Wild and Cultivated Species of Rice Have Distinctive Proteomic Responses to Drought
Abstract
1. Introduction
2. Results
2.1. Australian Rice Maintains Higher Leaf Water Potentials under Drought
2.2. Initial Proteome Characterization of Leaf Tissue from Plants Grown under Control and Drought Stress Conditions
2.3. Proteins Exclusively Identified in Droughted and Well-Watered Leaf Tissues
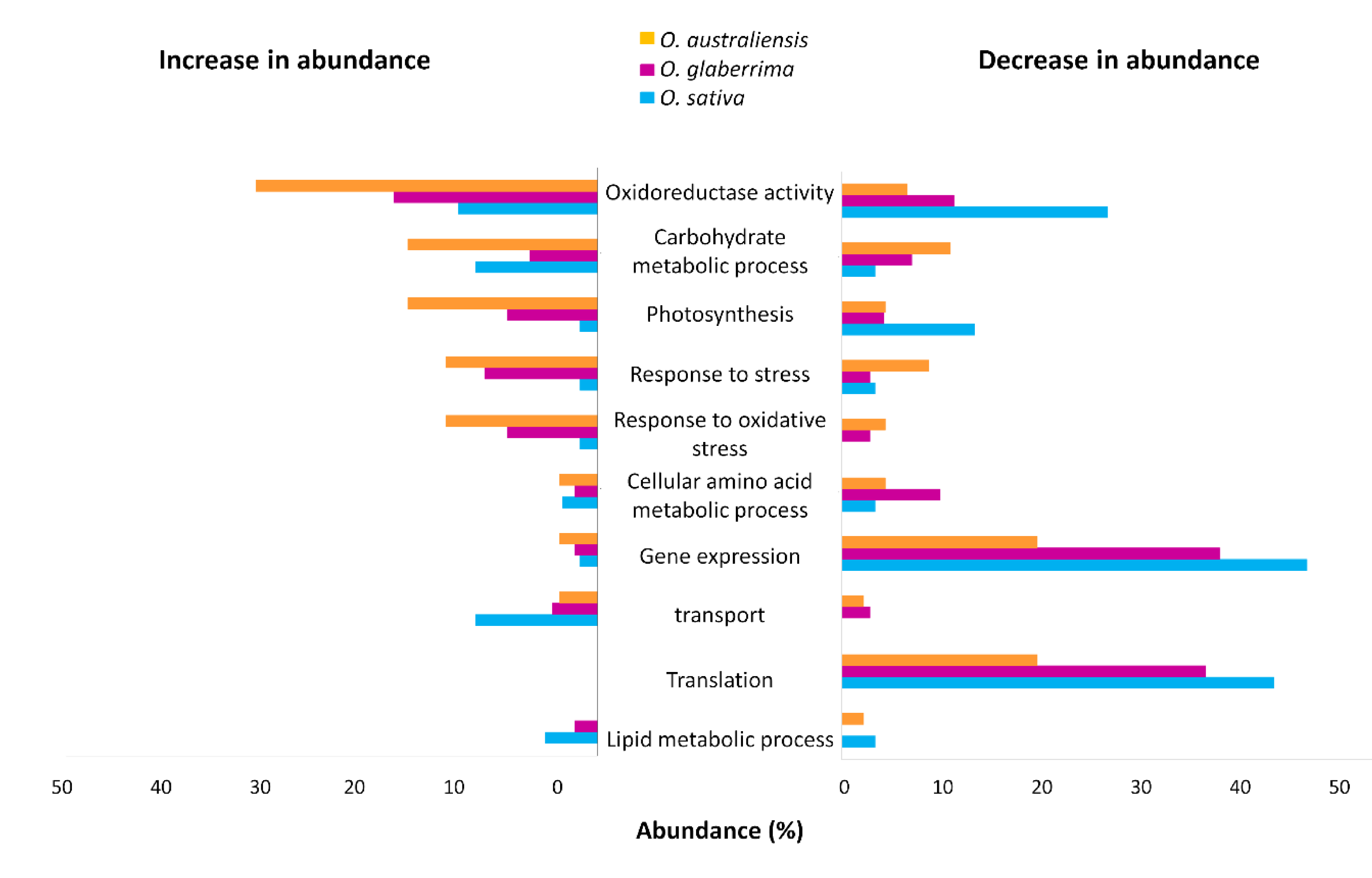
2.4. Gene Ontology Functional Classification of Significantly Altered Proteins in Response to Drought Stress
2.5. GO Functional Classification of Significantly Altered Proteins in Response to Drought Stress
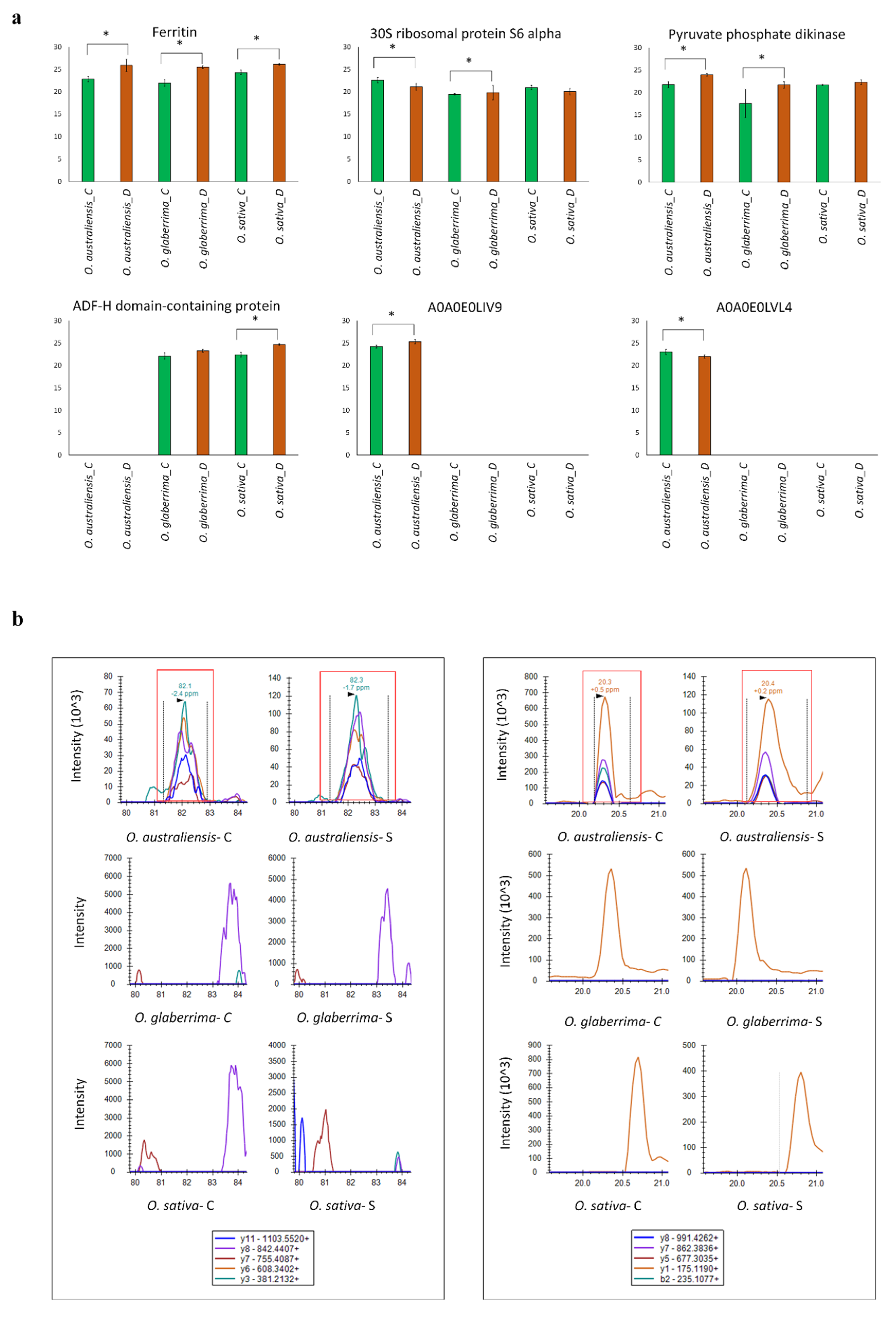
2.6. Parallel Reaction Monitoring (PRM) Validation
3. Discussion
3.1. Is Oryza australiensis a Source of Unique Drought-Stress Responsive Markers?
3.2. Photosynthetic Function Appears to Be Resilient under Drought in O. australiensis
3.3. Stress-Response Proteins Are Relatively More Abundant in O. glaberrima and O. australiensis than in O. sativa
3.4. Ribosomal Proteins Show Reduced Accumulation in Response to Drought Stress
4. Materials and Methods
4.1. Plant Material and Sample Preparation
4.2. Leaf Water Potential (LWP) Measurements
4.3. Protein Extraction and Protein Assay
4.4. In-Solution Digestion and Peptide Extraction
4.5. Nano LC–MS/MS
4.6. Parallel Reaction Monitoring (PRM) Analysis
4.7. Protein Identification and MS Data Analysis
4.8. Statistical Analysis
4.9. Functional Protein Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Access
Abbreviations
| FC | Field capacity |
| GO | Gene ontology |
| LFQ | Label-free quantitation |
| LWP | Leaf water potential |
| nLC–MS/MS | nanoflow liquid chromatography–tandem mass spectrometry |
| PRM | Parallel reaction monitoring |
| nsLTP | Non-specific lipid transfer protein |
| ROS | Reactive oxygen species |
| RP | Ribosomal protein |
| RSD | Relative standard deviation |
References
- Atwell, B.J.; Wang, H.; Scafaro, A.P. Could abiotic stress tolerance in wild relatives of rice be used to improve Oryza sativa? Plant Sci. 2014, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Marathi, B.; Ramos, J.; Hechanova, S.L.; Oane, R.H.; Jena, K.K. SNP genotyping and characterization of pistil traits revealing a distinct phylogenetic relationship among the species of Oryza. Euphytica 2014, 201, 131–148. [Google Scholar] [CrossRef]
