Beyond What Your Retina Can See: Similarities of Retinoblastoma Function between Plants and Animals, from Developmental Processes to Epigenetic Regulation
Abstract
:1. Introduction
2. Structure of the Retinoblastoma Protein
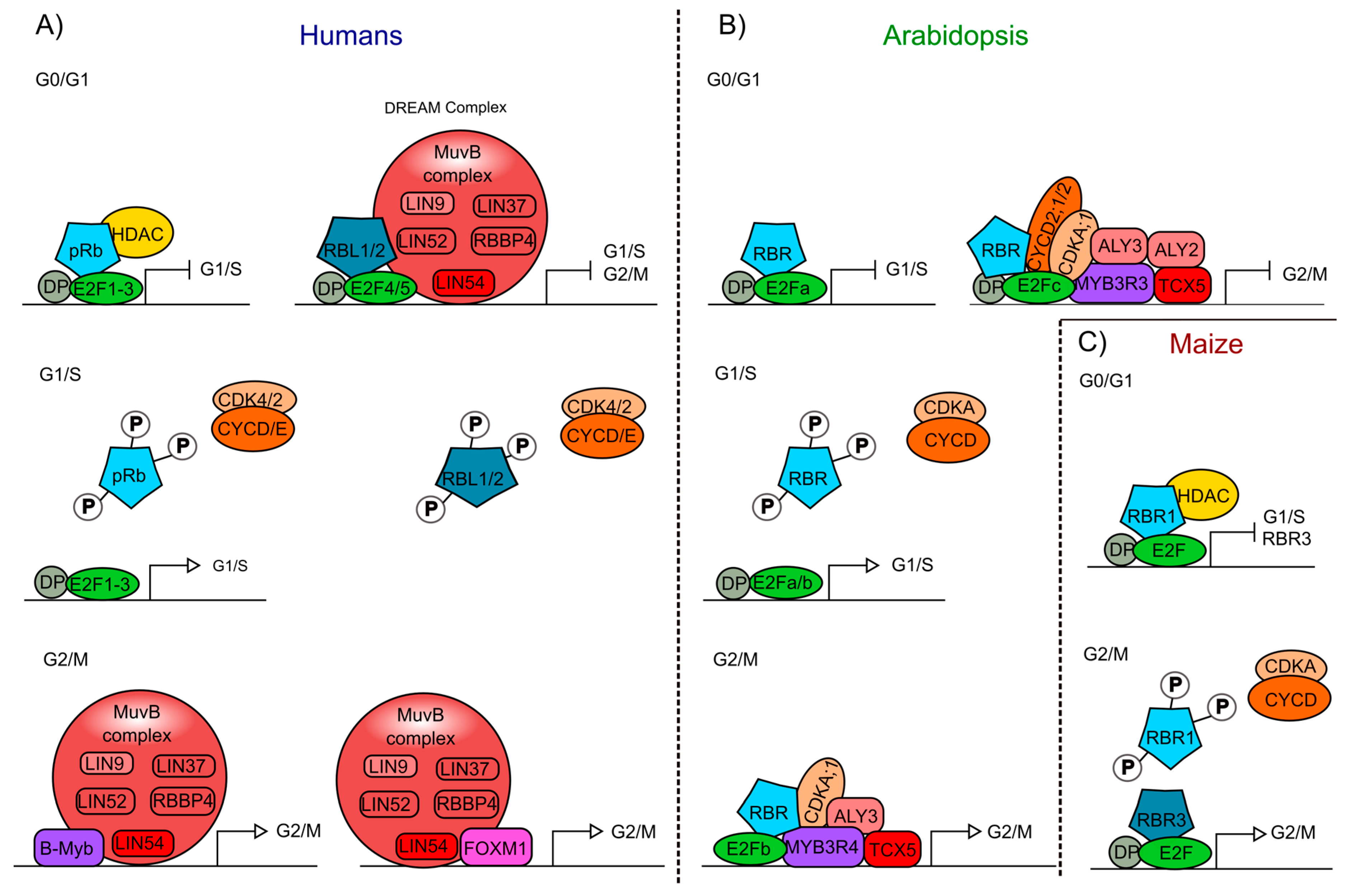
3. Cell Cycle Control through Retinoblastoma
4. The Roles of Retinoblastoma in Cell Differentiation
5. Retinoblastoma Function in Stem Cells Homeostasis
6. Function of Retinoblastoma in Epigenetic Modifications, Chromatin Regulation, and DNA Damage
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Koonin, E.V. The Incredible Expanding Ancestor of Eukaryotes. Cell 2010, 140, 606–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010, 11, 209. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, M.A.; Leger, M.M.; Wideman, J.G.; Ruiz-Trillo, I. Concepts of the last eukaryotic common ancestor. Nat. Ecol. Evol. 2019, 3, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Krinsky, B.H.; Long, M. New genes as drivers of phenotypic evolution. Nat. Rev. Genet. 2013, 14, 645–660. [Google Scholar] [CrossRef] [Green Version]
- Meyerowitz, E.M. Plants, animals and the logic of development. Trends Cell Biol. 1999, 9, M65–M68. [Google Scholar] [CrossRef]
- Meyerowitz, E.M. Comparative genomics. Plants compared to animals: The broadest comparative study of development. Science 2002, 295, 1482–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosberg, R.K.; Strathmann, R.R. The Evolution of Multicellularity: A Minor Major Transition? Annu. Rev. Ecol. Evol. Syst. 2007, 38, 621–654. [Google Scholar] [CrossRef] [Green Version]
- West, M.; Harada, J.J. Embryogenesis in Higher Plants: An Overview. Plant Cell 1993, 5, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M.; Chory, J.; Dangl, J.L.; Estelle, M.; Jacobsen, S.E.; Meyerowitz, E.M.; Nordborg, M.; Weigel, D. The Impact of Arabidopsis on Human Health: Diversifying Our Portfolio. Cell 2008, 133, 939–943. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.M.; Møller, S.G. The value of Arabidopsis research in understanding human disease states. Curr. Opin. Biotechnol. 2011, 22, 300–307. [Google Scholar] [CrossRef]
- Spampinato, C.P.; Gomez-Casati, D.F. Research on Plants for the Understanding of Diseases of Nuclear and Mitochondrial Origin. J. Biomed. Biotechnol. 2012, 2012, 836196. [Google Scholar] [CrossRef]
- Vergara, D.; de Domenico, S.; Maffia, M.; Piro, G.; Di Sansebastiano, G. Pietro Transgenic plants as low-cost platform for chemotherapeutic drugs screening. Int. J. Mol. Sci. 2015, 16, 2174–2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadia, P.; Barozzi, F.; Hoeschele, J.D.; Piro, G.; Margiotta, N.; Di Sansebastiano, G. Pietro Cisplatin, oxaliplatin, and kiteplatin subcellular effects compared in a plant model. Int. J. Mol. Sci. 2017, 18, 306. [Google Scholar] [CrossRef] [Green Version]
- Francke, U.; Kung, F. Sporadic bilateral retinoblastoma and 13q-chromosomal deletion. Med. Pediatric Oncol. 1976, 2, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Knudson, A.G. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knudson, A.G.; Meadows, A.T.; Nichols, W.W.; Hill, R. Chromosomal Deletion and Retinoblastoma. N. Engl. J. Med. 1976, 295, 1120–1123. [Google Scholar] [CrossRef]
- Friend, S.H.; Bernards, R.; Rogelj, S.; Weinberg, R.A.; Rapaport, J.M.; Albert, D.M.; Dryja, T.P. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 1986, 323, 643–646. [Google Scholar] [CrossRef]
- McGee, T.L.; Yandell, D.W.; Dryja, T.P. Structure and partial genomic sequence of the human retinoblastoma susceptibility gene. Gene 1989, 80, 119–128. [Google Scholar] [CrossRef]
- Sellers, W.R.; Kaelin, W.G. Role of the retinoblastoma protein in the pathogenesis of human cancer. J. Clin. Oncol. 1997, 15, 3301–3312. [Google Scholar] [CrossRef]
- Burkhart, D.L.; Sage, J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 2008, 8, 671–682. [Google Scholar] [CrossRef]
- Giacinti, C.; Giordano, A. RB and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, G.D.; Cook, J.G. The temporal regulation of s phase proteins during G1. Adv. Exp. Med. Biol. 2017, 1042, 335–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knudsen, E.S.; Wang, J.Y.J. Targeting the RB-pathway in cancer therapy. Clin. Cancer Res. 2010, 16, 1094–1099. [Google Scholar] [CrossRef] [Green Version]
- Knudsen, E.S.; Pruitt, S.C.; Hershberger, P.A.; Witkiewicz, A.K.; Goodrich, D.W. Cell Cycle and Beyond: Exploiting New RB1 Controlled Mechanisms for Cancer Therapy. Trends Cancer 2019, 5, 308–324. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, U.M.; O’Brien, J.M. Understanding pRb: Toward the necessary development of targeted treatments for retinoblastoma. J. Clin. Investig. 2012, 122, 425–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarang, S.; Pyakurel, U.; Weston, M.D.; Vijayakumar, S.; Jones, T.; Wagner, K.U.; Rocha-Sanchez, S.M. Spatiotemporally controlled overexpression of cyclin D1 triggers generation of supernumerary cells in the postnatal mouse inner ear. Hear. Res. 2020, 390. [Google Scholar] [CrossRef]
- Henry, D.; Brumaire, S.; Hu, X. Involvement of pRb-E2F pathway in green tea extract-induced growth inhibition of human myeloid leukemia cells. Leuk. Res. 2019, 77, 34–41. [Google Scholar] [CrossRef]
- Li, T.; Xiong, Y.; Wang, Q.; Chen, F.; Zeng, Y.; Yu, X.; Wang, Y.; Zhou, F.; Zhou, Y. Ribociclib (LEE011) suppresses cell proliferation and induces apoptosis of MDA-MB-231 by inhibiting CDK4/6-cyclin D-Rb-E2F pathway. Artif. Cells Nanomed. Biotechnol. 2019, 47, 4001–4011. [Google Scholar] [CrossRef] [Green Version]
- Kwapisz, D. Cyclin-dependent kinase 4/6 inhibitors in breast cancer: Palbociclib, ribociclib, and abemaciclib. Breast Cancer Res. Treat. 2017, 166, 41–54. [Google Scholar] [CrossRef]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldi, A.; Boccia, V.; Claudio, P.P.; De Luca, A.; Giordano, A. Genomic structure of the human retinoblastoma-related Rb2/p130 gene. Proc. Natl. Acad. Sci. USA 1996, 93, 4629–4632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claudio, P.P.; Tonini, T.; Giordano, A. The retinoblastoma family: Twins or distant cousins? Genome Biol. 2002, 3, 3012. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K.; Hanafusa, H.; Takimoto, H.; Ohgama, Y.; Akagi, T.; Shimizu, K. Structure of the human retinoblastoma-related p107 gene and its intragenic deletion in a B-cell lymphoma cell line. Gene 2000, 251, 37–43. [Google Scholar] [CrossRef]
- Chinnam, M.; Goodrich, D.W. RB1, Development, and Cancer. Curr. Top. Dev. Biol. 2011, 94, 129–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulligan, G.; Jacks, T. The retinoblastoma gene family: Cousins with overlapping interests. Trends Genet. 1998, 14, 223–229. [Google Scholar] [CrossRef]
- Dick, F.A. Structure-function analysis of the retinoblastoma tumor suppressor protein—Is the whole a sum of its parts? Cell Div. 2007, 2, 26. [Google Scholar] [CrossRef] [Green Version]
- Dick, F.A.; Rubin, S.M. Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 2013, 14, 297–306. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.O.; Russo, A.A.; Pavletich, N.P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 1998, 391, 859–865. [Google Scholar] [CrossRef]
- Morris, E.J.; Dyson, N.J. Retinoblastoma protein partners. Adv. Cancer Res. 2001, 82, 1–54. [Google Scholar] [CrossRef]
- Sanidas, I.; Morris, R.; Fella, K.A.; Rumde, P.H.; Boukhali, M.; Tai, E.C.; Ting, D.T.; Lawrence, M.S.; Haas, W.; Dyson, N.J. A Code of Mono-phosphorylation Modulates the Function of RB. Mol. Cell 2019, 73, 985–1000.e6. [Google Scholar] [CrossRef] [Green Version]
