Transcriptome Analysis and Expression of Selected Cationic Amino Acid Transporters in the Liver of Broiler Chicken Fed Diets with Varying Concentrations of Lysine
Abstract
:1. Introduction
2. Results
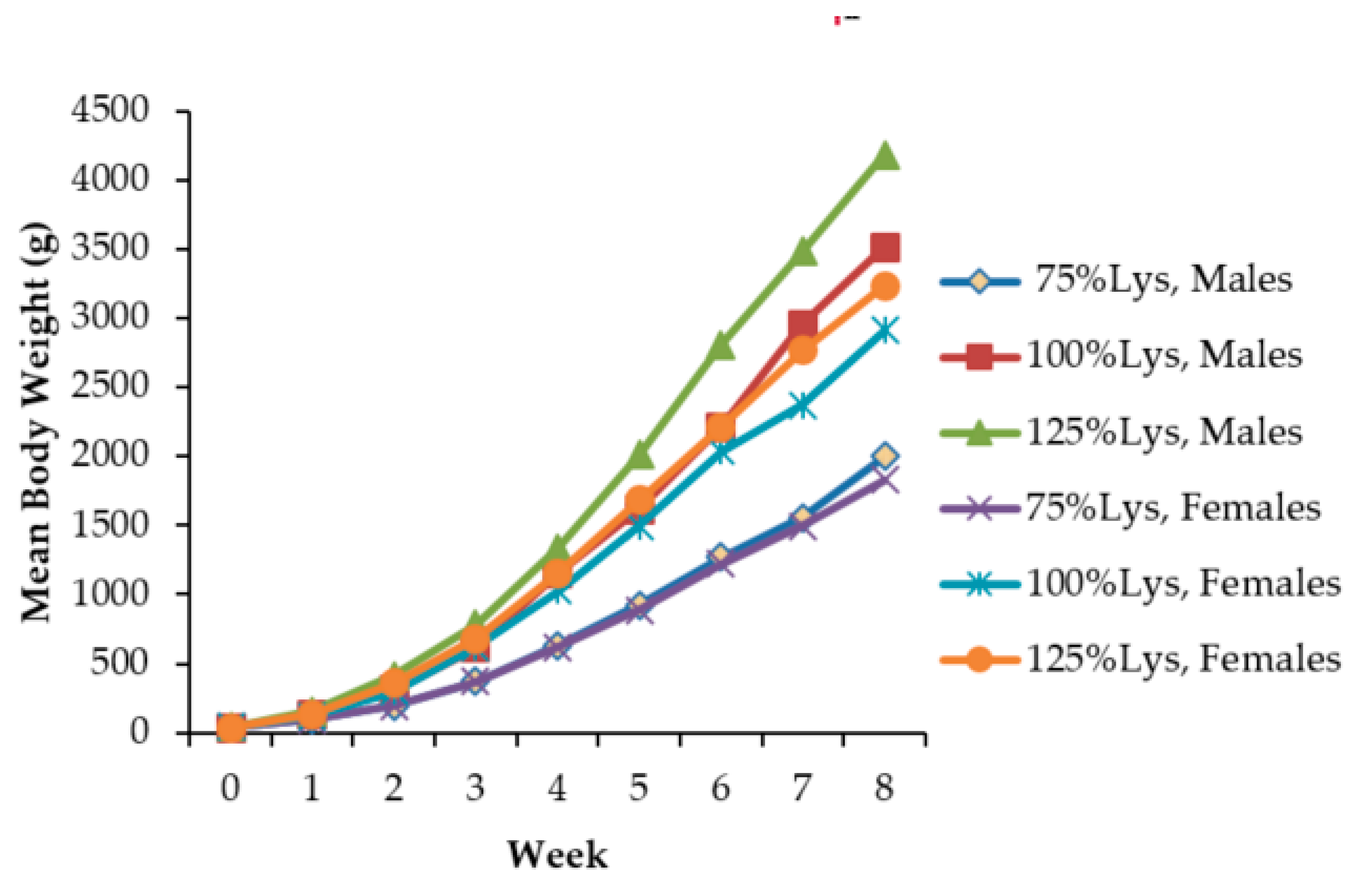
2.1. Growth Performance
2.2. Sequencing Data and Quality Control
2.3. Gene Expression Profile and Differential Expression Analysis
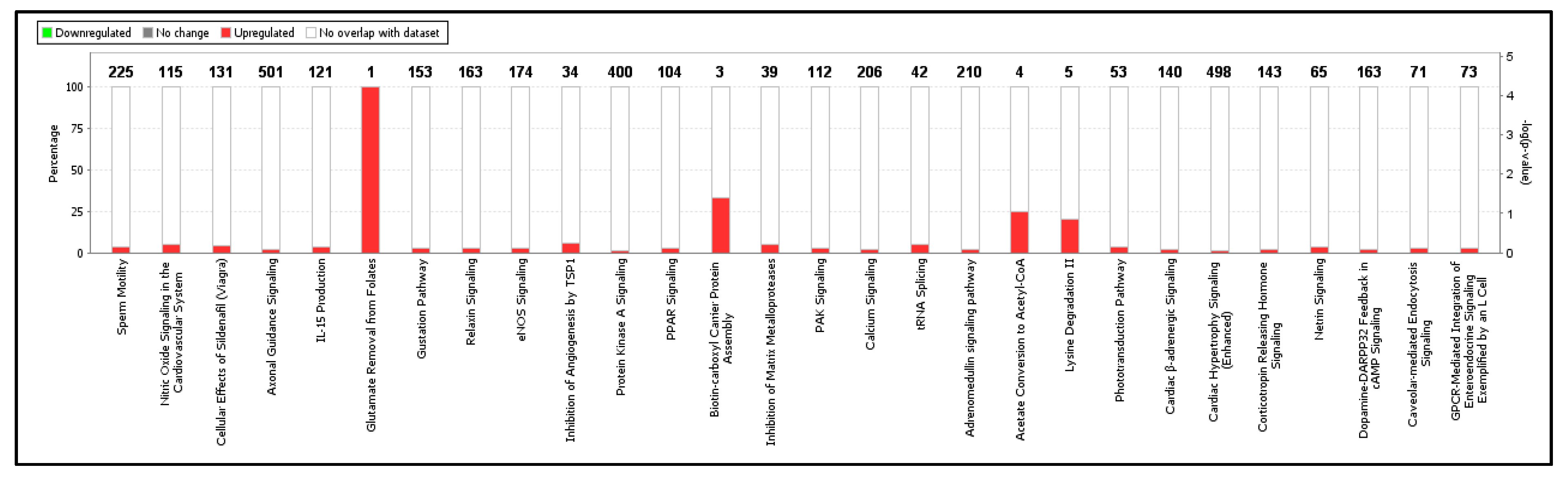
2.4. GO Enrichment and KEGG Pathway Analysis of DEGs
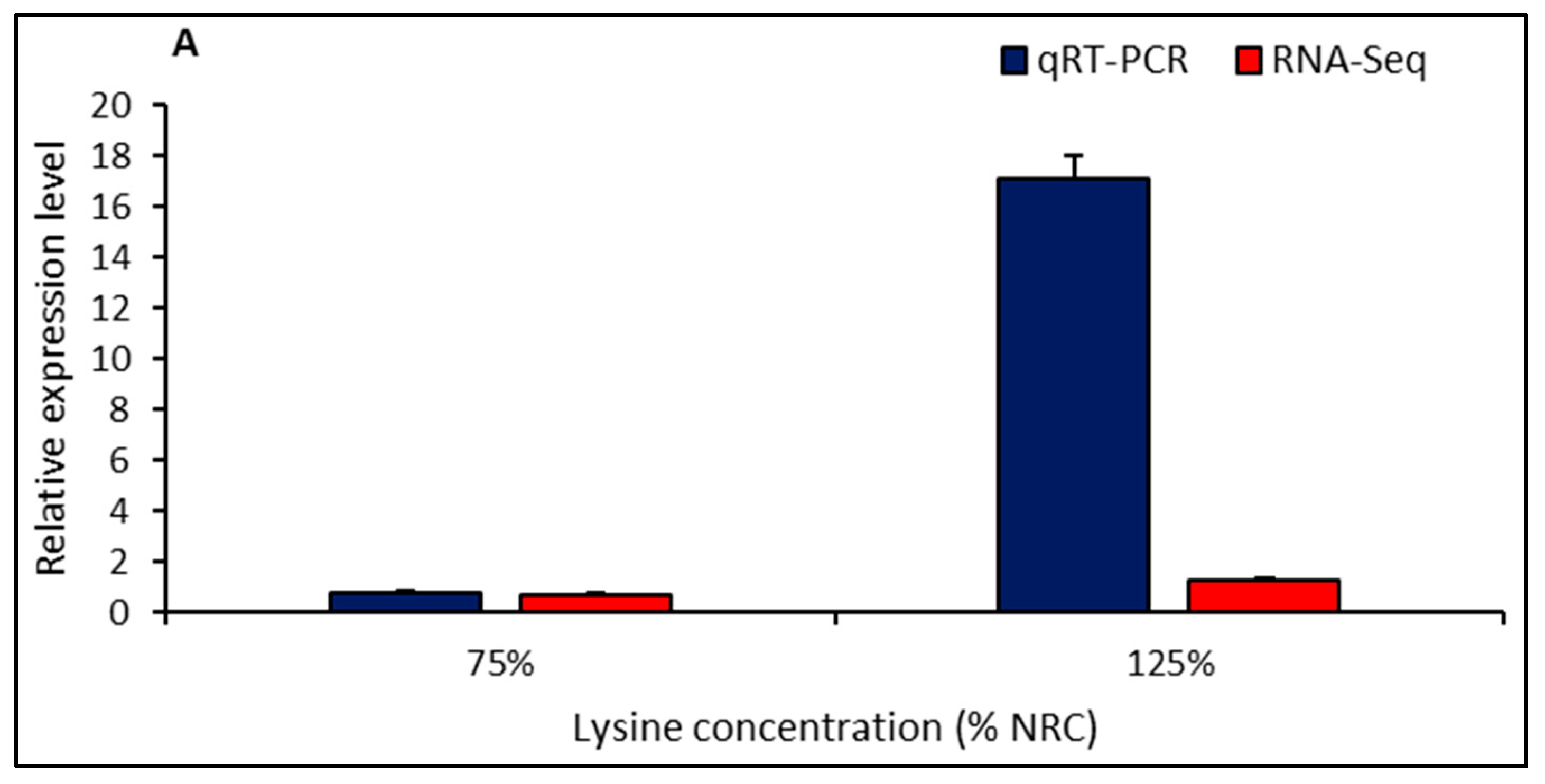
2.5. Validation of DEGs by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Management of Experimental Birds and Dietary Treatments Design
4.3. Tissue Sample Collection
4.4. Total RNA Extraction and RNA-Seq Library Construction
4.5. Quality Control and Comparative Analysis
4.6. Differential Expression Analysis
4.7. GO and KEGG Pathway Enrichment Analysis
4.8. Functional Analysis of DEGs Using Ingenuity Pathway Analysis (IPA)
4.9. Validation of RNA-Seq Results
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DEGs | Differentially expressed genes |
| SLC | Solute carrier |
| CATs | Cationic amino acid transports |
| BWG | Body weight gain |
| FCR | Feed conversion ratio |
| FI | Feed intake |
| FDR | False discovery rate |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| qRT-PCR | Quantitative real-time PCR |
| IPA | Ingenuity pathway analysis |
| RNA-Seq | RNA sequencing |
| FPKM | Fragments per kilobase of exon per million fragments |
References
- Tesseraud, S.; Peresson, R.; Lopes, J.; Chagneau, A.M. Dietary Lysine Deficiency Greatly Affects Muscle and Liver Protein Turnover in Growing Chickens. Br. J. Nutr. 1996, 75, 853–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Yin, Y.-L.; Li, D.; Kim, S.W.; Wu, G. Amino Acids and Immune Function. Br. J. Nutr. 2007, 98, 237–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasr, J.; Kheiri, F. Effect of Different Lysine Levels on Arian Broiler Performances. Italian J. Anim. Sci. 2011, 10, e32. [Google Scholar] [CrossRef] [Green Version]
- Khwatenge, C.N. Dietary Lysine: Effects on Lysine Homeostasis and Performance of Broiler Chickens. ETD Collect. Tenn. State Univ. 2015, 1–71. [Google Scholar]
- Bhogoju, S.; Nahashon, S.N.; Donkor, J.; Kimathi, B.; Johnson, D.; Khwatenge, C.; Bowden-Taylor, T. Effect of Varying Dietary Concentrations of Lysine on Growth Performance of the Pearl Grey Guinea Fowl. Poult. Sci. 2017, 96, 1306–1315. [Google Scholar] [CrossRef]
