE lines and WT under 150 mM NaCl. (F) The transcription levels of stress-related genes in GmbZIP19-OE lines and the WT under 150 mM NaCl. Three technical replicates were performed for each of the three independent biological replicates. The error bars indicate ±SD (n = 3 replicates). Asterisks indicate significant differences for the indicated comparisons based on a Student’s t-test (** p < 0.01; * p < 0.05).
With the treatment of 150 mM NaCl, the majority of stomata of WT leaves were completely closed, while the stomata of the GmbZIP19-OE lines did not close completely (Figure 4C). The stomatal aperture (width/length) of GmbZIP19-OE was extremely higher than that in WT under salt treatment, while there was nearly no difference under the control condition (Figure 4D). The expression levels of WRKY54 and WRKY70, two marker genes negatively regulating the stomatal closure [25], increased significantly in GmbZIP19-OE lines compared with that in WT under 150 mM NaCl treatment, while the expression levels of these two genes showed no significant difference in GmbZIP19-OE lines and WT under control also confirmed that the stomata of GmbZIP19-OE plants was defected. The defective stomatal aperture may be associated with the reduced tolerance of GmbZIP19-OE plants to salt.
To evaluate the involvement of plant hormones in the sensitivity of GmbZIP19-OE lines to salt, the relative expression levels of stress-responsive genes AtPR1, AtABI5, AtLOX4 and AtWRKY26 related to SA, ABA, JA and abiotic stress respectively, were determined by RT-qPCR. The results showed that the expression of these genes was higher in GmbZIP19-OE plants than WT at the normal condition. However, the expression levels of these four genes decreased dramatically in GmbZIP19-OE plants in contrast to the significantly increased expression in WT when subjected to 150 mM NaCl treatment stress for 12 h (Figure 4F). These results together indicated that Arabidopsis plants overexpressing GmbZIP19 were less tolerant to salt stress than WT plants and implied the involvement of plant hormone in salinity tolerance.
2.6. Overexpression GmbZIP19 Arabidopsis Exhibits Sensitivity to Drought (osmotic stress)
To further analyze the functions of GmbZIP19 in plant response to osmotic stress, WT and three GmbZIP19-OE Arabidopsis lines were subjected to 0, 300 and 400 mM mannitol. WT and GmbZIP19-OE plants have no significant difference in germination rate root length and fresh weight when grown in half strength MS medium without mannitol (Figure 5A,B). However, when subjected to drought stress with 300 and 400 mM mannitol, a considerable reduction in seed germination, root length and fresh weight were observed in both GmbZIP19-OE and WT plants, and the reduction rate was more severe in GmbZIP19-OE lines than WT (Figure 5A,B). These results suggested that overexpression of GmbZIP19 in Arabidopsis reduced plant drought tolerance. The reduced drought tolerance in GmbZIP19-OE plants was associated with defects in stomatal closure (Figure 5C–E), suggesting that stomatal closure may contribute to resistance to drought.
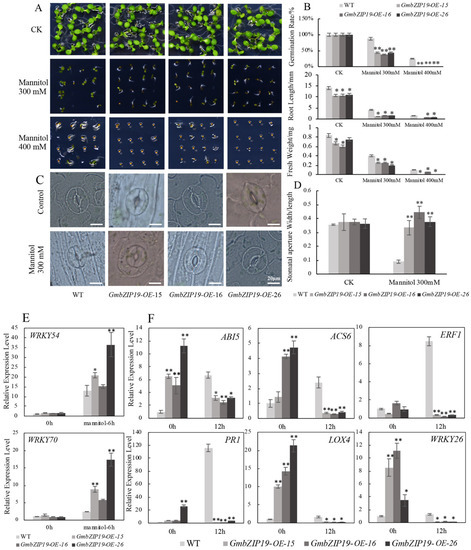
Figure 5.
GmbZIP19-overexpressed plants display sensitivity to drought. (A) The phenotype of GmbZIP19-OE transgenic lines and WT under 300 and 400 mM mannitol. (B) Quantification of the germination rate, root length and fresh weight of GmbZIP19-OE transgenic lines and WT under 300 and 400 mM mannitol. The error bars indicate ±SD (n > 10 seedlings). (C) The stomatal aperture phenotype of GmbZIP19-OE transgenic lines and WT under 300 mM Mannitol. Bar = 20 μm. (D) The quantitative analysis of stomatal aperture of GmbZIP19-OE transgenic lines and WT under 300 mannitol. (E) The expression levels of stomata movement marker genes in GmbZIP19-OE lines and WT under 300 mM mannitol. (F) The transcription levels of stress-related genes in GmbZIP19-OE lines and WT under 300 mM Mannitol. The error bars indicate ±SD (n = 3 replicates). Asterisks indicate significant differences for the indicated comparisons based on a Student’s t-test (** p < 0.01; * p < 0.05).
Moreover, we examined the relative expression levels of stress-responsive marker genes AtABI5, AtACS6, AtLOX4, AtERF1, AtPR1 and AtWRKY26 related to ABA (AtABI5, AtACS6), JA (AtLOX4), ETH (AtERF1), SA (AtPR1) and abiotic stress (AtWRKY26) respectively, and found that the expression of these marker genes in GmbZIP19-OE plants declined significantly after 12 h drought stress treatment. In contrast, increased marker genes’ expression was detected in WT upon drought stress (Figure 5F). These data together indicated that GmbZIP19 overexpression caused drought sensitivity in transgenic Arabidopsis.
2.7. Overexpression GmbZIP19 Arabidopsis Exhibits Sensitivity to Hormones
To further assess the response of GmbZIP19 to plant hormone, we treated the GmbZIP19-OE and WT plants with 450 μM ETH, 200 μM JA and 2.5 μM ABA, respectively. Comparable seed germination rate, root length and fresh weight were examined between the GmbZIP19-OE lines and WT grown on MS medium free of exogenous hormones (Figure S4B). The seed germination rate showed no obvious difference upon ETH and JA treatment compared to WT but reduced with JA supplement compared to WT (Figure S4B). Significantly reduced root length and fresh weight were detected in GmbZIP19-OE lines compared with WT when they were grown on the medium supplemented with ETH, JA and ABA (Figure S4B). These results showed that GmbZIP19-OE lines were sensitive to plant hormones.