- Macdonald, J.I.; Dick, F.A. Posttranslational modifications of the retinoblastoma tumor suppressor protein as determinants of function. Genes Cancer 2012, 3, 619–633. [Google Scholar] [CrossRef] [Green Version]
- Dyson, N.; Guida, P.; McCall, C.; Harlow, E. Adenovirus E1A makes two distinct contacts with the retinoblastoma protein. J. Virol. 1992, 66, 4606–4611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, H.M.; Krstic-Demonacos, M.; Smith, L.; Demonacos, C.; La Thangue, N.B. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol. 2001, 3, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.; Huang, J.; Wu, S.Q.; Hauser, P.; Reznikoff, C.A. Role of sv40 t antigen binding to prb and p53 in multistep transformation in vitro of human uroepithelial cells. Carcinogenesis 1993, 14, 2297–2302. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.R.; Deshong, A.J.; Pelton, J.G.; Rubin, S.M. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J. Biol. Chem. 2010, 285, 16286–16293. [Google Scholar] [CrossRef] [Green Version]
- Rubin, S.M.; Gall, A.L.; Zheng, N.; Pavletich, N.P. Structure of the Rb C-terminal domain bound to E2F1-DP1: A mechanism for phosphorylation-induced E2F release. Cell 2005, 123, 1093–1106. [Google Scholar] [CrossRef] [Green Version]
- Rubin, S.M. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem. Sci. 2013, 38, 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickard, A.; Wong, P.P.; McCance, D.J. Acetylation of Rb by PCAF is required for nuclear localization and keratinocyte differentiation. J. Cell Sci. 2010, 123, 3718–3726. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.Y.; Wang, D.-H.; Campbell, M.; Huerta, S.B.; Shevchenko, B.; Izumiya, C.; Izumiya, Y. PRMT4-mediated arginine methylation negatively regulates retinoblastoma tumor suppressor protein and promotes E2F-1 dissociation. Mol. Cell. Biol. 2015, 35, 238–248. [Google Scholar] [CrossRef] [Green Version]
- Saddic, L.A.; West, L.E.; Aslanian, A.; Yates, J.R.; Rubin, S.M.; Gozani, O.; Sage, J. Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 2010, 285, 37733–37740. [Google Scholar] [CrossRef] [Green Version]
- Munro, S.; Khaire, N.; Inche, A.; Carr, S.; La Thangue, N.B. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene 2010, 29, 2357–2367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledl, A.; Schmidt, D.; Müller, S. Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 2005, 24, 3810–3818. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.; Qian, J.; Yue, H.; Li, X.; Xue, K. SUMOylation of Rb enhances its binding with CDK2 and phosphorylation at early G1 phase. Cell Cycle 2016, 15, 1724–1732. [Google Scholar] [CrossRef]
- Miwa, S.; Uchida, C.; Kitagawa, K.; Hattori, T.; Oda, T.; Sugimura, H.; Yasuda, H.; Nakamura, H.; Chida, K.; Kitagawa, M. Mdm2-mediated pRB downregulation is involved in carcinogenesis in a p53-independent manner. Biochem. Biophys. Res. Commun. 2006, 340, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Burke, J.R.; Hura, G.L.; Rubin, S.M. Structures of inactive retinoblastoma protein reveal multiple mechanisms for cell cycle control. Genes Dev. 2012, 26, 1156–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamber, E.P.; Beuron, F.; Morris, E.P.; Svergun, D.I.; Mittnacht, S. Structural Insights into the Mechanism of Phosphoregulation of the Retinoblastoma Protein. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Carr, S.M.; La Thangue, N.B. Diversity within the pRb pathway: Is there a code of conduct? Oncogene 2012, 31, 4343–4352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, F.D.; Huang, H.J.S.; To, H.; Young, L.J.S.; Oro, A.; Bookstein, R.; Lee, E.Y.H.P.; Lee, W.H. Structure of the human retinoblastoma gene. Proc. Natl. Acad. Sci. USA 1989, 86, 5502–5506. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Carneiro, N.; Torres-Jerez, I.; Stevenson, B.; McCreery, T.; Helentjaris, T.; Baysdorfer, C.; Almira, E.; Ferl, R.J.; Habben, J.E. Partial sequencing and mapping of clones from two maize cDNA libraries. Plant Mol. Biol. 1994, 26, 1085–1101. [Google Scholar] [CrossRef]
- Xie, Q.; Sanz-Burgos, A.P.; Hannon, G.J.; Gutiérrez, C. Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 1996, 15, 4900–4908. [Google Scholar] [CrossRef]
- Xie, Q.; Suárez-López, P.; Gutiérrez, C. Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: Requirement for efficient viral DNA replication. EMBO J. 1995, 14, 4073–4082. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Sekine, M.; Murakami, H.; Shinmyo, A. Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J. Cell Mol. Biol. 1999, 18, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.-J. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 2000, 19, 3485–3495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lendvai, A.; Pettko-Szandtner, A.; Csordas-Toth, E.; Miskolczi, P.; Horvath, G.V.; Gyorgyey, J.; Dudits, D. Dicot and monocot plants differ in retinoblastoma-related protein subfamilies. J. Exp. Bot. 2007, 58, 1663–1675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durfee, T.; Feiler, H.S.; Gruissem, W. Retinoblastoma-related proteins in plants: Homologues or orthologues of their metazoan counterparts? Plant Mol. Biol. 2000, 43, 635–642. [Google Scholar] [CrossRef]
- Kuwabara, A.; Gruissem, W. Arabidopsis Retinoblastoma-related and Polycomb group proteins: Cooperation during plant cell differentiation and development. J. Exp. Bot. 2014, 65, 2667–2676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jager, S.M.; Murray, J.A.H. Retinoblastoma proteins in plants. Plant Mol. Biol. 1999, 41, 295–299. [Google Scholar] [CrossRef]
- Desvoyes, B.; De Mendoza, A.; Ruiz-Trillo, I.; Gutierrez, C. Novel roles of plant RETINOBLASTOMA-RELATED (RBR) protein in cell proliferation and asymmetric cell division. J. Exp. Bot. 2014, 65, 2657–2666. [Google Scholar] [CrossRef]
- Gutzat, R.; Borghi, L.; Gruissem, W. Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends Plant Sci. 2012, 17, 139–148. [Google Scholar] [CrossRef]
- Reiland, S.; Messerli, G.; Baerenfaller, K.; Gerrits, B.; Endler, A.; Grossmann, J.; Gruissem, W.; Baginsky, S. Large-scale arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks1[w]. Plant Physiol. 2009, 150, 889–903. [Google Scholar] [CrossRef] [Green Version]
- Magyar, Z.; Horváth, B.; Khan, S.; Mohammed, B.; Henriques, R.; De Veylder, L.; Bakó, L.; Scheres, B.; Bögre, L. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 2012, 31, 1480–1493. [Google Scholar] [CrossRef] [Green Version]
- Ábrahám, E.; Miskolczi, P.; Ayaydin, F.; Yu, P.; Kotogány, E.; Bakó, L.; Ötvös, K.; Horváth, G.V.; Dudits, D. Immunodetection of retinoblastoma-related protein and its phosphorylated form in interphase and mitotic alfalfa cells. J. Exp. Bot. 2011, 62, 2155–2168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boniotti, M.B.; Gutierrez, C. A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 2001, 28, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Kawamura, K.; Sugisaka, K.; Sekine, M.; Shinmyo, A. Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 2002, 14, 1847–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, C.; Ramirez-Parra, E.; Castellano, M.M.; Sanz-Burgos, A.P.; Luque, A.; Missich, R. Geminivirus DNA replication and cell cycle interactions. Vet. Microbiol. 2004, 98, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, C. Geminiviruses and the plant cell cycle. Plant Mol. Biol. 2000, 43, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Peng, B.; Yao, L.; Zhang, X.; Sun, K.; Yang, X.; Yu, L. The ancient function of RB-E2F Pathway: Insights from its evolutionary history. Biol. Direct 2010, 5, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Harashima, H.; Dissmeyer, N.; Schnittger, A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013, 23, 345–356. [Google Scholar] [CrossRef]