- Khwatenge, C.N.; Kimathi, B.M.; Taylor-Bowden, T.; Nahashon, S.N. Expression of Lysine-Mediated Neuropeptide Hormones Controlling Satiety and Appetite in Broiler Chickens. Poult. Sci. 2020, 99, 1409–1420. [Google Scholar] [CrossRef]
- Ruemmele, F.M.; Garnier-Lengliné, H. Why Are Genetics Important for Nutrition? Lessons from Epigenetic Research. Ann. Nutr. Metab. 2012, 60, 38–43. [Google Scholar] [CrossRef]
- Kilberg, M.S.; Pan, Y.-X.; Chen, H.; Leung-Pineda, V. NUTRITIONAL CONTROL OF GENE EXPRESSION: How Mammalian Cells Respond to Amino Acid Limitation. Annu. Rev. Nutr. 2005, 25, 59–85. [Google Scholar] [CrossRef] [Green Version]
- Higashino, K.; Fujioka, M.; Yamamura, Y. The Conversion of L-Lysine to Saccharopine and Alpha-Aminoadipate in Mouse. Arch. Biochem. Biophys. 1971, 142, 606–614. [Google Scholar] [CrossRef]
- Struys, E.A.; Jakobs, C. Metabolism of Lysine in α-Aminoadipic Semialdehyde Dehydrogenase-Deficient Fibroblasts: Evidence for an Alternative Pathway of Pipecolic Acid Formation. FEBS Lett. 2010, 584, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Payne, A.; Wang, X.; Ivy, M.T.; Stewart, A.; Nelson, K.; Darris, C.; Nahashon, S.N. Lysine Mediation of Neuroendocrine Food Regulation in Guinea Fowl. Poult. Sci. 2016, 95, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Hyde, R.; Taylor, P.M.; Hundal, H.S. Amino Acid Transporters: Roles in Amino Acid Sensing and Signalling in Animal Cells. Biochem. J. 2003, 373, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fotiadis, D.; Kanai, Y.; Palacín, M. The SLC3 and SLC7 Families of Amino Acid Transporters. Mol. Aspects Med. 2013, 34, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, J.; Lundeberg, J. The Plasticity of the Mammalian Transcriptome. Genomics 2010, 95, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, X.; Ba, Y.; Zhuang, Q.; Zhong, G. RNA-Seq Technology and Its Application in Fish Transcriptomics. Omics J. Integr. Biol. 2014, 18, 98–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Bai, Y.; Sartor, M.; Cavalcoli, J. Current Status and Future Perspectives for Sequencing Livestock Genomes. J. Anim. Sci. Biotechnol. 2012, 3, 8. [Google Scholar] [CrossRef] [Green Version]