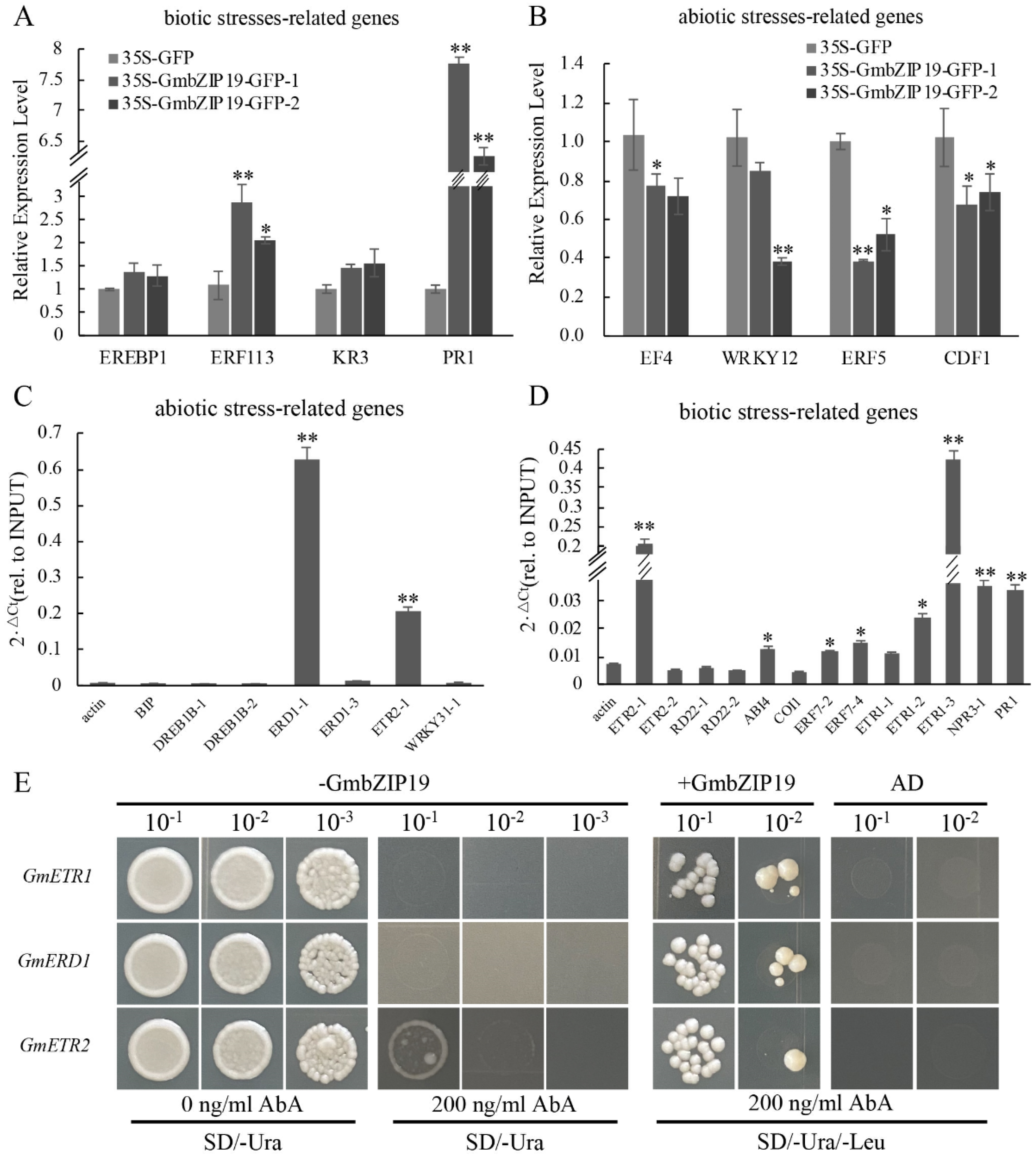
2.8. Consistent Effects on the Expression of Biotic and Abiotic Stress-Related Genes by Transient Overexpression of GmbZIP19 in Soybean
In order to infer the potential role of GmbZIP19 in response to biotic and abiotic stresses in soybean, we transiently overexpressed GmbZIP19-GFP fusion driven by 35S promoter (35S-GmbZIP19-GFP) in soybean leaf and evaluated the relative expression levels of biotic and abiotic stress-related genes in the 35S-GmbZIP19-GFP and 35S-GFP control soybean leaf. As shown in Figure 6A, the relative expression levels of biotic stress-related genes including EREBP1, ERF113, KR3 and PR1 were higher in 35S-GmbZIP19-GFP soybean leaf than that in the 35S-GFP control, coinciding with the activated stress-related gene expression in GmbZIP19-OE Arabidopsis plants upon pathogen infection (Figure 3). These results suggested that the activated biotic stress response was induced by transient overexpression of GmbZIP19 in soybean. In addition, similar to the decreased expression of the stress-responsive genes in GmbZIP19-OE Arabidopsis plants upon salt and drought stresses (Figure 4 and Figure 5), reduced expression of abiotic stress-related genes including EF4, WRKY12, ERF5 and CDF1 was examined in 35S-GmbZIP19-GFP soybean leaf as compared to 35S-GFP leaf (Figure 6B). These results suggested that transient overexpression of GmbZIP19 in soybean may also cause enhanced sensitivity to abiotic stress as it does in Arabidopsis.