- Cubry, P.; Tranchant-Dubreuil, C.; Thuillet, A.-C.; Monat, C.; Ndjiondjop, M.-N.; Labadie, K.; Cruaud, C.; Engelen, S.; Scarcelli, N.; Rhoné, B.; et al. The Rise and Fall of African Rice Cultivation Revealed by Analysis of 246 New Genomes. Curr. Boil. 2018, 28, 2274–2282.e6. [Google Scholar] [CrossRef] [PubMed]
- Manful, J.T.; Graham-Acquaah, S. African rice (Oryza glaberrima): A brief history and its growing importance in current rice breeding efforts. In Reference Module in Food Science.; Smithers, G., Ed.; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar] [CrossRef]
- Linares, O.F. African rice (Oryza glaberrima): History and future potential. Proc. Natl. Acad. Sci. USA 2002, 99, 16360–16365. [Google Scholar] [CrossRef]
- Wang, M.; Yu, Y.; Haberer, G.; Marri, P.R.; Fan, C.; Goicoechea, J.L.; Zuccolo, A.; Song, X.; Kudrna, D.; Ammiraju, J.S.S.; et al. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 2014, 46, 982–988. [Google Scholar] [CrossRef]
- Palmgren, M.; Edenbrandt, A.K.; Vedel, S.E.; Andersen, M.M.; Landes, X.; Østerberg, J.T.; Falhof, J.; Olsen, L.I.; Christensen, S.B.; Sandøe, P.; et al. Are we ready for back-to-nature crop breeding? Trends Plant Sci. 2015, 20, 155–164. [Google Scholar] [CrossRef]
- Jena, K.K. The species of the genus Oryza and transfer of useful genes from wild species into cultivated rice, O. sativa. Breed. Sci. 2010, 60, 518–523. [Google Scholar] [CrossRef]
- Hu, J.; Xiao, C.; Cheng, M.-X.; Gao, G.-J.; Zhang, Q.-L.; He, Y.-Q. A new finely mapped Oryza australiensis-derived QTL in rice confers resistance to brown planthopper. Gene 2015, 561, 132–137. [Google Scholar] [CrossRef]
- Van Mai, T.; Fujita, D.; Matsumura, M.; Yoshimura, A.; Yasui, H. Genetic basis of multiple resistance to the brown planthopper (Nilaparvata lugens stal) and the green rice leafhopper (Nephotettix cincticeps uhler) in the rice cultivar ‘asd7’ (Oryza sativa l. Ssp. Indica). Breed. Sci. 2015, 65, 420–429. [Google Scholar] [CrossRef]
- Ammiraju, J.S.; Lu, F.; Sanyal, A.; Yu, Y.; Song, X.; Jiang, N.; Pontaroli, A.C.; Rambo, T.; Currie, J.; Collura, K.; et al. Dynamic Evolution of Oryza Genomes Is Revealed by Comparative Genomic Analysis of a Genus-Wide Vertical Data Set. Plant Cell 2008, 20, 3191–3209. [Google Scholar] [CrossRef]
- Henry, R.; Rice, N.; Waters, D.L.E.; Kasem, S.; Ishikawa, R.; Hao, Y.; Dillon, S.; Crayn, D.; Wing, R.A.; Vaughan, D. Australian Oryza: Utility and Conservation. Rice 2009, 3, 235–241. [Google Scholar] [CrossRef]