- Costa, V.; Angelini, C.; De Feis, I.; Ciccodicola, A. Uncovering the Complexity of Transcriptomes with RNA-Seq. Available online: https://www.hindawi.com/journals/bmri/2010/853916 (accessed on 26 March 2020).
- Nagalakshmi, U.; Waern, K.; Snyder, M. RNA-Seq: A Method for Comprehensive Transcriptome Analysis. Curr. Protoc. Mol. Biol. 2010, 89, 4–11. [Google Scholar] [CrossRef]
- Dekkers, J.C.M. Application of Genomics Tools to Animal Breeding. Curr. Genom. 2012, 13, 207–212. [Google Scholar] [CrossRef] [Green Version]
- Lockhart, D.J.; Winzeler, E.A. Genomics, Gene Expression and DNA Arrays. Nature 2000, 405, 827–836. [Google Scholar] [CrossRef]
- Wang, A.; Liu, F.; Chen, S.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Sun, K.; Wu, Y.; Chen, X.; et al. Transcriptome Analysis and Identification of Differentially Expressed Transcripts of Immune-Related Genes in Spleen of Gosling and Adult Goose. Int. J. Mol. Sci. 2015, 16, 22904–22926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, W.; Rengaraj, D.; Kil, D.-Y.; Kim, H.; Lee, H.-K.; Song, K.-D. RNA-Seq Analysis of the Kidneys of Broiler Chickens Fed Diets Containing Different Concentrations of Calcium. Sci. Rep. 2017, 7, 11740. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Han, R.; Wang, Y.; Li, G.; Liu, X.; Tian, Y.; Kang, X.; Li, Z. Transcriptome Analysis of the Pectoral Muscles of Local Chickens and Commercial Broilers Using Ribo-Zero Ribonucleic Acid Sequencing. PLoS ONE 2017, 12, e0184115. [Google Scholar] [CrossRef] [Green Version]
- Na, W.; Wu, Y.-Y.; Gong, P.-F.; Wu, C.-Y.; Cheng, B.-H.; Wang, Y.-X.; Wang, N.; Du, Z.-Q.; Li, H. Embryonic Transcriptome and Proteome Analyses on Hepatic Lipid Metabolism in Chickens Divergently Selected for Abdominal Fat Content. BMC Genom. 2018, 19, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Wang, X.; Fu, Y.; Zhang, C.; Cao, Y.; Wang, J.; Zhang, Y.; Li, Y.; Chen, Y.; Li, Z.; et al. Transcriptome Analysis of the Breast Muscle of Xichuan Black-Bone Chickens Under Tyrosine Supplementation Revealed the Mechanism of Tyrosine-Induced Melanin Deposition. Front. Genet. 2019, 10, 457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latshaw, J.D. Dietary Lysine Concentrations from Deficient to Excessive and the Effects on Broiler Chicks. Br. Poult. Sci. 1993, 34, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Yoritaka, A.; Noriko, H.; Takanori, N.; Akira, Y. Effects of Some Potassium Compounds on Liver Lipid Accumulation Induced by Excess Dietary Lysine. Comp. Biochem. Physiol. Part A Physiol. 1994, 107, 227–231. [Google Scholar] [CrossRef]