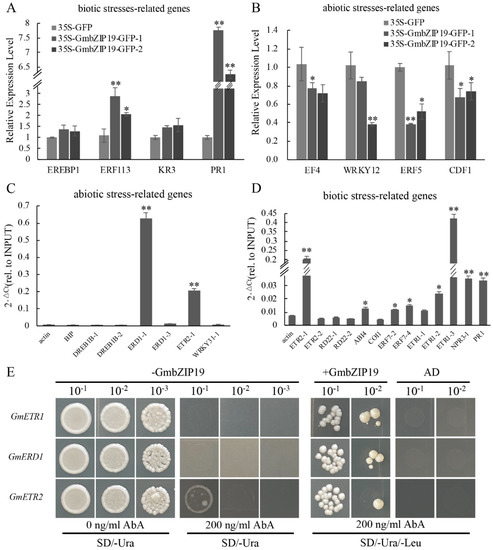
Figure 6.
The ChIP result of 35S-GmbZIP19–GFP transient expressing soybean. (A) qRT-PCR analysis of biotic stress-related genes in 35S-GmbZIP19–GFP transient expressing soybean. (B) qRT-PCR analysis of abiotic stress-related genes in 35S-GmbZIP19–GFP transient expressing soybean. (C) ChIP-qPCR analysis of GmbZIP19 binding to abiotic stress-related genes using GFP antibody and 35S-GmbZIP19–GFP transient expressing soybean. (D) ChIP-qPCR analysis of GmbZIP19 binding to biotic stress-related genes using GFP antibody and 35S-GmbZIP19–GFP transient expressing soybean. Three independent biological replicates were performed. The error bars indicate ± SD (n = 3 replicates). Asterisks indicate significant differences for the indicated comparisons based on a Student’s t-test (** p < 0.01; * p < 0.05). (E) The yeast one-hybrid result of GmbZIP19. Three independent biological replicates were performed.
2.9. Identification of GmbZIP19 Target Genes
To identify the possible target genes of GmbZIP19, Chromatin immunoprecipitation (ChIP) was performed by using 35S-GmbZIP19-GFP transient expressing soybean. The ChIP assay was performed by using a GFP antibody to pull down the putative GmbZIP19-bound DNA from the leaf tissues of 35S-GmbZIP19-GFP transient expressing soybean. ChIP-qPCR showed enrichment of GmbZIP19 in the promoters of some abiotic stress-related genes, including GmERD1 and GmETR2 (Figure 6C), and the promoters of biotic stress-related genes, such as GmETR2, GmABI4, GmERF7, GmETR1, GmNPR3 and GmPR1, which were also related to ETH (GmETR1, GmETR2, GmERF7), ABA (GmABI4), JA (GmNPR3) and SA (GmPR1) signaling pathways, respectively (Figure 6D). Moreover, three target genes (GmETR1, GmERD1 and GmETR2) with the most enrichment of GmbZIP19 were chosen to verify the direct interaction between GmbZIP19 and these genes using yeast one-hybrid. 70 bp original fragments (named as GmETR1, GmERD1, GmETR2) containing the cis-element were used as bait and cloned into the pABAi vector, while GmbZIP19 was used as a prey. As shown in Figure 6E, the yeast cells of original and mutational fragment were co-transformed with or without GmbZIP19. Results showed that, when bait vector transformed alone into yeast, they grew normally in screening medium (SD/-Ura/-Leu) but were inhibited by 200 ng/mL AbA (Aureobasidin A), and the bait vector co-transformed with GmbZIP19 could survive under 200 ng/mL, which suggested that GmbZIP19 could directly bind to the GmETR1, GmERD1 and GmETR2 promoters.
3. Discussion
The bZIP TF family is a plant-specific transcription factor family and is involved in diverse signaling pathways underlying biotic and abiotic stress responses and biological processes. Although the genome-wide analysis of the bZIP transcription factor family has been conducted in soybean [17] and there are several bZIP genes’ functions reported in plants [26,27], little is known about the functions of bZIP genes in soybean. In our study, GmbZIP19 was isolated from full-length soybean cDNA. The multiple alignment analysis showed that GmbZIP19 contains a typically conserved DNA binding domain (N-x7-R/K-x9) and a DOG1 structural domain of 79 amino acids among different plants (Figure S1). The subcellular localization indicated that GmbZIP19 was localized in the nucleus (Figure S2). Moreover, numerous stress-related cis-elements, including G-box, MYB, CGTCA-motif and TCA-motif were present in the 2000 bp promoter region of the GmbZIP19 gene (Figure S3). The MYB transcription factor can combine with MYBRS and is involved in ABA signaling pathway and abiotic stress responses [28]. Some genes can confer abiotic tolerance through their G-box [29]. Such an abundance of stress responsive cis-elements in the GmbZIP19 promoter suggested a potential role of this gene in stress responses. Therefore, in order to explore the potential functions of GmbZIP19 in response to different stresses, we overexpressed GmbZIP19 in Arabidopsis and revealed that GmbZIP19-OE Arabidopsis exhibited significantly increased resistance to S. sclerotiorum and Pseudomonas syringae (pst. DC3000) but sensitivity to abiotic stresses, including salinity and drought.