- Zhu, Q.; Zheng, X.; Luo, J.; Gaut, B.S.; Ge, S. Multilocus Analysis of Nucleotide Variation of Oryza sativa and Its Wild Relatives: Severe Bottleneck during Domestication of Rice. Mol. Boil. Evol. 2007, 24, 875–888. [Google Scholar] [CrossRef]
- Brar, D.; Khush, G. Alien introgression in rice. Plant Mol. Boil. 1997, 35, 35–47. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Renaut, J. Plant proteome changes under abiotic stress — Contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011, 74, 1301–1322. [Google Scholar] [CrossRef] [PubMed]
- Hamzelou, S.; Pascovici, D.; Kamath, K.S.; Amirkhani, A.; McKay, M.J.; Mirzaei, M.; Atwell, B.J.; Haynes, P.A. Proteomic Responses to Drought Vary Widely Among Eight Diverse Genotypes of Rice (Oryza sativa). Int. J. Mol. Sci. 2020, 21, 363. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.L.; Sanchez, P.L.; Yu, S.; Lorieux, M.; Eizenga, G.C. Chromosome Segment Substitution Lines: A Powerful Tool for the Introgression of Valuable Genes from Oryza Wild Species into Cultivated Rice (O. sativa). Rice 2010, 3, 218–234. [Google Scholar] [CrossRef]
- Pearson, W.R. An Introduction to Sequence Similarity (“Homology”) Searching. Curr. Protoc. Bioinform. 2013, 42, 3.1.1–3.1.8. [Google Scholar] [CrossRef]
- Shi, T.; Song, E.; Nie, S.; Rodland, K.D.; Liu, T.; Qian, W.-J.; Smith, R.D. Advances in targeted proteomics and applications to biomedical research. Proteomics 2016, 16, 2160–2182. [Google Scholar] [CrossRef]
- Nada, R.M.; Abo-Hegazy, S.E.; Budran, E.G.; Abogadallah, G.M. The interaction of genes controlling root traits is required for the developmental acquisition of deep and thick root traits and improving root architecture in response to low water or nitrogen content in rice (Oryza sativa L.) cultivars. Plant Physiol. Biochem. 2019, 141, 122–132. [Google Scholar] [CrossRef]
- Mirzaei, M.; Soltani, N.; Sarhadi, E.; George, I.S.; Neilson, K.; Pascovici, D.; Shahbazian, S.; Haynes, P.A.; Atwell, B.J.; Salekdeh, G.H. Manipulating Root Water Supply Elicits Major Shifts in the Shoot Proteome. J. Proteome Res. 2013, 13, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.; Pascovici, D.; Keighley, T.; George, I.S.; Voelckel, C.; Heenan, P.B.; Haynes, P.A. Shotgun proteomic profiling of five species of New Zealand Pachycladon. Proteomics 2010, 11, 166–171. [Google Scholar] [CrossRef]