- Fisher, C. Lysine: Amino Acid Requirements of Broiler Breeders. Poult. Sci. 1998, 77, 124–133. [Google Scholar] [CrossRef]
- Leclercq, B. Lysine: Specific Effects of Lysine on Broiler Production: Comparison with Threonine and Valine. Poult. Sci. 1998, 77, 118–123. [Google Scholar] [CrossRef]
- Henry, M.H.; Pesti, G.M.; Bakalli, R.; Lee, J.; Toledo, R.T.; Eitenmiller, R.R.; Phillips, R.D. The Performance of Broiler Chicks Fed Diets Containing Extruded Cottonseed Meal Supplemented with Lysine. Poult. Sci. 2001, 80, 762–768. [Google Scholar] [CrossRef]
- Waguespack, A.M.; Powell, S.; Bidner, T.D.; Payne, R.L.; Southern, L.L. Effect of Incremental Levels of L-Lysine and Determination of the Limiting Amino Acids in Low Crude Protein Corn-Soybean Meal Diets for Broilers. Poult. Sci. 2009, 88, 1216–1226. [Google Scholar] [CrossRef] [PubMed]
- Dozier, W.A.; Corzo, A.; Kidd, M.T.; Tillman, P.B.; McMurtry, J.P.; Branton, S.L. Digestible Lysine Requirements of Male Broilers from 28 to 42 Days of Age1. Poult. Sci. 2010, 89, 2173–2182. [Google Scholar] [CrossRef] [PubMed]
- Cemin, H.S.; Vieira, S.L.; Stefanello, C.; Kipper, M.; Kindlein, L.; Helmbrecht, A. Digestible Lysine Requirements of Male Broilers from 1 to 42 Days of Age Reassessed. PLoS ONE 2017, 12, e0179665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agostini, P.S.; Santos, R.R.; Khan, D.R.; Siebert, D.; van der Aar, P. The Optimum Valine: Lysine Ratios on Performance and Carcass Traits of Male Broilers Based on Different Regression Approaches. Poult. Sci. 2019, 98, 1310–1320. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Poultry, 9th ed.; National Academy Press: Washington, DC, USA, 1994; pp. 1–156.
- Morales, A.; Barrera, M.A.; Araiza, A.B.; Zijlstra, R.T.; Bernal, H.; Cervantes, M. Effect of Excess Levels of Lysine and Leucine in Wheat-Based, Amino Acid-Fortified Diets on the MRNA Expression of Two Selected Cationic Amino Acid Transporters in Pigs: Expression of Amino Acid Transporters in Pigs. J. Anim. Physiol. Anim. Nutr. 2013, 97, 263–270. [Google Scholar] [CrossRef]