Plants have a wide range of mechanisms to cope with many stresses in their natural environments [30]. In terms of disease defense, plants can recruit an inducible defense system to resist the infection of certain pathogens. Previous studies have shown that phytohormones, including ABA, SA, JA and ETH play crucial roles in biotic stress signaling following pathogen infection [31,32,33,34]. Overexpression of ERF1 in Arabidopsis increased tolerances to some necrotrophic pathogens [35]. Overexpression of GmKR3 in soybean enhanced virus tolerance through affecting ABA signaling [36]. In the current study, the expression of GmbZIP19 in wild-type soybean was dramatically induced by S. sclerotiorum (Figure 1A), and GUS activity of pGmbZIP19-GUS transgenic leaves was also induced by inoculation of S. sclerotiorum (Figure 2G,H). Furthermore, the expression of GmbZIP19 was activated by diverse hormones including JA, SA, ETH, ABA and BR (Figure 1B–F), indicating that GmbZIP19 may be involved in pathogen defense which is related to different hormone signaling pathways. In addition, a significant upregulation in the expression levels of several marker genes responding to ABA, JA, SA and ETH was detected in the transgenic GmbZIP19-OE lines exposed to S. sclerotiorum compared to WT (Figure 3D), which further confirmed the involvement of phytohormone signaling in regulation of GmbZIP19-mediated pathogen resistance. GmbZIP19 confers similar resistance to pst. DC3000 but only JA- and SA-dependent genes are detected to be functioning in pst. DC3000 resistance (Figure 3E,F). According to previous studies, PR2, a SA-responsive gene [37], is a key component in the SA defense signaling pathway. The increased expression of PR2 in GmbZIP19-OE lines upon pathogen infection suggested that the SA signaling pathway may participate in pathogen tolerance of GmbZIP19-OE transgenic Arabidopsis. Moreover, the signaling pathways mediated by JA and ETH usually respond to necrotrophic pathogens, insects, herbivores and injury [38]. LOX4 and ERF1, which participate in the JA and ETH signaling pathways respectively, were upregulated in GmbZIP19-OE leaves upon S. sclerotiorum inoculation. The induced expression of the two marker genes by pathogen infection may indicate that JA and ETH are involved in the regulation of GmbZIP19 to disease defense as well.
Previously, studies showed that various bZIP genes were involved in regulating plant response to abiotic stresses such as salt, drought and low temperature, etc. [13,39]. Overexpression of TabZIP60 in wheat confers multiple abiotic stress tolerances [13], and overexpression of TaWRKY2 could increase drought tolerance of transgenic wheat [40]. Different hormones are essential in the regulation of abiotic stress resistance. OsbZIP72 serves as a positive regulator of ABA response and confers drought tolerance in transgenic rice [41]. Additionally, OsJAZ1 alternates drought resistance by regulating JA and ABA signaling in rice [42]. In our study, the expression of GmbZIP19 was reduced by salt and drought (Figure 1G,H), and the GUS activity of pGmbZIP19-GUS transgenic seedlings after abiotic stresses was also dramatically decreased (Figure 2E,F), suggesting that GmbZIP19 is involved in abiotic response. Furthermore, our study showed that the salt and drought tolerance were significantly decreased in GmbZIP19-OE transgenic Arabidopsis compared to WT (Figure 4A,B and Figure 5A,B). Moreover, stomatal closure was defective in GmbZIP19-OE Arabidopsis with salt or drought treatment (Figure 4C–E and Figure 5C–E). Previous studies indicated that stomatal closure was associated with the ABA signaling pathway and therefore affected drought tolerance [43,44]. It is possible that the drought and salt sensitivity of GmbZIP19-OE transgenic lines was due to the defective stomatal aperture (Figure 4C–E and Figure 5C–E). Because the transcription factors may interact with the cis-elements in the promoter regions of many abiotic stress-related genes, thus affecting the expression of these stress-related genes, resulting in defective abiotic stress tolerance [45], it is necessary to detect the expression levels of stress-related marker genes. In the current study, the decreased expression of WRKY26, which is reported to be induced by various abiotic stresses such as salt, heat and cold treatment [46], directly confirmed the sensitivity of GmbZIP19-OE transgenic plants to abiotic stresses. In addition, the hormone-related marker genes involved in ABA, SA, JA and ETH signaling pathways were expressed at lower levels in GmbZIP19-OE plants than in the WT after salt or drought treatment (Figure 4F and Figure 5F), demonstrating that the hormone-related marker genes are involved in the abiotic stress tolerance. These results suggested that GmbZIP19 affects the abiotic tolerances by being involved in the hormone responsive pathways.
Although GmbZIP19-OE plants were more sensitive to exogenous hormones than wild-type (Figure S4), the expression of GmbZIP19 was induced by ABA, SA, JA and ETH (Figure 1B–E). The results implied the interlink between phytohormone signaling and GmbZIP19. ChIP-qPCR analysis showed that GmbZIP19 could bind to the promoter of many hormone-associated stress-related genes (Figure 6C,D). The yeast one-hybrid assay confirmed the result (Figure 6E). These findings further confirmed that phytohormone signaling pathways are involved in GmbZIP19 regulated biological processes.
In animals, there are several TFs that serve either as transcriptional activators or repressors [47]. In plants, few transcription factors with dual functions have been characterized. In Arabidopsis, AtWRKY33 can activate the expression of CYP71A13 and PAD3, while transcript levels of NCED3 and NCED5 increased simultaneously in the wrky33 mutant after B. cinerea infections, suggesting that it acts as a repressor as well [48]. Also, ATAF2, a NAC domain transcription factor, acts as transcriptional activator or repressor dependent on promoter context [49]. In our study, GmbZIP19 is a multi-functional TF that can regulate not only the disease defense but also the abiotic stress tolerance by participating in phytohormone signaling pathways. However, the more precise mechanism of GmbZIP19 function and how GmbZIP19 acts as an activator of biotic stress and a repressor of abiotic stress still need to be confirmed and the interaction of GmbZIP19 with its homologous genes in Arabidopsis in stress responses continues to attract our attention.
In general, our study provides vital insights into the mechanism underlying the response of Arabidopsis and soybean to biotic and abiotic stresses and offer clues for brand new soybean designing, which can be more accessible to negative environments.
4. Material and Methods
4.1. Plant Materials and Growth Condition
Arabidopsis thaliana (Columbia ecotype) seeds were surface-sterilized with absolute ethyl alcohol for 5 min and 75% ethanol for 15 min, followed by washing three or four times with sterilized distilled water under aseptic conditions. The sterilized Arabidopsis seeds were placed on the Petri plates containing half-strength Murashige and Skoog (MS) medium. The plates were kept in the growth incubator for germination and development (22 °C, 16 h light/8 h dark). After seven days, the seedlings were transferred into soil and allowed to grow under control environmental conditions (22 °C, 16 h light/8 h dark).