- Budirahardja, Y.; Gönczy, P. Coupling the cell cycle to development. Development 2009, 136, 2861–2872. [Google Scholar] [CrossRef] [Green Version]
- Cooper, G.M. The Eukaryotic Cell Cycle. In The Cell: A Molecular Approach; Sinauer Associates: Washington, DC, USA, 2004; pp. 591–594. [Google Scholar]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef]
- Bertoli, C.; Skotheim, J.M.; de Bruin, R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blagosklonny, M.V.; Pardee, A.B. The Restriction Point of the Cell Cycle. Cell Cycle 2002, 1, 102–109. [Google Scholar] [CrossRef] [Green Version]
- Komaki, S.; Sugimoto, K. Control of the Plant Cell Cycle by Developmental and Environmental Cues. Plant Cell Physiol. 2012, 53, 953–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherr, C.J.; McCormick, F. The RB and p53 pathways in cancer. Cancer Cell 2002, 2, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Takaki, T.; Fukasawa, K.; Suzuki-Takahashi, I.; Hirai, H. Cdk-mediated phosphorylation of pRB regulates HDAC binding in vitro. Biochem. Biophys. Res. Commun. 2004, 316, 252–255. [Google Scholar] [CrossRef]
- Schaal, C.; Pillai, S.; Chellappan, S.P. The Rb-E2F transcriptional regulatory pathway in tumor angiogenesis and metastasis. Adv. Cancer Res. 2014, 121, 147–182. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.L.; Wenzel, P.L.; Sáenz-Robles, M.T.; Nair, V.; Ferrey, A.; Hagan, J.P.; Gomez, Y.M.; Sharma, N.; Chen, H.Z.; Ouseph, M.; et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature 2009, 462, 930–934. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.-K.; Bhinge, A.A.; Iyer, V.R. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res. 2011, 39, 3558–3573. [Google Scholar] [CrossRef] [Green Version]
- Komori, H.; Goto, Y.; Kurayoshi, K.; Ozono, E.; Iwanaga, R.; Bradford, A.P.; Araki, K.; Ohtani, K. Differential requirement for dimerization partner DP between E2F-dependent activation of tumor suppressor and growth-related genes. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Logan, N.; Graham, A.; Zhao, X.; Fisher, R.; Maiti, B.; Leone, G.; La Thangue, N.B. E2F-8: An E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene 2005, 24, 5000–5004. [Google Scholar] [CrossRef] [Green Version]
- Lammens, T.; Li, J.; Leone, G.; De Veylder, L. Atypical E2Fs: New players in the E2F transcription factor family. Trends Cell Biol. 2009, 19, 111–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Z.H.; Han, Z.C. The transcription factor E2F: A crucial switch in the control of homeostasis and tumorigenesis. Histol. Histopathol. 2006, 21, 403–413. [Google Scholar] [PubMed]
- Magae, J.; Wu, C.L.; Illenye, S.; Harlow, E.; Heintz, N.H. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J. Cell Sci. 1996, 109, 1717–1726. [Google Scholar] [PubMed]
- Hsu, J.; Sage, J. Novel functions for the transcription factor E2F4 in development and disease. Cell Cycle 2016, 15, 3183–3190. [Google Scholar] [CrossRef] [PubMed]
- Iaquinta, P.J.; Lees, J.A. Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 2007, 19, 649–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuitiño, M.C.; Pécot, T.; Sun, D.; Kladney, R.; Okano-Uchida, T.; Shinde, N.; Saeed, R.; Perez-Castro, A.J.; Webb, A.; Liu, T.; et al. Two Distinct E2F Transcriptional Modules Drive Cell Cycles and Differentiation. Cell Rep. 2019, 27, 3547–3560.e5. [Google Scholar] [CrossRef] [Green Version]
- Scott, M.C.; Sarver, A.L.; Tomiyasu, H.; Cornax, I.; Van Etten, J.; Varshney, J.; O’Sullivan, M.G.; Subramanian, S.; Modiano, J.F. Aberrant retinoblastoma (RB)-E2F transcriptional regulation defines molecular phenotypes of osteosarcoma. J. Biol. Chem. 2015, 290, 28070–28083. [Google Scholar] [CrossRef] [Green Version]
- Dowdy, S.F.; Hinds, P.W.; Louie, K.; Reed, S.I.; Arnold, A.; Weinberg, R.A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell 1993, 73, 499–511. [Google Scholar] [CrossRef]
- Kato, J.; Matsushime, H.; Hiebert, S.W.; Ewen, M.E.; Sherr, C.J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993, 7, 331–342. [Google Scholar] [CrossRef] [Green Version]
- Gillett, C.; Fantl, V.; Smith, R.; Fisher, C.; Bartek, J.; Dickson, C.; Barnes, D.; Peters, G. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994, 54, 1812–1817. [Google Scholar]
- Rizzolio, F.; Tuccinardi, T.; Caligiuri, I.; Lucchetti, C.; Giordano, A. CDK Inhibitors: From the Bench to Clinical Trials. Curr. Drug Targets 2010, 11, 279–290. [Google Scholar] [CrossRef]
- Markey, M.P.; Bergseid, J.; Bosco, E.E.; Stengel, K.; Xu, H.; Mayhew, C.N.; Schwemberger, S.J.; Braden, W.A.; Jiang, Y.; Babcock, G.F.; et al. Loss of the retinoblastoma tumor suppressor: Differential action on transcriptional programs related to cell cycle control and immune function. Oncogene 2007, 26, 6307–6318. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Müller, G.A. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 638–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadasivam, S.; DeCaprio, J.A. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 2013, 13, 585–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guiley, K.Z.; Liban, T.J.; Felthousen, J.G.; Ramanan, P.; Litovchick, L.; Rubin, S.M. Structural mechanisms of DREAM complex assembly and regulation. Genes Dev. 2015, 29, 961–974. [Google Scholar] [CrossRef] [Green Version]
- Sadasivam, S.; Duan, S.; DeCaprio, J.A. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012, 26, 474–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahl, M.; Meskiene, I.; Bogre, L.; Ha, D.T.; Swoboda, I.; Hubmann, R.; Hirt, H.; Heberle-Bors, E. The D-type alfalfa cyclin gene cycMs4 complements G1 cyclin-deficient yeast and is induced in the G1 phase of the cell cycle. Plant Cell 1995, 7, 1847–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntley, R.; Healy, S.; Freeman, D.; Lavender, P.; de Jager, S.; Greenwood, J.; Makker, J.; Walker, E.; Jackman, M.; Xie, Q.; et al. The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol. Biol. 1998, 37, 155–169. [Google Scholar] [CrossRef]
- Soni, R.; Carmichael, J.P.; Shah, Z.H.; Murray, J.A. A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 1995, 7, 85–103. [Google Scholar] [CrossRef]
- Ach, R.A.; Durfee, T.; Miller, A.B.; Taranto, P.; Hanley-Bowdoin, L.; Zambryski, P.C.; Gruissem, W. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 1997, 17, 5077–5086. [Google Scholar] [CrossRef] [Green Version]
- Ebel, C.; Mariconti, L.; Gruissem, W. Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 2004, 429, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Johnston, A.J.; Matveeva, E.; Kirioukhova, O.; Grossniklaus, U.; Gruissem, W. A Dynamic Reciprocal RBR-PRC2 Regulatory Circuit Controls Arabidopsis Gametophyte Development. Curr. Biol. 2008, 18, 1680–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borghi, L.; Gutzat, R.; Fütterer, J.; Laizet, Y.; Hennig, L.; Gruissem, W. Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 2010, 22, 1792–1811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Ramírez, A.; Díaz-Triviño, S.; Wachsman, G.; Du, Y.; Arteága-Vázquez, M.; Zhang, H.; Benjamins, R.; Blilou, I.; Neef, A.B.; Chandler, V.; et al. A SCARECROW-RETINOBLASTOMA Protein Network Controls Protective Quiescence in the Arabidopsis Root Stem Cell Organizer. PLoS Biol. 2013, 11. [Google Scholar] [CrossRef] [Green Version]