- Dave, M.H.; Schulz, N.; Zecevic, M.; Wagner, C.A.; Verrey, F. Expression of Heteromeric Amino Acid Transporters along the Murine Intestine. J. Physiol. 2004, 558, 597–610. [Google Scholar] [CrossRef]
- Liao, S.F.; Vanzant, E.S.; Harmon, D.L.; McLeod, K.R.; Boling, J.A.; Matthews, J.C. Ruminal and Abomasal Starch Hydrolysate Infusions Selectively Decrease the Expression of Cationic Amino Acid Transporter MRNA by Small Intestinal Epithelia of Forage-Fed Beef Steers. J. Dairy Sci. 2009, 92, 1124–1135. [Google Scholar] [CrossRef] [Green Version]
- Kerr, B.J.; Kidd, M.T.; Halpin, K.M.; McWard, G.W.; Quarles, C.L. Lysine Level Increases Live Performance and Breast Yield in Male Broilers. J. Appl. Poult. Res. 1999, 8, 381–390. [Google Scholar] [CrossRef]
- Corzo, A.; Moran, E.T.; Hoehler, D. Lysine Need of Heavy Broiler Males Applying the Ideal Protein Concept. Poult. Sci. 2002, 81, 1863–1868. [Google Scholar] [CrossRef]
- Wu, P.; Dai, G.; Chen, F.; Chen, L.; Zhang, T.; Xie, K.; Wang, J.; Zhang, G. Transcriptome Profile Analysis of Leg Muscle Tissues between Slow- and Fast-Growing Chickens. PLoS ONE 2018, 13, e0206131. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; Ward, W.F. PGC-1α: A Key Regulator of Energy Metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef]
- Pagel-Langenickel, I.; Bao, J.; Joseph, J.J.; Schwartz, D.R.; Mantell, B.S.; Xu, X.; Raghavachari, N.; Sack, M.N. PGC-1α Integrates Insulin Signaling, Mitochondrial Regulation, and Bioenergetic Function in Skeletal Muscle. J. Biol. Chem. 2008, 283, 22464–22472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, H.; Iskender, A.U.; Srikanth, K.; Kim, H.; Zhunushov, A.T.; Chooq, H.; Jang, G.W.; Lim, Y.; Song, K.D.; Park, J.E. Transcriptome of Chicken Liver Tissues Reveals the Candidate Genes and Pathways Responsible for Adaptation into Two Different Climatic Conditions. Animals 2019, 9, 1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, B.D.; Stephensen, C.B.; Calvert, C.C.; Klasing, K.C. Lysine Deficiency and Feed Restriction Independently Alter Cationic Amino Acid Transporter Expression in Chickens (Gallus Gallus Domesticus). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2006, 143, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Azizi, S.; Nematollahi, M.A.; Amiri, B.M.; Vélez, E.J.; Lutfi, E.; Navarro, I.; Capilla, E.; Gutiérrez, J. Lysine and Leucine Deficiencies Affect Myocytes Development and IGF Signaling in Gilthead Sea Bream (Sparus Aurata). PLoS ONE 2016, 11, e0147618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goberdhan, D.C.I.; Wilson, C.; Harris, A.L. Amino Acid Sensing by MTORC1: Intracellular Transporters Mark the Spot. Cell Metab. 