- Nishikawa, T.; Vaughan, D.A.; Kadowaki, K.-I. Phylogenetic analysis of Oryza species, based on simple sequence repeats and their flanking nucleotide sequences from the mitochondrial and chloroplast genomes. Theor. Appl. Genet. 2005, 110, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Roulin, A.; Piegu, B.; Wing, R.A.; Panaud, O. Evidence of multiple horizontal transfers of the long terminal repeat retrotransposon RIRE1 within the genus Oryza. Plant J. 2007, 53, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Ribeiro, C.W.; Korbes, A.P.; Garighan, J.; Jardim-Messeder, D.; Carvalho, F.E.; Sousa, R.H.V.; Caverzan, A.; Teixeira, F.K.; Silveira, J.A.G.; Margis-Pinheiro, M. Rice peroxisomal ascorbate peroxidase knockdown affects ROS signaling and triggers early leaf senescence. Plant Sci. 2017, 263, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Boil. 2012, 35, 1011–1019. [Google Scholar] [CrossRef]
- Hao, Z.; Wang, X.; Zong, Y.; Wen, S.; Cheng, Y.; Li, H. Enzymatic activity and functional analysis under multiple abiotic stress conditions of a dehydroascorbate reductase gene derived from Liriodendron Chinense. Environ. Exp. Bot. 2019, 167, 103850. [Google Scholar] [CrossRef]
- Filiz, E.; Ozyigit, I.I.; Saracoglu, I.A.; Uras, M.E.; Sen, U.; Yalcin, B. Abiotic stress-induced regulation of antioxidant genes in different Arabidopsis ecotypes: Microarray data evaluation. Biotechnol. Biotechnol. Equip. 2019, 33, 128–143. [Google Scholar] [CrossRef]
- Chowdhury, S.; Basu, A.; Kundu, S. Overexpression of a New Osmotin-Like Protein Gene (SindOLP) Confers Tolerance against Biotic and Abiotic Stresses in Sesame. Front. Plant Sci. 2017, 8, 410. [Google Scholar] [CrossRef]
- Li, F.; Asami, T.; Wu, X.; Tsang, E.W.; Cutler, A.J. A Putative Hydroxysteroid Dehydrogenase Involved in Regulating Plant Growth and Development. Plant Physiol. 2007, 145, 87–97. [Google Scholar] [CrossRef]
- Maguire, B.A.; Zimmermann, R.A. The Ribosome in Focus. Cell 2001, 104, 813–816. [Google Scholar] [CrossRef]
- Naora, H.; Naora, H. Involvement of ribosomal proteins in regulating cell growth and apoptosis: Translational modulation or recruitment for extraribosomal activity? Immunol. Cell Boil. 1999, 77, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Das, S.; Moin, M.; Dutta, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Genome-Wide Identification and Comprehensive Expression Profiling of Ribosomal Protein Small Subunit (RPS) Genes and their Comparative Analysis with the Large Subunit (RPL) Genes in Rice. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, K.B.; Bonham-Smith, P.C. Ribosomal protein gene regulation: What about plants? Can. J. Bot. 2006, 84, 342–362. [Google Scholar] [CrossRef]