- Wildwater, M.; Campilho, A.; Perez-Perez, J.M.; Heidstra, R.; Blilou, I.; Korthout, H.; Chatterjee, J.; Mariconti, L.; Gruissem, W.; Scheres, B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 2005, 123, 1337–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desvoyes, B.; Ramirez-Parra, E.; Xie, Q.; Chua, N.H.; Gutierrez, C. Cell type-specific role of the retinoblastoma/E2F pathway during arabidopsis leaf development. Plant Physiol. 2006, 140, 67–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabelli, P.A.; Dante, R.A.; Leiva-Neto, J.T.; Jung, R.; Gordon-Kamm, W.J.; Larkins, B.A. RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 13005–13012. [Google Scholar] [CrossRef] [Green Version]
- Sabelli, P.A.; Liu, Y.; Dante, R.A.; Lizarraga, L.E.; Nguyen, H.N.; Brown, S.W.; Klingler, J.P.; Yu, J.; LaBrant, E.; Layton, T.M.; et al. Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proc. Natl. Acad. Sci. USA 2013, 110. [Google Scholar] [CrossRef] [Green Version]
- Grafi, G.; Burnett, R.J.; Helentjaris, T.; Larkins, B.A.; Decaprio, J.A.; Sellers, W.R.; Kaelin, W.G. A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proc. Natl. Acad. Sci. USA 1996, 93, 8962–8967. [Google Scholar] [CrossRef] [Green Version]
- Rossi, V.; Locatelli, S.; Lanzanova, C.; Boniotti, M.B.; Varotto, S.; Pipal, A.; Goralik-Schramel, M.; Lusser, A.; Gatz, C.; Gutierrez, C.; et al. A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol. Biol. 2003, 51, 401–413. [Google Scholar] [CrossRef]
- Sabelli, P.A.; Larkins, B.A. Grasses Like Mammals? Redundancy and Compensatory Regulation within the Retinoblastoma Protein Family. Cell Cycle 2006, 5, 352–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villajuana-Bonequi, M.; Matei, A.; Ernst, C.; Hallab, A.; Usadel, B.; Doehlemann, G. Cell type specific transcriptional reprogramming of maize leaves during Ustilago maydis induced tumor formation. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Sabelli, P.A.; Hoerster, G.; Lizarraga, L.E.; Brown, S.W.; Gordon-Kamm, W.J.; Larkins, B.A. Positive regulation of minichromosome maintenance gene expression, DNA replication, and cell transformation by a plant retinoblastoma gene. Proc. Natl. Acad. Sci. USA 2009, 106, 4042–4047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon-Kamm, W.; Dilkes, B.P.; Lowe, K.; Hoerster, G.; Sun, X.; Ross, M.; Church, L.; Bunde, C.; Farrell, J.; Hill, P.; et al. Stimulation of the cell cycle and maize transformation by disruption of the plant retinoblastoma pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 11975–11980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sage, J.; Miller, A.L.; Pérez-Mancera, P.A.; Wysocki, J.M.; Jacks, T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 2003, 424, 223–228. [Google Scholar] [CrossRef]
- Gutierrez, C. The Arabidopsis Cell Division Cycle. Arab. Book 2009, 7, e0120. [Google Scholar] [CrossRef]
- Wang, G.; Kong, H.; Sun, Y.; Zhang, X.; Zhang, W.; Altman, N.; DePamphilis, C.W.; Ma, H. Genome-wide analysis of the cyclin family in arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 2004, 135, 1084–1099. [Google Scholar] [CrossRef] [Green Version]
- Quandt, E.; Ribeiro, M.P.C.; Clotet, J. Atypical cyclins: The extended family portrait. Cell. Mol. Life Sci. 2020, 77, 231–242. [Google Scholar] [CrossRef] [Green Version]
- Magyar, Z.; De Veylder, L.; Atanassova, A.; Bakó, L.; Inzé, D.; Bögre, L. The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 2005, 17, 2527–2541. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Parra, E.; López-Matas, M.A.; Fründt, C.; Gutierrez, C. Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell 2004, 16, 2350–2363. [Google Scholar] [CrossRef] [Green Version]
- Vandepoele, K.; Vlieghe, K.; Florquin, K.; Hennig, L.; Beemster, G.T.S.; Gruissem, W.; Van De Peer, Y.; Inzé, D.; De Veylder, L. Genome-wide identification of potential plant E2F target genes. Plant Physiol. 2005, 139, 316–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oszi, E.; Papdi, C.; Mohammed, B.; Petkó-Szandtner, A.; Leviczky, T.; Molnár, E.; Galvan-Ampudia, C.; Khan, S.; Juez, E.L.; Horváth, B.; et al. E2FB interacts with RETINOBLASTOMA RELATED and regulates cell proliferation during leaf development. Plant Physiol. 2020, 182, 518–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudolf, V.; Barrôco, R.; De Engler, J.A.; Verkest, A.; Beeckman, T.; Naudts, M.; Inzé, D.; De Veylder, L. B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 2004, 16, 945–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scofield, S.; Jones, A.; Murray, J.A.H. The plant cell cycle in context. J. Exp. Bot. 2014, 65, 2557–2562. [Google Scholar] [CrossRef]
- Caldon, C.E.; Musgrove, E.A. Distinct and redundant functions of cyclin E1 and cyclin E2 in development and cancer. Cell Div. 2010, 5, 2. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, L.K.; Reed, S.I. Cyclin E deregulation and genomic instability. Adv. Exp. Med. Biol. 2017, 1042, 527–547. [Google Scholar] [CrossRef]
- Fischer, M.; DeCaprio, J.A. Does Arabidopsis thaliana DREAM of cell cycle control? EMBO J. 2015, 34, 1987–1989. [Google Scholar] [CrossRef] [Green Version]
- Magyar, Z.; Bögre, L.; Ito, M. DREAMs make plant cells to cycle or to become quiescent. Curr. Opin. Plant Biol. 2016, 34, 100–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K.; Suzuki, T.; Iwata, E.; Magyar, Z.; Bögre, L.; Ito, M. MYB3Rs, plant homologs of Myb oncoproteins, control cell cycle-regulated transcription and form DREAM-like complexes. Transcription 2015, 6, 106–111. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Sijacic, P.; Xu, P.; Lian, H.; Liu, Z. Arabidopsis TSO1 and MYB3R1 form a regulatory module to coordinate cell proliferation with differentiation in shoot and root. Proc. Natl. Acad. Sci. USA 2018, 115, E3045–E3054. [Google Scholar] [CrossRef] [Green Version]
- Bouyer, D.; Heese, M.; Chen, P.; Harashima, H.; Roudier, F.; Grüttner, C.; Schnittger, A. Genome-wide identification of RETINOBLASTOMA RELATED 1 binding sites in Arabidopsis reveals novel DNA damage regulators. PLoS Genet. 2018, 14, 1–35. [Google Scholar] [CrossRef]
- Jiang, C.; Gu, J.; Chopra, S.; Gu, X.; Peterson, T. Ordered origin of the typical two- and three-repeat Myb genes. Gene 2004, 326, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Suzuki, T.; Iwata, E.; Nakamichi, N.; Suzuki, T.; Chen, P.; Ohtani, M.; Ishida, T.; Hosoya, H.; Müller, S.; et al. Transcriptional repression by MYB 3R proteins regulates plant organ growth. EMBO J. 2015, 34, 1992–2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araki, S.; Ito, M.; Soyano, T.; Nishihama, R.; Machida, Y. Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J. Biol. Chem. 2004, 279, 32979–32988. [Google Scholar] [CrossRef] [Green Version]
- Del Pozo, J.C.; Boniotti, M.B.; Gutierrez, C. Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell 2002, 14, 3057–3071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, S.A. Cell differentiation: What have we learned in 50 years? J. Theor. Biol. 2020, 485, 110031. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Alvarado, A.; Yamanaka, S. Rethinking Differentiation: Stem Cells, Regeneration, and Plasticity. Cell 2014, 157, 110–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Falco, G.; Comes, F.; Simone, C. pRb: Master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene 2006, 25, 5244–5249. [Google Scholar] [CrossRef] [Green Version]
- Goodrich, D.W. The retinoblastoma tumor-suppressor gene, the exception that proves the rule. Oncogene 2006, 25, 5233–5243. [Google Scholar] [CrossRef] [Green Version]