Seeds of soybean (YC03-3) were sown in soil and grown at 25 °C with a 16 h light/8 h dark (normal condition). Two-week-old soybean plants were infected with S. sclerotiorum and treated with 100 μM JA, 1 mM SA, 100 μM ABA, 1 mM ETH, 100 μM BR, 100 mM NaCl for salt condition, 250 mM mannitol for drought condition and 4 °C (placed the soybean in a 4 °C refrigerator and used plant growth light to keep the light condition the same as other plants) for low temperature, respectively. Normal condition or control means there is no treatment with the Arabidopsis or soybean.
4.2. GmbZIP19 Sequence Analysis
The GmbZIP19 (Glyma11G16050) cDNA was identified from Soybean cDNA libraries (https://phytozome.jgi.doe.gov/pz/portal.html) and the specific primers were designed, which are listed in Table S1. Homologous sequences of GmbZIP19 in other species (Arabidopsis and rice) were downloaded from phytozome databases and sequence alignment was performed with DNAMAN.
4.3. The RNA Extraction and RT-qPCR Analysis
Total RNA used for the expression profile of GmbZIP19 was isolated from leaves of wild-type soybean grown under normal growth conditions and different stress conditions, including salinity (NaCl), drought (mannitol), cold (4 °C), JA, ABA, SA, BR and ETH at 2, 6, 12, 24 and 48 h after treatment. In addition, total RNA of different tissues of wild-type soybean, including leaf, stem, root and inflorescence were isolated using a plant RNA extraction kit (OMEGA, China). During the extraction of RNA, RNase-free DNase (Promega, Madison, WI, USA) was used to avoid DNA contamination, and then the first-strand cDNA synthesis was carried out with approximately 1 mg RNA using the Transgene First Strand cDNA Synthesis Kit. The cDNAs were diluted five times to conduct RT-qPCR. Soybean ACTIN was used as reference gene (primers are listed in Table S2).
Total RNA used for transcription analyses of GmbZIP19-OE Atabidopsis was isolated from leaves of three-week-old WT and GmbZIP19-OE Arabidopsis plants under different stress conditions: 150 mM NaCl (0, 6, 12, 24, 48 h), 300 mM mannitol (0, 6, 12, 24, 48 h). Total RNA extraction and cDNA synthesis were carried out using methods described in the above sections. qPCR was performed to examine the relative expression levels of stress-related genes (the name and specific primers of these stress-related genes are listed in Table S2). In each case, all the experiments were repeated three times with all three technical replicates. Arabidopsis HK2 was used as reference gene (primers are listed in Table S2).
To examine the specific expression of different materials, RT-qPCR was performed using SYBR Premix Ex Taq (TaKaRa, Toyoto, Japan). Reaction mixtures were prepared with a total volume of 20 μL each, containing: 1 μL of template, 8.2 μL of RNase-free water, 10 μL of 2× SYBR Premix and 0.4 μL of each specific primer according to the Bio-Rad Real-time PCR system (Foster city, CA, USA) and the SYBR Premix Ex II system (TakaRa Perfect Real Time). The RT-qPCR program was: 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 34 s, and 95 °C for 15 s. The primers used for RT-qPCR are listed in Table S2. In each case, three technical replicates were performed for each of the three independent biological replicates. Only Ct values less than 40 were used to calculate correlation coefficients (R2 values) and amplification efficiencies (E) from the slope generated in Microsoft Excel 2013, based on the equation: E = [10-(1/slope) − 1] × 100%. All PCR assays showed efficiency values between 95% and 110% [50].
4.4. Transformation and Generation of Transgenic Arabidopsis Plants and Stress Treatments
A 723 bp fragment of GmbZIP19 (coding sequence) was amplified by PCR from the cDNA of wild-type soybean leaf with specific primers (shown in Table S1) and the promoter of GmbZIP19 was cloned from the DNA of wild-type soybean with specific primers (shown in Table S1). The PCR products were cloned into pENTER/D-TOPO vector (Invitrogen) and then the recombinants were further fused into the destination vector pGWB605 and pGWB633 vector respectively, using Gateway LR Clonase II enzyme mix (Invitrogen). The 35S:GmbZIP19-GFP and 35S:pGmbZIP19-GUS recombinant vector was transformed into wild-type Arabidopsis (Columbia ecotype) using the Agrobacterium-mediated floral dip method [51]. Then, one-week-old T1 generation seedlings were sprayed with 0.4 mg/mL basta for selection of positively transformed plants and surviving seedlings were subsequently transferred to soil. The homozygous T1 generation seeds were harvested individually to obtain T2 generation seeds of different lines.
WT and transgenic Arabidopsis seeds were harvested at the same time. For salt, drought, ABA, JA and ETH, GmbZIP19-OE Arabidopsis and wild-type seeds were germinated on different types of 1/2 MS medium supplemented with 150 and 200 mM NaCl, 300 and 400 mM mannitol, 0.5 μM ABA, 200 μM JA and 450 μM ETH, respectively. In addition, seeds of pGmbZIP19-GUS Arabidopsis were planted on 1/2 MS medium with 150 mM NaCl and 300 mM mannitol, respectively. We used 1/2 MS medium without any supplementation as a control to germinate WT and transgenic Arabidopsis seeds. Plates were placed under controlled environmental conditions (22 ℃) for 4–7 days. The germination rate, root length and fresh weight of wild-type and transgenic Arabidopsis were measured and statistically analyzed. Three independent biological replicates were performed, and Student’s t-test was used for statistical analysis.
4.5. Subcellular Localization of GmbZIP19
The recombinant vector 35S-GmbZIP19-GFP was used for the subcellular localization study. To confirm the localization of GmbZIP19 fusion protein in cells, the 35S-GmbZIP19-GFP and 35S-GFP (vector control) recombinant fused plasmids were transformed into tobacco leaf cells using Agrobacterium tumefaciens strain GV3101.Transfected tobacco leaves were incubated in a greenhouse at 22 °C for more than 36 h. The fluorescence signals were monitored using confocal laser-scanning microscopy (SP5, Leica, Germany) with GFP fluorescence detection at 488 nm [52].