- Fromont-Racine, M.; Senger, B.; Saveanu, C.; Fasiolo, F. Ribosome assembly in eukaryotes. Gene 2003, 313, 17–42. [Google Scholar] [CrossRef]
- Kressler, D.; Hurt, E.; Bassler, J. Driving ribosome assembly. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1803, 673–683. [Google Scholar] [CrossRef]
- Thomson, E.; Ferreira-Cerca, S.; Hurt, E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013, 126, 4815–4821. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Expression Profiling of Ribosomal Protein Gene Family in Dehydration Stress Responses and Characterization of Transgenic Rice Plants Overexpressing RPL23A for Water-Use Efficiency and Tolerance to Drought and Salt Stresses. Front. Chem. 2017, 5, 97. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Reddy, M.K.; Singla-Pareek, S.L.; Sopory, S.K. Transcriptional Downregulation of Rice rpL32 Gene under Abiotic Stress Is Associated with Removal of Transcription Factors within the Promoter Region. PLoS ONE 2011, 6, e28058. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Omranian, N.; Ahmadi, F.S.; Nikoloski, Z.; Mueller-Roeber, B. Effect of salt stress on genes encoding translation-associated proteins in Arabidopsis thaliana. Plant Signal. Behav. 2012, 7, 1095–1102. [Google Scholar] [CrossRef]
- Faghani, E.; Gharechahi, J.; Komatsu, S.; Mirzaei, M.; Khavarinejad, R.A.; Najafi, F.; Farsad, L.K.; Salekdeh, G.H. Comparative physiology and proteomic analysis of two wheat genotypes contrasting in drought tolerance. J. Proteom. 2015, 114, 1–15. [Google Scholar] [CrossRef]
- Fotovat, R.; Alikhani, M.; Valizadeh, M.; Mirzaei, M.; Salekdeh, G.H. A Proteomics Approach to Discover Drought Tolerance Proteins in Wheat Pollen Grain at Meiosis Stage. Protein Pept. Lett. 2016, 24, 26–36. [Google Scholar] [CrossRef] [PubMed]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, J.S. Protein identification using MS/MS data. J. Proteom. 2011, 74, 1842–1851. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, 11033. [Google Scholar] [CrossRef]
- Pascovici, D.; Keighley, T.; Mirzaei, M.; Haynes, P.A.; Cooke, B. PloGO: Plotting gene ontology annotation and abundance in multi-condition proteomics experiments. Proteomics 2012, 12, 406–410. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
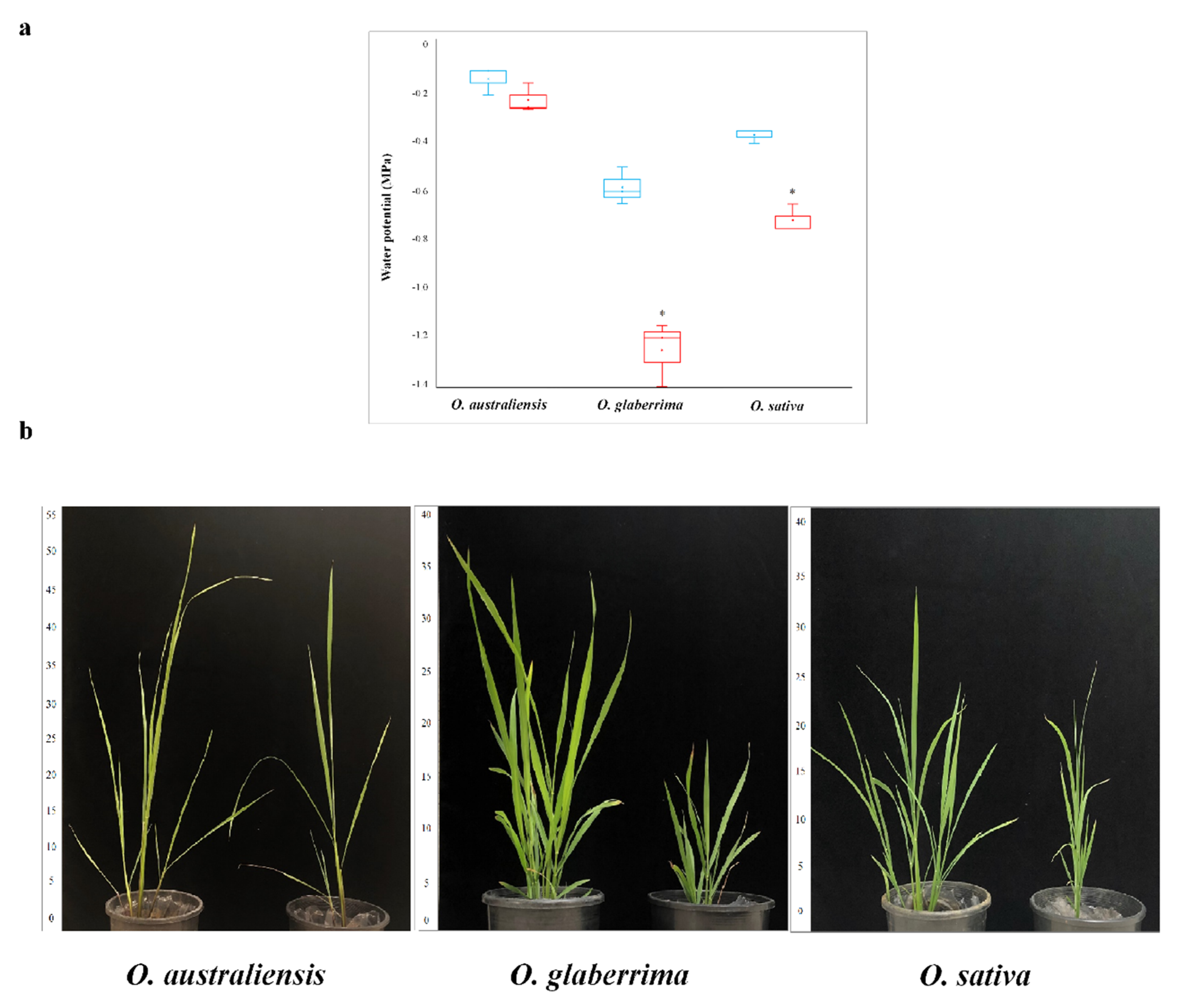



| Species and Status | Shoot Fresh Mass, Average ± SD (g) | Shoot Dried Mass, Average ± SD (g) | Root Dried Mass, Average ± SD (g) | Shoot Height, Average ± SD (cm) |