- Hatzistergos, K.E.; Williams, A.R.; Dykxhoorn, D.; Bellio, M.A.; Yu, W.; Hare, J.M. Tumor Suppressors RB1 and CDKN2a Cooperatively Regulate Cell-Cycle Progression and Differentiation During Cardiomyocyte Development and Repair. Circ. Res. 2019, 124, 1184–1197. [Google Scholar] [CrossRef]
- Khidr, L.; Chen, P.-L. RB, the conductor that orchestrates life, death and differentiation. Oncogene 2006, 25, 5210–5219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Narayanan, C.; Bian, J.; Sambo, D.; Brickler, T.; Zhang, W.; Chetty, S. A transient DMSO treatment increases the differentiation potential of human pluripotent stem cells through the Rb family. PLoS ONE 2018, 13, e0208110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipinski, M.M.; Jacks, T. The retinoblastoma gene family in differentiation and development. Oncogene 1999, 18, 7873–7882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busch, M.; Philippeit, C.; Weise, A.; Dünker, N. Re-characterization of established human retinoblastoma cell lines. Histochem. Cell Biol. 2014, 143, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Dyer, M.A.; Bremner, R. The search for the retinoblastoma cell of origin. Nat. Rev. Cancer 2005, 5, 91–101. [Google Scholar] [CrossRef]
- Nair, R.M.; Vemuganti, G.K. Transgenic Models in Retinoblastoma Research. Ocul. Oncol. Pathol. 2015, 1, 207–213. [Google Scholar] [CrossRef] [Green Version]
- Clarke, A.R.; Maandag, E.R.; Van Roon, M.; Van Der Lugt, N.M.T.; Van Der Valk, M.; Hooper, M.L.; Berns, A.; Te Rielef, H. Requirement for a functional Rb-1 gene in murine development. Nature 1992, 359, 328–330. [Google Scholar] [CrossRef]
- Jacks, T.; Fazeli, A.; Schmitt, E.M.; Bronson, R.T.; Goodell, M.A.; Weinberg, R.A. Effects of an Rb mutation in the mouse. Nature 1992, 359, 295–300. [Google Scholar] [CrossRef]
- Lee, E.Y.H.P.; Chang, C.Y.; Hu, N.; Wang, Y.C.J.; Lai, C.C.; Herrup, K.; Lee, W.H.; Bradley, A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 1992, 359, 288–294. [Google Scholar] [CrossRef]
- Morgenbesser, S.D.; Williams, B.O.; Jacks, T.; Depinho, R.A. P53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 1994, 371, 72–74. [Google Scholar] [CrossRef]
- Zhang, J.; Gray, J.; Wu, L.; Leone, G.; Rowan, S.; Cepko, C.L.; Zhu, X.; Craft, C.M.; Dyer, M.A. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat. Genet. 2004, 36, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Robanus-Maandag, E.; Dekker, M.; Van Der Valk, M.; Carrozza, M.L.; Jeanny, J.C.; Dannenberg, J.H.; Berns, A.; Riele, H. Te p107 is a suppressor of retinoblastoma development in pRB-deficient mice. Genes Dev. 1998, 12, 1599–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fajas, L.; Landsberg, R.L.; Huss-Garcia, Y.; Sardet, C.; Lees, J.A.; Auwerx, J. E2Fs regulate adipocyte differentiation. Dev. Cell 2002, 3, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Flowers, S.; Xu, F.; Moran, E. Cooperative activation of tissue-specific genes by pRB and E2F1. Cancer Res. 2013, 73, 2150–2158. [Google Scholar] [CrossRef] [Green Version]
- Julian, L.M.; Liu, Y.; Pakenham, C.A.; Dugal-Tessier, D.; Ruzhynsky, V.; Bae, S.; Tsai, S.Y.; Leone, G.; Slack, R.S.; Blais, A. Tissue-specific targeting of cell fate regulatory genes by E2f factors. Cell Death Differ. 2016, 23, 565–575. [Google Scholar] [CrossRef] [Green Version]
- Müller, H.; Bracken, A.P.; Vernell, R.; Moroni, M.C.; Christians, F.; Grassilli, E.; Prosperini, E.; Vigo, E.; Oliner, J.D.; Helin, K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001, 15, 267–285. [Google Scholar] [CrossRef] [Green Version]
- Sellers, W.R.; Novitch, B.G.; Miyake, S.; Heith, A.; Otterson, G.A.; Kaye, F.J.; Lassar, A.B.; Kaelin, W.G. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998, 12, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Cecchini, M.J.; Thwaites, M.J.; Talluri, S.; MacDonald, J.I.; Passos, D.T.; Chong, J.-L.; Cantalupo, P.; Stafford, P.M.; Saenz-Robles, M.T.; Francis, S.M.; et al. A Retinoblastoma Allele That Is Mutated at Its Common E2F Interaction Site Inhibits Cell Proliferation in Gene-Targeted Mice. Mol. Cell. Biol. 2014, 34, 2029–2045. [Google Scholar] [CrossRef] [Green Version]
- Wyrzykowska, J.; Schorderet, M.; Pien, S.; Gruissem, W.; Fleming, A.J. Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiol. 2006, 141, 1338–1348. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Bramsiepe, J.; Van Durme, M.; Komaki, S.; Prusicki, M.A.; Maruyama, D.; Forner, J.; Medzihradszky, A.; Wijnker, E.; Harashima, H.; et al. RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science 2017, 356. [Google Scholar] [CrossRef]
- Perilli, S.; Perez-Perez, J.M.; Di Mambro, R.; Peris, C.L.; Díaz-Triviño, S.; Del Bianco, M.; Pierdonati, E.; Moubayidin, L.; Cruz-Ramírez, A.; Costantino, P.; et al. RETINOBLASTOMA-RELATED protein stimulates cell differentiation in the Arabidopsis root meristem by interacting with cytokinin signaling. Plant Cell 2013, 25, 4469–4478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moubayidin, L.; Perilli, S.; Dello Ioio, R.; Di Mambro, R.; Costantino, P.; Sabatini, S. The rate of cell differentiation controls the arabidopsis root meristem growth phase. Curr. Biol. 2010, 20, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Fambrini, M.; Pugliesi, C. The Dynamic Genetic-Hormonal Regulatory Network Controlling the Trichome Development in Leaves. Plants 2019, 8, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morohashi, K.; Grotewold, E. A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 2009, 5, e1000396. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Avci, U.; Grant, E.H.; Haigler, C.H.; Beers, E.P. XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J. 2008, 53, 425–436. [Google Scholar] [CrossRef]
- Zhao, C.; Lasses, T.; Bako, L.; Kong, D.; Zhao, B.; Chanda, B.; Bombarely, A.; Cruz-Ramírez, A.; Scheres, B.; Brunner, A.M.; et al. XYLEM NAC DOMAIN1, an angiosperm NAC transcription factor, inhibits xylem differentiation through conserved motifs that interact with RETINOBLASTOMA-RELATED. New Phytol. 2017, 1. [Google Scholar] [CrossRef] [Green Version]
- Putarjunan, A.; Torii, K.U. Stomagenesis versus myogenesis: Parallels in intrinsic and extrinsic regulation of transcription factor mediated specialized cell-type differentiation in plants and animals. Dev. Growth Differ. 2016, 58, 341–354. [Google Scholar] [CrossRef] [Green Version]
- Matos, J.L.; Bergmann, D.C. Convergence of stem cell behaviors and genetic regulation between animals and plants: Insights from the Arabidopsis thaliana stomatal lineage. F1000Prime Rep. 2014, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Le Grand, F.; Rudnicki, M.A. Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell Biol. 2007, 19, 628–633. [Google Scholar] [CrossRef] [Green Version]
- Novitch, B.G.; Mulligan, G.J.; Jacks, T.; Lassar, A.B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 1996, 135, 441–456. [Google Scholar] [CrossRef]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Rigby, P.W.J. Gene Regulatory Networks and Transcriptional Mechanisms that Control Myogenesis. Dev. Cell 2014, 28, 225–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumont, N.A.; Wang, Y.X.; Rudnicki, M.A. Intrinsic and extrinsic mechanisms regulating satellite cell function. Development 2015, 142, 1572–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelison, D.D.W.; Olwin, B.B.; Rudnicki, M.A.; Wold, B.J. MyoD(-/-) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev. Biol. 2000, 224, 122–137. [Google Scholar] [CrossRef] [Green Version]