4.6. GUS Staining
Inflorescence samples were fixed in prechilled 90% acetone for 20 min and washed with distilled water. After brief vacuum infiltration, the inflorescences were incubated in β-glucuronidase (GUS) staining buffer overnight at 37 °C. After being cleared in 20% lactic acid/20% glycerol solution, the inflorescences were observed under a Leica (M205 FA) microscope.
4.7. S. sclerotiorum Infection and Staining
Three-week-old plants of WT and GmbZIP19-OE Arabidopsis were inoculated with S. sclerotiorum. The similar detached leaves of WT and GmbZIP19-OE lines are inoculated with the same size of S. sclerotiorum. The inoculated leaves were collected at 12 h post-infection. The inoculated leaves were stained with 1 mg/mL DAB (diaminobenzidin) for 8 h and then boiled in 75% ethanol for approximately 10 min to decolorize. The detailed methods of inoculation and disease scoring were the same as those described previously [53].
To verify the expression profile of stress-related genes under pathogen stress, we inoculated S. sclerotiorum to the three-week-old leaves and isolated inoculated leaves after 0, 6, and 12 h for total RNA extraction for qPCR (the stress-related genes are: AtPR2, AtABI5, AtLOX4 and AtERF1 and specific primers are listed in Table S2).
4.8. Pseudomonas Syringae Infection and Staining
Pst DC3000 was grown overnight in King’s B (KB) liquid medium containing 50 μg/mL rifampicin. The medium with bacteria was centrifuged at 2800 rpm for 10 min to collect bacteria. The pellet was resuspended in 10 mM MgCl2 and diluted to get OD600 = 0.001–0.002 for plant inoculation. Three-week-old WT and GmbZIP19-OE Arabidopsis plants were vacuum-infiltrated with suspension of Pseudomonas syringae. The inoculated leaves were sprayed with water and kept in a sealed container to ensure high humidity and the disease progress was observed daily [54]. Inoculated leaves were collected after 48 h post inoculation and stained with trypan blue, as mentioned below (trypan blue staining solution contains: 10 mL lactic acid, 10 mL glycerol, 10 g phenol, 10 mL distilled water, 40 mL 75% ethanol and 2 mL trypan blue). The leaves were boiled for 5 min in the trypan blue staining solution and then decolorized in chloral hydrate (0.5 g chloral hydrate dissolved in 1 mL of 75% ethanol) for at least 30 min and then viewed under a microscope with bright light [53].
4.9. Stomatal Aperture Observation under Different Stresses
Four-week-old WT and GmbZIP19-OE Arabidopsis were used to observe and measure the stomatal aperture conditions. Leaves of similar size, age and side branches of stems were selected to compare stomatal aperture. At first, the leaves and stems were infused in stomatal solution containing: 50 mM CaCl2, 10 mM MES, 5 mM KCl, pH 6.15 and exposed to light for approximately 2 h to ensure all the stomatal had opened. Subsequently, 150 mM NaCl and 300 mM mannitol were added to the solutions to induce salt and drought stress conditions, respectively. After 2 h of treatments, leaves and stems of WT and GmbZIP19-OE Arabidopsis were blended and observed with a digital microscope (Leica). The length and width of stomata were measured using image analysis (Adobe photoshop CC 2019) computer software [55].
To confirm the phenotype and quantitative result, we examined the expression levels of marker genes of stomatal movement and closure using GsmbZIP19-OE plants under salt or drought treatment. The stomatal movement marker genes are: WRKY54 and WRKY70, the primers are listed in Table S2.
4.10. Transient Expression of GmbZIP19 and Chromatin Immunoprecipitation
Two-week-old soybean cultivar Huachun6 (YC03-3) was used to achieve the transient expression of GmbZIP19. The Agrobacterium containing recombinant vector 35S-GmbZIP19-GFP and 35S-GFP were grown overnight in LB liquid medium containing 50 μg/mL of spectinomycin and rifampicin. The media were centrifuged at 4000 rpm for 10 min. Pellets were resuspended in 3% sugar aqueous solution (M/V) with 0.1% silweet-77 (V/V). The leaves of soybean were infiltrated with Agrobacterium solutions. Two days after inoculation, the GFP signal of inoculated leaves was observed with a Leica TCS SP8X DLS confocal laser scanning microscope to confirm that the transient expression had been achieved successfully. The qPCR was performed by primers in Table S3 with GmbZIP19 transient expressing soybean. Three technical replicates were performed for each of the three independent biological replicates.
For each chromatin immunoprecipitation (ChIP) experiment, 4 g of leaves were used. Crosslinked chromatin was fragmented with 0.2 units of micrococcal nuclease in 1 mL of MNase digestion buffer (10 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM-mercaptoethanol, 0.1% NP40, 1 mM CaCl2, and 1× protease inhibitor cocktail). Digestion was stopped using 5 mM EDTA. ChIP was performed using a GFP polyclonal antibody. Relative enrichment of associated DNA fragments was analyzed by qPCR. All oligonucleotide sequences used in the ChIP experiments are given in Table S3. Each ChIP experiment was repeated twice, and the presented data are from one representative experiment. In each case, three technical replicates were performed for each of the three independent biological replicates.
4.11. Yeast One-Hybrid System
GmbZIP19 CDS without stop codon was amplified and then integrated into pGADT7 by the In-fusion cloning technique (Clontch, Takara) to form a pGADT7-GmbZIP19 effect vector. At the same time, the predicted binding sites were synthesized by DNA synthesis technology (the primers are listed in Table S4), and cloned into pABAi vector by In-fusion cloning technology to form pABAi-GmETR1, pABAi-GmETR2, pABAi-GmERD1, pABAi-GmNPR3 and pABAi-GmPR1 bait report vectors, respectively.