2016, 23, 580–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bröer, S.; Bröer, A. Amino Acid Homeostasis and Signalling in Mammalian Cells and Organisms. Biochem. J. 2017, 474, 1935–1963. [Google Scholar] [CrossRef] [Green Version]
- Graber, T.G.; Borack, M.S.; Reidy, P.T.; Volpi, E.; Rasmussen, B.B. Essential Amino Acid Ingestion Alters Expression of Genes Associated with Amino Acid Sensing, Transport, and MTORC1 Regulation in Human Skeletal Muscle. Nutr. Metab. 2017, 14, 35. [Google Scholar] [CrossRef]
- He, X.-D.; Gong, W.; Zhang, J.-N.; Nie, J.; Yao, C.-F.; Guo, F.-S.; Lin, Y.; Wu, X.-H.; Li, F.; Li, J.; et al. Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations. Cell Metab. 2018, 27, 151–166. [Google Scholar] [CrossRef] [Green Version]
- Taylor, P.M. Role of Amino Acid Transporters in Amino Acid Sensing. Am. J. Clin. Nutr. 2014, 99, 223S–230S. [Google Scholar] [CrossRef] [Green Version]
- Rebsamen, M.; Pochini, L.; Stasyk, T.; De Araújo, M.E.G.; Galluccio, M.; Kandasamy, R.K.; Snijder, B.; Fauster, A.; Rudashevskaya, E.L.; Bruckner, M.; et al. SLC38A9 Is a Component of the Lysosomal Amino Acid Sensing Machinery That Controls MTORC1. Nature 2015, 519, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Mackey, K. Short Technical Reports. Modification of the TRI Reagent Procedure for Isolation of RNA from Polysaccharide- and Proteoglycan-Rich Sources. BioTechniques 1995, 19, 942–945. [Google Scholar] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential Analysis of Gene Regulation at Transcript Resolution with RNA-Seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M. The KEGG Resource for Deciphering the Genome. Nucleic Acids Research 2004, 32, 277D–280D. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Sławińska, A.; D’Andrea, M.; Pilla, F.; Bednarczyk, M.; Siwek, M. Expression Profiles of Toll-like Receptors 1, 2 and 5 in Selected Organs of Commercial and Indigenous Chickens. J. Appl. Genet. 2013, 54, 489–492. [Google Scholar] [CrossRef] [Green Version]
- SAS Institute. SAS/STAT User’s Guide, Version 9.3; SAS Institute Inc.: Cary, NC, USA, 2011.