- Endo, T.; Goto, S. Retinoblastoma gene product Rb accumulates during myogenic differentiation and is deinduced by the expression of SV40 large T antigen. J. Biochem. 1992, 112, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Huh, M.S.; Parker, M.H.; Scimè, A.; Parks, R.; Rudnicki, M.A. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 2004, 166, 865–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapscott, S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005, 132, 2685–2695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, W.; Schneider, J.W.; Condorelli, G.; Kaushal, S.; Mahdavi, V.; Nadal-Ginard, B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell 1993, 72, 309–324. [Google Scholar] [CrossRef]
- Smialowski, P.; Singh, M.; Mikolajka, A.; Majumdar, S.; Joy, J.K.; Nalabothula, N.; Krajewski, M.; Degenkolbe, R.; Bernard, H.-U.; Holak, T.A. NMR and mass spectrometry studies of putative interactions of cell cycle proteins pRb and CDK6 with cell differentiation proteins MyoD and ID-2. Biochim. Biophys. Acta 2005, 1750, 48–60. [Google Scholar] [CrossRef]
- Magenta, A.; Cenciarelli, C.; De Santa, F.; Fuschi, P.; Martelli, F.; Caruso, M.; Felsani, A. MyoD Stimulates RB Promoter Activity via the CREB/p300 Nuclear Transduction Pathway. Mol. Cell. Biol. 2003, 23, 2893–2906. [Google Scholar] [CrossRef] [Green Version]
- Martelli, F.; Cenciarelli, C.; Santarelli, G.; Polikar, B.; Felsani, A.; Caruso, M. MyoD induces retinoblastoma gene expression during myogenic differentiation. Oncogene 1994, 9, 3579–3590. [Google Scholar]
- Puri, P.L.; Sartorelli, V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J. Cell. Physiol. 2000, 185, 155–173. [Google Scholar] [CrossRef]
- Novitch, B.G.; Spicer, D.B.; Kim, P.S.; Cheung, W.L.; Lassar, A.B. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr. Biol. 1999, 9, 449–459. [Google Scholar] [CrossRef] [Green Version]
- Knight, J.D.R.; Kothary, R. The myogenic kinome: Protein kinases critical to mammalian skeletal myogenesis. Skelet. Muscle 2011, 1, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajabi, H.N.; Takahashi, C.; Ewen, M.E. Retinoblastoma protein and MyoD function together to effect the repression of Fra-1 and in turn cyclin D1 during terminal cell cycle arrest associated with myogenesis. J. Biol. Chem. 2014, 289, 23417–23427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skapek, S.X.; Rhee, J.; Kim, P.S.; Novitch, B.G.; Lassar, A.B. Cyclin-mediated inhibition of muscle gene expression via a mechanism that is independent of pRB hyperphosphorylation. Mol. Cell. Biol. 1996, 16, 7043–7053. [Google Scholar] [CrossRef] [Green Version]
- Ruijtenberg, S.; van den Heuvel, S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 2016, 15, 196–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, A.; Wang, Q.; Goebl, M.G.; Harrington, M.A. Phosphorylation of Nuclear MyoD Is Required for Its Rapid Degradation. Mol. Cell. Biol. 1998, 18, 4994–4999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.M.; Wei, Q.; Zhao, X.; Paterson, B.M. Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with cdk4. EMBO J. 1999, 18, 926–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puri, P.L.; Iezzi, S.; Stiegler, P.; Chen, T.T.; Schiltz, R.L.; Muscat, G.E.O.; Giordano, A.; Kedes, L.; Wang, J.Y.J.; Sartorelli, V. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell 2001, 8, 885–897. [Google Scholar] [CrossRef]
- Simone, C.; Forcales, S.V.; Hill, D.A.; Imbalzano, A.N.; Latella, L.; Puri, P.L. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 2004, 36, 738–743. [Google Scholar] [CrossRef] [Green Version]
- Pillitteri, L.J.; Dong, J. Stomatal Development in Arabidopsis. Arab. Book 2013, 11, e0162. [Google Scholar] [CrossRef] [Green Version]
- Vatén, A.; Bergmann, D.C. Mechanisms of stomatal development: An evolutionary view. EvoDevo 2012, 3, 11. [Google Scholar] [CrossRef] [Green Version]
- Gray, J.E. Plant Development: Three Steps for Stomata. Curr. Biol. 2007, 17, R213–R215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanaoka, M.M.; Pillitteri, L.J.; Fujii, H.; Yoshida, Y.; Bogenschutz, N.L.; Takabayashi, J.; Zhu, J.K.; Torii, K.U. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 2008, 20, 1775–1785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillitteri, L.J.; Torii, K.U. Breaking the silence: Three bHLH proteins direct cell-fate decisions during stomatal development. BioEssays 2007, 29, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Han, S.K.; Qi, X.; Sugihara, K.; Dang, J.H.; Endo, T.A.; Miller, K.L.; Kim, E.D.; Miura, T.; Torii, K.U. MUTE Directly Orchestrates Cell-State Switch and the Single Symmetric Division to Create Stomata. Dev. Cell 2018, 45, 303–315.e5. [Google Scholar] [CrossRef] [Green Version]
- Ohashi-Ito, K.; Bergmann, D.C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 2006, 18, 2493–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoulias, N.; Harrison, E.L.; Casson, S.A.; Gray, J.E. Molecular control of stomatal development. Biochem. J. 2018, 475, 441–454. [Google Scholar] [CrossRef] [Green Version]
- Matos, J.L.; Lau, O.S.; Hachez, C.; Cruz-Ramírez, A.; Scheres, B.; Bergmann, D.C. Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. eLife 2014, 3, 1–15. [Google Scholar] [CrossRef]
- Han, S.-K.; Torii, K.U. Linking cell cycle to stomatal differentiation. Curr. Opin. Plant Biol. 2019, 51, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.Z.; Jiang, M.; Wang, M.; Xue, S.; Zhu, L.L.; Wang, H.Z.; Zou, J.J.; Lee, E.K.; Sack, F.; Le, J. Phosphorylation of serine 186 of bHLH transcription factor SPEECHLESS promotes stomatal development in arabidopsis. Mol. Plant 2015, 8, 783–795. [Google Scholar] [CrossRef] [Green Version]
- Nowack, M.K.; Harashima, H.; Dissmeyer, N.; Zhao, X.; Bouyer, D.; Weimer, A.K.; De Winter, F.; Yang, F.; Schnittger, A. Genetic Framework of Cyclin-Dependent Kinase Function in Arabidopsis. Dev. Cell 2012, 22, 1030–1040. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Lucas, J.R.; Sack, F.D. Deep functional redundancy between FAMA and FOUR LIPS in stomatal development. Plant J. 2014, 78, 555–565. [Google Scholar] [CrossRef]
- Weimer, A.K.; Matos, J.L.; Sharma, N.; Patell, F.; Murray, J.A.H.; Dewitte, W.; Bergmann, D.C. Lineage- and stage-specific expressed CYCD7;1 coordinates the single symmetric division that creates stomatal guard cells. Development (Camb. Engl.) 2018, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamora-Zaragoza, J.; Scheres, B. Tuning Division and Differentiation in Stomata: How to Silence a MUTE. Dev. Cell 2018, 45, 282–283. [Google Scholar] [CrossRef]
- Hachez, C.; Ohashi-Ito, K.; Dong, J.; Bergmann, D.C. Differentiation of arabidopsis guard cells: Analysis of the networks incorporating the basic helix-loop-helix transcription factor, FAMA. Plant Physiol. 2011, 155, 1458–1472. [Google Scholar] [CrossRef] [Green Version]
- Hecker, L.; Jagirdar, R.; Jin, T.; Thannickal, V.J. Reversible differentiation of myofibroblasts by MyoD. Exp. Cell Res. 2011, 317, 1914–1921. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wu, Z.; Hou, S. SPEECHLESS Speaks Loudly in Stomatal Development. Front. Plant Sci. 2020, 11, 114. [Google Scholar] [CrossRef]
- Badodi, S.; Baruffaldi, F.; Ganassi, M.; Battini, R.; Molinari, S. Phosphorylation-dependent degradation of MEF2C contributes to regulate G2/M transition. Cell Cycle (Georget. Tex.) 2015, 14, 1517–1528. [Google Scholar] [CrossRef] [Green Version]