|---|---|---|---|---|
| O. australiensis - C | 3.91 ± 0.43 | 0.57 ± 0.09 | 0.21 ± 0.06 | 48.67 ± 3.51 |
| O. australiensis - D | 1.62 ± 0.85 | 0.35 ± 0.11 | 0.23 ± 0.06 | 42.33 ± 11.93 |
| O. glaberrima - C | 3.60 ± 1.85 | 0.68 ± 0.31 | 0.52 ± 0.06 | 28.00 ± 4.00 |
| O. glaberrima - D | 2.38 ± 0.56 | 0.65 ± 0.14 | 0.57 ± 0.13 | 24.67 ± 4.04 |
| O. sativa - C | 3.19 ± 1.50 | 0.68 ± 0.34 | 0.60 ± 0.33 | 29.67 ± 4.04 |
| O. sativa - D | 1.87 ± 0.25 | 0.55 ± 0.05 | 0.30 ± 0.06 | 27.33 ± 2.52 |
| Row | Rice Species- Treatment | Proteins | Peptides | Protein RSD b (%) | Peptide RSD (%) | Reproducibly Identified Proteins | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| R1 a | R2 | R3 | R1 | R2 | R3 | |||||
| 1 | O. australiensis- C c | 1411 | 1324 | 1219 | 9341 | 8755 | 7821 | 7.3 | 8.9 | 1033 |
| 2 | O. australiensis- D | 1299 | 1128 | 1272 | 8667 | 7625 | 8580 | 7.5 | 7.0 | 934 |
| 3 | O. glaberrima- C | 1574 | 1513 | 1656 | 8860 | 8925 | 9342 | 4.5 | 2.9 | 1230 |
| 4 | O. glaberrima- D | 1522 | 1519 | 1626 | 8985 | 8829 | 9465 | 3.9 | 3.6 | 1213 |
| 5 | O. sativa- C | 1545 | 1463 | 1539 | 8285 | 8126 | 8435 | 3.2 | 1.9 | 1160 |
| 6 | O. sativa- D | 1541 | 1698 | 1697 | 8197 | 9698 | 9571 | 5.5 | 9.1 | 1354 |
| Row | Rice Species | Total | Unchanged | Increased | Decreased | %Changed |
|---|---|---|---|---|---|---|
| 1 | O. australiensis | 1122 | 1045 | 28 | 49 | 6.8 |
| 2 | O. glaberrima | 1369 | 1246 | 47 | 76 | 9.0 |
| 3 | O. sativa | 1429 | 1336 | 61 | 32 | 6.5 |
| Species | Row | UniProt ID | UniProt Protein Name | Phytozome Gene Name | Fold Change | Homologous Protein | Identity (%) | Functional Domain(s) |
|---|---|---|---|---|---|---|---|---|
| 1 | A0A0E0JFX3 | Uncharacterized protein | LOC_Os01g12830.1 erythronate-4-phosphate dehydrogenase domain-containing protein | 26.7 | Putative D-isomer specific 2-hydroxyacid dehydrogenase (Oryza sativa) | 97% | D-isomer specific 2-hydroxyacid dehydrogenase (2HADH), NAD-binding domain; 2HADH, catalytic domain | |
| 2 | D0TZD6 | Starch synthase | LOC_Os07g22930.2 starch synthase | 1.9 | - a | - | Glycosyl transferase group 1; Starch synthase catalytic domain | |
| 3 | A0A0E0LIV9 | Uncharacterized protein | LOC_Os07g13969.1 expressed protein | 1.7 | Thylakoid soluble phosphoprotein TSP9 (Carex littledalei) | 44% | Thylakoid soluble phosphoprotein TSP9 | |
| O. australiensis | 4 | A0A0E0KBQ8 | FBPase domain-containing protein | LOC_Os03g16050.1 fructose-1,6-bisphosphatase | 1.3 | Fructose-1,6-bisphosphatase | 97% | Fructose-1-6-bisphosphatase (FBPase), N-terminal domain; FBPase C-terminal domain |