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]


| Parameters | Dietary Lysine Concentration | SEM | p-Value | ||
|---|---|---|---|---|---|
| 75% | 100% | 125% | |||
| Initial body weight, g | 39 | 39 | 39 | 0.057 | p < 0.05 |
| 8-week body weight, g | 2002 c | 3513 b | 4180 a | 119.2 | p < 0.05 |
| 8-week body weight gain, g | 1983 c | 3493 b | 4147 a | 128.7 | p < 0.05 |
| Feed intake, g/8 weeks | 3860 c | 6277 b | 7132 a | 72.6 | p < 0.05 |
| 1 Feed conversion ratio | 1.98 a | 1.75 b | 1.63 b | 0.09 | p < 0.05 |
| Lysine consumption, g/8 weeks | 32.81 c | 71.56 b | 101.3 a | 19.83 | p < 0.05 |
| Percentage mortality | 21.43 a | 3.57 b | 2.91 b | - | p < 0.05 |
| Sample ID | Raw Reads | Clean Reads | Total Bases | Clean Bases | Mean Read Length (bp) | GC Content (%) | Final Library (%) |
|---|---|---|---|---|---|---|---|
| LR1-7233B | 3,668,185 | 3,165,966 | 9,941,1210 | 9,940,3150 | 144 | 52.46 | 86 |
| LR1-7224 | 3,250,491 | 2,816,687 | 444,050,107 | 395,722,688 | 158 | 44.76 | 87 |
| LR1-7233 | 1,399,447 | 644,481 | 65,085,876 | 54,302,460 | 94 | 52.29 | 87 |
| LR3-7278 | 3,795,682 | 3,186,700 | 399,444,431 | 363,388,261 | 125 | 48.91 | 89 |
| LR3-7345 | 3,627,938 | 2,804,016 | 343,715,544 | 318,661,316 | 122 | 48.21 | 77 |
| LR3-7320 | 4,876,392 | 4,241,423 | 589,052,379 | 551,541,058 | 139 | 49.82 | 51 |
| Term ID | Description | Genes | Frequency (%) | Uniqueness |
|---|---|---|---|---|
| GO:0016049 | Cell growth | ALCAM, EXFABP, ID2, LLPH, SEMA5A, SEMA5B, SHH, TTR, ULK1 | 1.71 | 0.85 |
| GO:0022610 | Biological adhesion | M2, TENM3, TGFBI, THY1, TINAG, TINAGL1, TNFSF8, UBASH3B, VCL, VWA2 | 5.68 | 0.99 |
| GO:0032502 | Developmental process | SAP, PTK2, PTK7, PTN, PUM1, PXN, PYGO1, RAB10, RAB18, RAB3A, RAB8A, RAC3 | 21.32 | 1 |
| GO:0040007 | Growth | FGF1, GAP43, ID2, LARGE1, LLPH, PCK1, PSAP, PTK7, SEMA5A, SEMA5B, SHH, TEC, TTR, ULK1 | 3.21 | 0.99 |
| GO:0007154 | Cell communication | RNF152, RPTOR, RRAGC, SESN1, SESN2, SESN3, SIK2, SLC12A4, SLC12A7, STK11, WDR45B, WIPI2 | 27.90 | 0.98 |
| GO:0017144 | Drug metabolic process | CYP1A1, CYP1A2 | 0.05 | 0.97 |
| GO:0008283 | Drug Cell proliferation | ACE, ACVR2A, ACVR2B, BMP4, BMPR1A, CD180, CD40LG, ENTPD5, EXFABP, ID2, TGFB2, TOP2A, TWIST1, USP28, WNT2B, ASCC3 | 6.48 | 0.96 |
| GO:0043567 | Regulation of insulin-like growth factor receptor signaling pathway | IGFBP1, IGFBP2, IGFBP3, IGFBP4, IGFBP5 | 0.07 | 0.88 |
| GO:0008219 | Cell death | ATCAY, BNIP2, BRINP1, CASP10, CASP18, CASP2, CASP3, CASP7, CASP8, CASP9, STK4, TGFB2, TGFBR2, TNFRSF21, WNT1, XKR4, XKR9 | 6.39 | 0.9 |
| GO:0006950 | Response to stress | RPS27A, RPS3, RPTOR, RRAGC, SAMHD1, SELENOK, SEM1, SERP1, SESN1, SESN2, SESN3, SFR1, SGMS1 | 11.91 | 0.92 |
| GO:0006869 | Lipid transport | SLC43A2, SLC6A9, SLC7A1, SLC7A2, SLC7A3, SLC7A5, SLC7A9 | 1.42 | 0.87 |
| GO:0006629 | Lipid metabolic process | ACAT1, ACAT2, ACER3, ORMDL2, ORMDL3, OXSM, PCK1, PDCD2 | 5.34 | 0.88 |
| GO:0009267 | Cellular response to starvation | ATG7, BECN1, BMT2, EIF2S1, GABARAPL1 | 0.46 | 0.85 |
| GO:0044248 | Cellular catabolic process | ADH5, ADRM1, AFMID, AGL, AGO3, AGO4 | 5.96 | 0.93 |
| GO:0071398 | Cellular response to fatty acid | EXFABP, PTGER2 | 0.13 | 0.87 |
| GO:0048468 | Cell development | BMPR1A, CITED2, EXFABP, GDF11, HS6ST1, ID2, INHBA | 7.41 | 0.77 |
| GO:0055088 | Lipid homeostasis | ABCG1, ACOX1, ACOX2, EXFABP, LIPG, LPL, MTTP, ORMDL1, ORMDL2, ORMDL3, PLA2G12B, PNPLA2 | 0.41 | 0.91 |