- Estrella, N.L.; Desjardins, C.A.; Nocco, S.E.; Clark, A.L.; Maksimenko, Y.; Naya, F.J. MEF2 transcription factors regulate distinct gene programs in mammalian skeletal muscle differentiation. J. Biol. Chem. 2015, 290, 1256–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potthoff, M.J.; Olson, E.N. MEF2: A central regulator of diverse developmental… [Development. 2007]—PubMed result. Development (Camb. Engl.) 2007, 134, 4131–4140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; De Pouplana, L.R.; Martinez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.-X.; Miao, Z.-Q.; Zhang, J.; Liu, Q.-Q.; Xiang, C.-B. MADS-box factor AGL16 negatively regulates drought resistance via stomatal density and stomatal movement. bioRxiv 2019, 723106. [Google Scholar] [CrossRef]
- Kutter, C.; Schöb, H.; Stadler, M.; Meins, F.; Si-Ammour, A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 2007, 19, 2417–2429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammad, K.; Dakik, P.; Medkour, Y.; Mitrofanova, D.; Titorenko, V.I. Quiescence entry, maintenance, and exit in adult stem cells. Int. J. Mol. Sci. 2019, 20, 2158. [Google Scholar] [CrossRef] [Green Version]
- Santoro, A.; Vlachou, T.; Carminati, M.; Pelicci, P.G.; Mapelli, M. Molecular mechanisms of asymmetric divisions in mammary stem cells. EMBO Rep. 2016, 17, 1700–1720. [Google Scholar] [CrossRef] [Green Version]
- Rumman, M.; Dhawan, J.; Kassem, M. Concise Review: Quiescence in Adult Stem Cells: Biological Significance and Relevance to Tissue Regeneration. Stem Cells 2015, 33, 2903–2912. [Google Scholar] [CrossRef]
- Mushtaq, M.; Gaza, H.V.; Kashuba, E.V. Role of the RB-Interacting Proteins in Stem Cell Biology. Adv. Cancer Res. 2016, 131, 133–157. [Google Scholar] [CrossRef]
- Kohno, S.; Kitajima, S.; Sasaki, N.; Takahashi, C. Retinoblastoma tumor suppressor functions shared by stem cell and cancer cell strategies. World J. Stem Cells 2016, 8, 170. [Google Scholar] [CrossRef]
- Sage, J. The retinoblastoma tumor suppressor and stem cell biology. Genes Dev. 2012, 26, 1409–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Livne-bar, I.; Vanderluit, J.L.; Slack, R.S.; Agochiya, M.; Bremner, R. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 2004, 5, 539–551. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, G.M.; Kong, E.; Sabbagh, Y.; Brown, N.E.; Lee, J.S.; Demay, M.B.; Thomas, D.M.; Hinds, P.W. Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1-/- mice. Proc. Natl. Acad. Sci. USA 2008, 105, 18402–18407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosoyama, T.; Nishijo, K.; Prajapati, S.I.; Li, G.; Keller, C. Rb1 gene inactivation expands satellite cell and postnatal myoblast pools. J. Biol. Chem. 2011, 286, 19556–19564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, S.; Santos, M.; Segrelles, C.; Leis, H.; Jorcano, J.L.; Berns, A.; Paramio, J.M.; Vooijs, M. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development (Camb. Engl.) 2004, 131, 2737–2748. [Google Scholar] [CrossRef] [Green Version]
- Vanderluit, J.L.; Wylie, C.A.; McClellan, K.A.; Ghanem, N.; Fortin, A.; Callaghan, S.; MacLaurin, J.G.; Park, D.S.; Slack, R.S. The Retinoblastoma family member p107 regulates the rate of progenitor commitment to a neuronal fate. J. Cell Biol. 2007, 178, 129–139. [Google Scholar] [CrossRef]
- Viatour, P.; Somervaille, T.C.; Venkatasubrahmanyam, S.; Kogan, S.; McLaughlin, M.E.; Weissman, I.L.; Butte, A.J.; Passegué, E.; Sage, J. Hematopoietic Stem Cell Quiescence Is Maintained by Compound Contributions of the Retinoblastoma Gene Family. Cell Stem Cell 2008, 3, 416–428. [Google Scholar] [CrossRef] [Green Version]
- Viatour, P.; Ehmer, U.; Saddic, L.A.; Dorrell, C.; Andersen, J.B.; Lin, C.; Zmoos, A.F.; Mazur, P.K.; Schaffer, B.E.; Ostermeier, A.; et al. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J. Exp. Med. 2011, 208, 1963–1976. [Google Scholar] [CrossRef] [Green Version]
- Takahira, T.; Oda, Y.; Tamiya, S.; Yamamoto, H.; Kobayashi, C.; Izumi, T.; Ito, K.; Iwamoto, Y.; Tsuneyoshi, M. Alterations of the RB1 gene in dedifferentiated liposarcoma. Mod. Pathol. 2005, 18, 1461–1470. [Google Scholar] [CrossRef] [Green Version]
- Jopling, C.; Boue, S.; Belmonte, J.C.I. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef]
- Bracken, A.P.; Pasini, D.; Capra, M.; Prosperini, E.; Colli, E.; Helin, K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003, 22, 5323–5335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kareta, M.S.; Gorges, L.L.; Hafeez, S.; Benayoun, B.A.; Marro, S.; Zmoos, A.F.; Cecchini, M.J.; Spacek, D.; Batista, L.F.Z.; O’Brien, M.; et al. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell 2015, 16, 39–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Lee, W.S.; Kim, S.-H. Hormonal regulation of stem cell maintenance in roots. J. Exp. Bot. 2013, 64, 1153–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, K.; Feldman, L.J. REGULATION OF ROOT APICAL MERISTEM DEVELOPMENT. Annu. Rev. Cell Dev. Biol. 2005, 21, 485–509. [Google Scholar] [CrossRef]
- Timilsina, R.; Kim, J.H.; Nam, H.G.; Woo, H.R. Temporal changes in cell division rate and genotoxic stress tolerance in quiescent center cells of Arabidopsis primary root apical meristem. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wachsman, G.; Heidstra, R.; Scheres, B. Distinct cell-autonomous functions of RETINOBLASTOMA-RELATED in Arabidopsis stem cells revealed by the brother of Brainbow Clonal analysis system. Plant Cell 2011, 23, 2581–2591. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Ramírez, A.; Díaz-Triviño, S.; Blilou, I.; Grieneisen, V.A.; Sozzani, R.; Zamioudis, C.; Miskolczi, P.; Nieuwland, J.; Benjamins, R.; Dhonukshe, P.; et al. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 2012, 150, 1002–1015. [Google Scholar] [CrossRef] [Green Version]
- Weimer, A.K.; Nowack, M.K.; Bouyer, D.; Zhao, X.; Harashima, H.; Naseer, S.; De Winter, F.; Dissmeyer, N.; Geldner, N.; Schnittger, A. RETINOBLASTOMA RELATED1 regulates asymmetric cell divisions in Arabidopsis. Plant Cell 2012, 24, 4083–4095. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A Jasmonate Signaling Network Activates Root Stem Cells and Promotes Regeneration. Cell 2019, 177, 942–956.e14. [Google Scholar] [CrossRef]
- Durand-Dubief, M.; Persson, J.; Norman, U.; Hartsuiker, E.; Ekwall, K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010, 29, 2126–2134. [Google Scholar] [CrossRef]
- Zhang, F.; Rothermund, K.; Gangadharan, S.B.; Pommier, Y.; Prochownik, E.V.; Lazo, J.S. Phenotypic screening reveals topoisomerase I as a breast cancer stem cell therapeutic target. Oncotarget 2012, 3, 998–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zheng, L.; Hong, J.H.; Gong, X.; Zhou, C.; Pérez-Pérez, J.M.; Xu, J. TOPOISOMERASE1α Acts through Two Distinct Mechanisms to Regulate Stele and Columella Stem Cell Maintenance. Plant Physiol. 2016, 171, 483–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Gao, L.; Dinh, T.T.; Shi, T.; Li, D.; Wang, R.; Gao, L.; Xiao, L.; Chen, X. DNA topoisomerase I affects polycomb group protein-mediated epigenetic regulation and plant development by altering nucleosome distribution in Arabidopsis. Plant Cell 2014, 26, 2803–2817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atlasi, Y.; Stunnenberg, H.G. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 2017, 18, 643–658. [Google Scholar] [CrossRef]
- Fiorentino, F.P.; Marchesi, I.; Giordano, A. On the role of retinoblastoma family proteins in the establishment and maintenance of the epigenetic landscape. J. Cell. Physiol. 2013, 228, 276–284. [Google Scholar] [CrossRef]
- Kalozoumi, G.; Tzimas, C.; Sanoudou, D. The expanding role of epigenetics. Glob. Cardiol. Sci. Pract. 2012, 2012, 7. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Uchida, C. Roles of pRB in the Regulation of Nucleosome and Chromatin Structures. Biomed Res. Int. 2016, 2016, 5959721. [Google Scholar] [CrossRef]