| 5 | A0A0E0KXS1 | Uncharacterized protein | LOC_Os05g02530.1 glutathione S-transferase, N-terminal domain-containing protein | 1.0 | Probable glutathione S-transferase DHAR1 (Oryza sativa) | 98% | Glutathione S-transferase (GST), N-terminal domain; GST, C-terminal domain | |
| 6 | A0A0E0JMC4 | Peroxiredoxin | LOC_Os01g48420.1 peroxiredoxin | 0.8 | - | - | Redoxin | |
| 7 | A0A0E0KKF2 | Uncharacterized protein | LOC_Os03g59100.1 pheophorbide a oxygenase, chloroplast precursor | 0.7 | Protochlorophyllide-dependent translocon component 52 (Oryza sativa) | 88% | Pheophorbide a oxygenase; Rieske [2Fe-2S] domain | |
| 8 | A0A0E0C629 | Peptidase A1 domain-containing protein | LOC_Os01g48740.1 aspartyl protease family protein | −0.7 | Aspartic proteinase (Oryza sativa) | 98% | Xylanase inhibitor N-terminal | |
| 9 | B0LT90 | Triosephosphate isomerase | LOC_Os01g05490.1 triosephosphate isomerase, cytosolic | −0.7 | - | - | Triosephosphate isomerase | |
| 10 | O04432 | Glycine-rich protein | LOC_Os12g43600.1 RNA recognition motif-containing protein | −1.4 | - | - | RNA recognition motif (RRM, RBD, or RNP domain) | |
| 11 | A0A0P0VIN1 | Os02g0452500 protein | LOC_Os02g25580.1 Sec1 family transport protein | −22.7 | Probable protein transport Sec1a (Oryza sativa) | 98% | Sec1 family | |
| 12 | A0A0D3G9Z3 | ATP-dependent 6-phospho-fructokinase | LOC_Os05g44922.1 6-phosphofructokinase | −24.1 | - | - | Phosphofructokinase | |
| 13 | Q69XJ9 | Os06g0602600 protein | LOC_Os06g40040.1 protein of unknown function domain-containing protein | −25.7 | Alba domain-containing protein (Cephalotus follicularis) | 78% | Alba | |
| 14 | A0A0E0EFX8 | Xylose isomerase | LOC_Os07g47290.1 xylose isomerase | −26.3 | - | - | Xylose isomerase-like | |
| 15 | A0A0E0LVL4 | Uncharacterized protein | LOC_Os08g35710.1 expressed protein | −26.7 | Ferredoxin-like protein (Striga asiatica) | 56% | ND b | |
| 16 | A0A0E0LDP3 | Sulfotransferase | LOC_Os06g42120.1 sulfotransferase domain-containing protein | −26.8 | - | - | Sulfotransferase domain | |
| 17 | A0A0E0L7D3 | Mg-por_mtran_C domain-containing protein | LOC_Os06g04150.1 magnesium-protoporphyrin O-methyltransferase | −27.3 | - | - | Magnesium-protoporphyrin IX methyltransferase C-terminus | |
| 18 | A0A0E0JLW2 | Uncharacterized protein | LOC_Os01g46600.1 seed maturation protein PM41 | −28.2 | Salt-tolerant correlative protein (Triticum aestivum) | 78% | ND | |