Yeast one-hybrid was carried out according to instructions provided by Clontech (Takara). Bait was transformed into Y1H gold yeast strain and cultured on SD/-Ura medium with or without 200 ng/mL Aureobasidin A (AbA) for 3 days. In addition, the yeast cells co-transformed by prey and bait were cultured on SD/-Leu medium containing 200 ng/mL AbA for 3 days.
4.12. Statistics Analysis
All experiments were carried out with three biological replicates and three technical replicates per biological replicate, and the data were shown as means ± standard errors (SD; n = 3). Asterisks indicate significant differences for the indicated comparisons based on a Student’s t-test (** p < 0.01; * p < 0.05). Three biological replicates were used for each of the genotypes (GUS staining, biotic and abiotic phenotype, stomatal aperture, DAB and trypan blue staining), including the wild-type and GmbZIP19-OE-15, GmbZIP19-OE-16 and GmbZIP19-OE-26 transgenic lines. For relative lesion area measurement, five biological replicates were performed for each line. For seed germination, root length and fresh weight measurement, more than ten biological replicates were performed. Asterisks indicate significant differences for the indicated comparisons based on a Student’s t-test (** p < 0.01; * p < 0.05).
5. Conclusions
In this study, we cloned and characterized soybean GmbZIP19. Overexpression of GmbZIP19 in Arabidopsis resulted in increased tolerance to pathogen and decreased tolerance to drought and salt stress. Our results indicated that GmbZIP19 increased plant pathogen tolerance and suppressed salt and drought stress response in association with plant hormones. Our findings obtained interesting basic scientific results that may shed light on the role of GmbZIP19 TF in biotic and abiotic stress response which might be crucial for developing environmental stress-resistant soybean varieties.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/13/4701/s1.
Author Contributions
Q.H. and Y.Q. initiated and designed the research; Q.H. performed the experiment; H.C. (Hanyang Cai) and M.B. performed the bioinformatic analyses; M.Z., S.V.G.N.P., F.C. and Y.H. carried out gene expression analyses; M.C., L.L., Y.L. and H.C. (Huihuang Chen) helped with a critical discussion on the work; Q.H. wrote the paper; Y.Q., H.C. (Hanyang Cai) and S.V.G.N.P. revised the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by NSFC (U1605212, 31970333) and a Guangxi Distinguished Experts Fellowship.
Acknowledgments
We would like to thank the reviewers for their helpful comments on the original manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Maruyama, K.; Ogata, T.; Kanamori, N.; Yoshiwara, K.; Goto, S.; Yamamoto, Y.Y.; Tokoro, Y.; Noda, C.; Takaki, Y.; Urawa, H.; et al. Design of an optimal promoter involved in the heat-induced transcriptional pathway in Arabidopsis, soybean, rice and maize. Plant. J. 2017, 89, 671–680. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant. Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant. J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Laxmi, A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant. Sci. 2015, 6, 895. [Google Scholar] [CrossRef]
- Gahlaut, V.; Jaiswal, V.; Kumar, A.; Gupta, P. Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (triticum aestivum L.). Theor. Appl. Genet. 2016, 129, 2019–2042. [Google Scholar] [CrossRef]
- Hurst, H.C. Transcription factors. 1: bZIP proteins. Protein Profile 1994, 1, 123–168. [Google Scholar]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant. Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Ellenberger, T.E.; Brandl, C.J.; Struhl, K.; Harrison, S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: Crystal structure of the protein-DNA complex. Cell 1992, 71, 1223–1237. [Google Scholar] [CrossRef]
- Izawa, T.; Foster, R.; Chua, N.H. Plant bZIP protein DNA binding specificity. J. Mol. Biol. 1993, 230, 1131–1144. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.F.; Zhang, L.X.; Yu, Y.W.; Quan, R.D.; Zhang, Z.J.; Zhang, H.W.; Huang, R.F. The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant. J. 2011, 68, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Liu, J.; Jia, J.; Kong, X. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plant. 2015, 153, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, J.; Zhang, B.; Vanitha, J.; Ramachandran, S.; Jiang, S.Y. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant. Biol. 2011, 53, 212–231. [Google Scholar] [CrossRef]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, Y.; Shi, H.; Guo, M.; Chai, M.; He, Q.; Yan, M.; Cao, D.; Zhao, L.; Cai, H.; et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genom. 2018, 19, 159. [Google Scholar] [CrossRef]
- Zhang, J.; Du, H.; Chao, M.; Yin, Z.; Yang, H.; Li, Y.; Huang, F.; Yu, D. Identification of two bZIP transcription factors interacting with the promoter of soybean rubisco activase gene (GmRCAalpha). Front. Plant. Sci. 2016, 7, 628. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Q.; Nan, H.; Li, X.; Lu, S.; Zhao, X.; Liu, B.; Guo, C.; Kong, F.; Cao, D. Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PLoS ONE 2017, 12, e0179554. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, T.F.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Wei, W.L.; Xu, Z.S. The Soybean bZIP Transcription Factor Gene GmbZIP2 Confers Drought and Salt Resistances in Transgenic Plants. Int. J. Mol. Sci. 2020, 21, 670. [Google Scholar] [CrossRef]
- Inaba, S.; Kurata, R.; Kobayashi, M.; Yamagishi, Y.; Mori, I.; Ogata, Y.; Fukao, Y. Identification of putative target genes of bZIP19, a transcription factor essential for Arabidopsis adaptation to Zn deficiency in roots. Plant. J. 2015, 84, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Assuncao, A.G.; Herrero, E.; Lin, Y.F.; Huettel, B.; Talukdar, S.; Smaczniak, C.; Immink, R.G.; Van Eldik, M.; Fiers, M.; Schat, H.; et al. Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc. Natl. Acad. Sci. USA 2010, 107, 10296–10301. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. The Microbial Opsin Homolog Sop1 is involved in Sclerotinia sclerotiorum Development and Environmental Stress Response. Front. Microbiol. 2015, 6, 1504. [Google Scholar] [CrossRef]