| GO:0050801 | Ion homeostasis | ANXA5, ANXA6, ANXA7, ATOX1, ATP12A, SLC24A3, SLC30A9, SLC31A1, SLC35G1, SLC9A2, SLC9A7, SLC9A8, SLC9A9 | 2.62 | 0.89 |
| GO:0019477 | Lysine catabolic process | AASS | 1.35 | 0.91 |
| GO:0019878 | Lysine biosynthetic process via Aminoadipic acid | AASDHPPT, AASS | 7.50 | 0.9 |
| Top Canonical Pathways | p-Value | Overlap | Ratio |
| Sperm Motility | 1.06 × 10−5 | 3.6% | 8/225 |
| Nitric Oxide Signaling in the Cardiovascular System | 1.65 × 10−5 | 5.2% | 6/115 |
| Cellular Effects of Sildenafil (Viagra) | 3.73 × 10−4 | 3.8% | 5/131 |
| Axonal Guidance signaling | 2.39 × 10−3 | 1.6% | 8/501 |
| IL-15 Production | 2.49 × 10−3 | 3.3% | 4/121 |
| Top Upstream Regulators | p-value of overlap | Predicted activation | |
| TGFB1 | 3.63 × 10−8 | Activated | |
| ROCK2 | 5.12 × 10−7 | - | |
| PPARGC1A | 2.24 × 10−6 | - | |
| IGFBP2 | 2.69 × 10−6 | Activated | |
| KLF4 | 2.73 × 10−6 | Activated | |
| Top Molecular and Cellular Functions | p-value | # Genes | |
| Cellular Movement | 3.87 × 10−3–9.67 × 10−7 | 39 | |
| Cellular Assembly and Organization | 3.30 × 10−3–1.39 × 10−6 | 31 | |
| Cellular Function and Maintenance | 3.30 × 10−3–1.39 × 10−6 | 37 | |
| Cellular Development | 3.49 × 10−3–8.20 × 10−6 | 42 | |
| Cellular Growth and Proliferation | 3.49 × 10−3–8.20 × 10−6 | 33 | |
| Physiological System Development and Function | p-value range | # Genes | |
| Organismal Development | 3.80 × 10−3–2.28 × 10−7 | 58 | |
| Tissue Development and Function | 3.80 × 10−3–1.12 × 10−6 | 47 | |
| Respiratory System Development and Function | 6.94 × 10−3–1.85 × 10−6 | 15 | |
| Cardiovascular System Development and Function | 3.80 × 10−3–6.23 × 10−6 | 43 | |
| Tissue Development | 3.80 × 10−3–6.50 × 10−6 | 33 | |
| Top Up-regulated genes | Low vs High lysine | Top Down-regulated genes | Low vs High lysine |
| ANO6 | 8.426 | LRRC75B | −7.410 |
| ADAMTS2 | 8.185 | ANKRD22 | −7.359 |
| SEMA3C | 7.651 | ANGPTL4 | −3.017 |
| PRKAR2B | 7.599 | ADAMTS20 | −2.87 |
| UPK1B | 7.567 | MAGI2 | −2.43 |
| CYBRD1 | 7.489 | KIF21A | −2.33 |
| ADTRP | 7.370 | CCDC77 | −2.30 |
| ROR2 | 7.319 | ABCC4 | −2.20 |
| ITGB6 | 7.267 | CACNA1C | −2.01 |
| PDE1C | 7.267 | LRP6 | −1.56 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khwatenge, C.N.; Kimathi, B.M.; Nahashon, S.N. Transcriptome Analysis and Expression of Selected Cationic Amino Acid Transporters in the Liver of Broiler Chicken Fed Diets with Varying Concentrations of Lysine. Int. J. Mol. Sci. 2020, 21, 5594. https://doi.org/10.3390/ijms21165594
Khwatenge CN, Kimathi BM, Nahashon SN. Transcriptome Analysis and Expression of Selected Cationic Amino Acid Transporters in the Liver of Broiler Chicken Fed Diets with Varying Concentrations of Lysine. International Journal of Molecular Sciences. 2020; 21(16):5594. https://doi.org/10.3390/ijms21165594
Chicago/Turabian StyleKhwatenge, Collins N., Boniface M. Kimathi, and Samuel N. Nahashon. 2020. "Transcriptome Analysis and Expression of Selected Cationic Amino Acid Transporters in the Liver of Broiler Chicken Fed Diets with Varying Concentrations of Lysine" International Journal of Molecular Sciences 21, no. 16: 5594. https://doi.org/10.3390/ijms21165594
APA StyleKhwatenge, C. N., Kimathi, B. M., & Nahashon, S. N. (2020). Transcriptome Analysis and Expression of Selected Cationic Amino Acid Transporters in the Liver of Broiler Chicken Fed Diets with Varying Concentrations of Lysine. International Journal of Molecular Sciences, 21(16), 5594. https://doi.org/10.3390/ijms21165594