- Gonzalo, S.; Blasco, M.A. Role of Rb Family in the Epigenetic Definition of Chromatin. Cell Cycle 2005, 4, 752–755. [Google Scholar] [CrossRef] [Green Version]
- Vélez-Cruz, R.; Johnson, D. The Retinoblastoma (RB) Tumor Suppressor: Pushing Back against Genome Instability on Multiple Fronts. Int. J. Mol. Sci. 2017, 18, 1776. [Google Scholar] [CrossRef]
- Aprelikova, O.N.; Fang, B.S.; Meissner, E.G.; Cotter, S.; Campbell, M.; Kuthiala, A.; Bessho, M.; Jensen, R.A.; Liu, E.T. BRCA1-associated growth arrest is RB-dependent. Proc. Natl. Acad. Sci. USA 1999, 96, 11866–11871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnaghi-Jaulin, L.; Groisman, R.; Naguibneva, I.; Robin, P.; Lorain, S.; Le Villain, J.P.; Troalen, F.; Trouche, D.; Harel-Bellan, A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 1998, 391, 601–605. [Google Scholar] [CrossRef]
- Robertson, K.D.; Ait-Si-Ali, S.; Yokochi, T.; Wade, P.A.; Jones, P.L.; Wolffe, A.P. DNMT1 forms a complex with RB, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000, 25, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Bourbon, H.-M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Y.; Cai, J.; Hou, Y.; Huang, Z.; Wang, Z. Role of EZH2 in cancer stem cells: From biological insight to a therapeutic target. Oncotarget 2017, 8, 37974–37990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishak, C.A.; Marshall, A.E.; Passos, D.T.; White, C.R.; Kim, S.J.; Cecchini, M.J.; Ferwati, S.; MacDonald, W.A.; Howlett, C.J.; Welch, I.D.; et al. An RB-EZH2 Complex Mediates Silencing of Repetitive DNA Sequences. Mol. Cell 2016, 64, 1074–1087. [Google Scholar] [CrossRef] [Green Version]
- Vélez-Cruz, R.; Manickavinayaham, S.; Biswas, A.K.; Clary, R.W.; Premkumar, T.; Cole, F.; Johnson, D.G. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev. 2016, 30, 2500–2512. [Google Scholar] [CrossRef] [Green Version]
- Sokka, M.; Parkkinen, S.; Pospiech, H.; Syväoja, J.E. Function of TopBP1 in genome stability. Sub-Cell. Biochem. 2010, 50, 119–141. [Google Scholar] [CrossRef]
- Jiang, Y.; Chu, W.K. Potential Roles of the Retinoblastoma Protein in Regulating Genome Editing. Front. Cell Dev. Biol. 2018, 6. [Google Scholar] [CrossRef]
- Dunaief, J.L.; Strober, B.E.; Guha, S.; Khavari, P.A.; Ålin, K.; Luban, J.; Begemann, M.; Crabtree, G.R.; Goff, S.P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 1994, 79, 119–130. [Google Scholar] [CrossRef]
- Marquez-Vilendrer, S.B.; Rai, S.K.; Gramling, S.J.; Lu, L.; Reisman, D.N. BRG1 and BRM loss selectively impacts RB and P53, respectively: BRG1 and BRM have differential functions in vivo. Oncoscience 2016, 3, 337–350. [Google Scholar] [CrossRef]
- Strobeck, M.W.; Knudsen, K.E.; Fribourg, A.F.; DeCristofaro, M.F.; Weissman, B.E.; Imbalzano, A.N.; Knudsen, E.S. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 2000, 97, 7748–7753. [Google Scholar] [CrossRef] [Green Version]
- Xiao, H.; Goodrich, D.W. The retinoblastoma tumor suppressor protein is required for efficient processing and repair of trapped topoisomerase II-DNA-cleavable complexes. Oncogene 2005, 24, 8105–8113. [Google Scholar] [CrossRef] [PubMed]
- Yarden, R.I.; Brody, L.C. BRCA1 interacts with components of the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 1999, 96, 4983–4988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, R.; Zoumpoulidou, G.; Luczynski, M.T.; Rieger, S.; Moquet, J.; Spanswick, V.J.; Hartley, J.A.; Rothkamm, K.; Huang, P.H.; Mittnacht, S. Direct involvement of retinoblastoma family proteins in DNA repair by non-homologous end-joining. Cell Rep. 2015, 10, 2006–2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desvoyes, B.; Fernández-Marcos, M.; Sequeira-Mendes, J.; Otero, S.; Vergara, Z.; Gutierrez, C. Looking at plant cell cycle from the chromatin window. Front. Plant Sci. 2014, 5, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Paz Sanchez, M.; Aceves-García, P.; Petrone, E.; Steckenborn, S.; Vega-León, R.; Álvarez-Buylla, E.R.; Garay-Arroyo, A.; García-Ponce, B. The impact of Polycomb group (PcG) and Trithorax group (TrxG) epigenetic factors in plant plasticity. New Phytol. 2015, 208, 684–694. [Google Scholar] [CrossRef]
- Jullien, P.E.; Mosquna, A.; Ingouff, M.; Sakata, T.; Ohad, N.; Berger, F. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 2008, 6, 1693–1705. [Google Scholar] [CrossRef]
- Feil, R.; Berger, F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007, 23, 192–199. [Google Scholar] [CrossRef]
- Gutzat, R.; Borghi, L.; Fütterer, J.; Bischof, S.; Laizet, Y.; Hennig, L.; Feil, R.; Lunn, J.; Gruissem, W. Retinoblastoma-related protein controls the transition to autotrophic plant development. Development 2011, 138, 2977–2986. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Lucas, J.R.; Goodrich, J.; Sack, F.D. Arabidopsis guard cell integrity involves the epigenetic stabilization of the FLP and FAMA transcription factor genes. Plant J. 2014, 78, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, S.; Harashima, H.; Chen, P.; Heese, M.; Bouyer, D.; Sofroni, K.; Schnittger, A. The retinoblastoma homolog RBR 1 mediates localization of the repair protein RAD 51 to DNA lesions in Arabidopsis. EMBO J. 2017, 36, 1279–1297. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.L.; Rodriguez, A.M.; Snyder, R.R.; Hankins, G.D.V.; Boehning, D. Structure-function of the tumor suppressor BRCA1. Comput. Struct. Biotechnol. J. 2012, 1, e201204005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horvath, B.M.; Kourova, H.; Nagy, S.; Nemeth, E.; Magyar, Z.; Papdi, C.; Ahmad, Z.; Sanchez-Perez, G.F.; Perilli, S.; Blilou, I.; et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 2017, 36, 1261–1278. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zluhan-Martínez, E.; Pérez-Koldenkova, V.; Ponce-Castañeda, M.V.; Sánchez, M.d.l.P.; García-Ponce, B.; Miguel-Hernández, S.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. Beyond What Your Retina Can See: Similarities of Retinoblastoma Function between Plants and Animals, from Developmental Processes to Epigenetic Regulation. Int. J. Mol. Sci. 2020, 21, 4925. https://doi.org/10.3390/ijms21144925
Zluhan-Martínez E, Pérez-Koldenkova V, Ponce-Castañeda MV, Sánchez MdlP, García-Ponce B, Miguel-Hernández S, Álvarez-Buylla ER, Garay-Arroyo A. Beyond What Your Retina Can See: Similarities of Retinoblastoma Function between Plants and Animals, from Developmental Processes to Epigenetic Regulation. International Journal of Molecular Sciences. 2020; 21(14):4925. https://doi.org/10.3390/ijms21144925
Chicago/Turabian StyleZluhan-Martínez, Estephania, Vadim Pérez-Koldenkova, Martha Verónica Ponce-Castañeda, María de la Paz Sánchez, Berenice García-Ponce, Sergio Miguel-Hernández, Elena R. Álvarez-Buylla, and Adriana Garay-Arroyo. 2020. "Beyond What Your Retina Can See: Similarities of Retinoblastoma Function between Plants and Animals, from Developmental Processes to Epigenetic Regulation" International Journal of Molecular Sciences 21, no. 14: 4925. https://doi.org/10.3390/ijms21144925
APA StyleZluhan-Martínez, E., Pérez-Koldenkova, V., Ponce-Castañeda, M. V., Sánchez, M. d. l. P., García-Ponce, B., Miguel-Hernández, S., Álvarez-Buylla, E. R., & Garay-Arroyo, A. (2020). Beyond What Your Retina Can See: Similarities of Retinoblastoma Function between Plants and Animals, from Developmental Processes to Epigenetic Regulation. International Journal of Molecular Sciences, 21(14), 4925. https://doi.org/10.3390/ijms21144925