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis thaliana-pseudomonas syringae interaction. Arab. Book 2002, 1, e0039. [Google Scholar] [CrossRef]
- Li, J.; Besseau, S.; Toronen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, E.T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013, 200, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Iwata, Y.; Koizumi, N. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 2005, 102, 5280–5285. [Google Scholar] [CrossRef]
- Joo, J.; Lee, Y.H.; Song, S.I. OsbZIP42 is a positive regulator of ABA signaling and confers drought tolerance to rice. Planta 2019, 249, 1521–1533. [Google Scholar] [CrossRef]
- Iturriaga, G.; Leyns, L.; Villegas, A.; Gharaibeh, R.; Salamini, F.; Bartels, D. A family of novel myb-related genes from the resurrection plant Craterostigma plantagineum are specifically expressed in callus and roots in response to ABA or desiccation. Plant. Mol. Biol. 1996, 32, 707–716. [Google Scholar] [CrossRef]
- Ahmad, A.; Niwa, Y.; Goto, S.; Ogawa, T.; Shimizu, M.; Suzuki, A.; Kobayashi, K.; Kobayashi, H. bHLH106 Integrates Functions of Multiple Genes through Their G-Box to Confer Salt Tolerance on Arabidopsis. PLoS ONE 2015, 10, e0126872. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Chen, X.; Xu, Z.; Guan, S.; Li, L.C.; Li, A.; Guo, J.; Mao, L.; Ma, Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J. Exp. Bot. 2008, 59, 4095–4107. [Google Scholar] [CrossRef]
- Adie, B.A.; Perez-Perez, J.; Perez-Perez, M.M.; Godoy, M.; Sanchez-Serrano, J.J.; Schmelz, E.A.; Solano, R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant. Cell 2007, 19, 1665–1681. [Google Scholar] [CrossRef]
- Mazarei, M.; Elling, A.A.; Maier, T.R.; Puthoff, D.P.; Baum, T.J. GmEREBP1 is a transcription factor activating defense genes in soybean and Arabidopsis. Mol. Plant. Microbe Interact. 2007, 20, 107–119. [Google Scholar] [CrossRef]
- Jing, Y.; Liu, J.; Liu, P.; Ming, D.; Sun, J. Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Luan, Q.; Chen, C.; Liu, M.; Li, Q.; Wang, L.; Ren, Z. CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant. Sci. 2019, 279, 59–69. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Molina, A.; Solano, R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant. J. 2002, 29, 23–32. [Google Scholar] [CrossRef]
- Xun, H.; Yang, X.; He, H.; Wang, M.; Guo, P.; Wang, Y.; Pang, J.; Dong, Y.; Feng, X.; Wang, S.; et al. Over-expression of GmKR3, a TIR-NBS-LRR type R gene, confers resistance to multiple viruses in soybean. Plant. Mol. Biol. 2019, 99, 95–111. [Google Scholar] [CrossRef]
- Alves, M.S.; Dadalto, S.P.; Goncalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef]
- Howe, G.A.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant. Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef]
- Huang, C.; Zhou, J.; Jie, Y.; Xing, H.; Zhong, Y.; Yu, W.; She, W.; Ma, Y.; Liu, Z.; Zhang, Y. A Ramie bZIP Transcription Factor BnbZIP2 Is Involved in Drought, Salt, and Heavy Metal Stress Response. DNA Cell Biol. 2016, 35, 776–786. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Y.; Xu, P.; Zhang, Z. Overexpression of a WRKY transcription factor tawrky2 enhances drought stress tolerance in transgenic wheat. Front. Plant. Sci. 2018, 9, 997. [Google Scholar] [CrossRef]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant. Sci. 2017, 8, 2108. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z.; et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2 O2 signalling in rice. Plant. Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef]
- He, F.; Wang, H.L.; Li, H.G.; Su, Y.; Li, S.; Yang, Y.; Feng, C.H.; Yin, W.; Xia, X. PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant. Biotechnol. J. 2018, 16, 1514–1528. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Jha, B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 2010, 54, 201–212. [Google Scholar] [CrossRef]
- Fu, Q.T.; Yu, D.Q. Expression profiles of AtWRKY25, AtWRKY26 and AtWRKY33 under abiotic stresses. Yi Chuan 2010, 32, 848–856. [Google Scholar] [CrossRef]
- Sakabe, N.J.; Aneas, I.; Shen, T.; Shokri, L.; Park, S.Y.; Bulyk, M.L.; Evans, S.M.; Nobrega, M.A. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum. Mol. Genet. 2012, 21, 2194–2204. [Google Scholar] [CrossRef]
- Liu, S.; Kracher, B.; Ziegler, J.; Birkenbihl, R.P.; Somssich, I.E. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 2015, 4, e07295. [Google Scholar] [CrossRef]
- Nagahage, I.S.P.; Sakamoto, S.; Nagano, M.; Ishikawa, T.; Kawai-Yamada, M.; Mitsuda, N.; Yamaguchi, M. An NAC domain transcription factor ATAF2 acts as transcriptional activator or repressor dependent on promoter context. Plant. Biotechnol. 2018, 35, 285–289. [Google Scholar] [CrossRef]
- Niu, X.; Qi, J.; Chen, M.; Zhang, G.; Tao, A.; Fang, P.; Xu, J.; Onyedinma, S.A.; Su, J. Reference genes selection for transcript normalization in kenaf (Hibiscus cannabinus L.) under salinity and drought stress. PeerJ 2015, 3, e1347. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant. J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Hou, L.; Xiao, P.; Guo, Y.; Deyholos, M.K.; Liu, X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant. Sci. 2019, 280, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Koch, E.; Slusarenko, A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant. Cell 1990, 2, 437–445. [Google Scholar]
- Li, X.; Huang, L.; Zhang, Y.; Ouyang, Z.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant. Biol. 2014, 14, 286. [Google Scholar] [CrossRef]
- Pei, Z.M.; Kuchitsu, K.; Ward, J.M.; Schwarz, M.; Schroeder, J.I. Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant. Cell 1997, 9, 409–423. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).