| 19 | A0A0D3GCD0 | H15 domain-containing protein | LOC_Os06g04020.1 histone H1 | 28.6 | - | - | Linker histone H1/H5 | |
| O. glaberrima | 20 | A0A0D3F0Q5 | PPM-type phosphatase | LOC_Os02g05630.1 protein phosphatase 2C | 25.7 | - | - | Protein phosphatase 2C |
| 21 | Q0JPF1 | Reticulon-like protein B1 | LOC_Os01g12650.1 reticulon domain-containing protein | −25.4 | - | - | Reticulon | |
| 22 | A0A0P0W8M8 | Probable aldo-keto reductase 1 | LOC_Os04g26870.1 oxidoreductase, aldo/keto reductase family protein | −26.6 | - | - | Aldo/keto reductase family | |
| O. sativa | 23 | I1PGT7 | ADF-H domain-containing protein | LOC_Os03g60580.1 actin-depolymerizing factor | 26.7 | - | - | Cofilin/tropomyosin-type actin-binding protein |
| 24 | A0A0E0MDF4 | Small nuclear ribonucleoprotein Sm D3 (Sm-D3) | LOC_Os02g01250.1 LSM domain-containing protein | 26.5 | - | - | LSM domain | |
| 25 | B8ATR3 | Uncharacterized protein | LOC_Os04g34600.1 abscisic stress-ripening protein | 1.9 | Glycine-rich cell wall structural protein 2-like isoform X1 (Panicum hallii) | 56% | ABA/WDS induced protein | |
| 26 | Q94HJ5 | Putative 3-beta hydroxysteroid dehydrogenase/isomerase protein | LOC_Os05g01970.4 NAD-dependent epimerase/dehydratase family protein | 0.8 | - | - | NAD(P)H-binding | |
| 27 | Q7XQC9 | Uncharacterized protein | LOC_Os04g02050.1 bifunctional 3-phosphoadenosine 5-phosphosulfate synthetase | −0.7 | ATP sulfurylase 2-like (Oryza brachyantha) | 96% | PUA-like domain; ATP-sulfurylase | |
| 28 | P41344 | Ferredoxin--NADP reductase, leaf isozyme 1, chloroplastic | LOC_Os06g01850.1 ferredoxin--NADP reductase, chloroplast precursor | −0.9 | - | - | Oxidoreductase NAD-binding domain; Oxidoreductase FAD-binding domain | |
| 29 | B9G6K2 | PMEI domain-containing protein | LOC_Os10g36500.2 invertase/pectin methylesterase inhibitor family protein | −25.7 | - | - | Plant invertase/pectin methylesterase inhibitor |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamzelou, S.; Kamath, K.S.; Masoomi-Aladizgeh, F.; Johnsen, M.M.; Atwell, B.J.; Haynes, P.A. Wild and Cultivated Species of Rice Have Distinctive Proteomic Responses to Drought. Int. J. Mol. Sci. 2020, 21, 5980. https://doi.org/10.3390/ijms21175980
Hamzelou S, Kamath KS, Masoomi-Aladizgeh F, Johnsen MM, Atwell BJ, Haynes PA. Wild and Cultivated Species of Rice Have Distinctive Proteomic Responses to Drought. International Journal of Molecular Sciences. 2020; 21(17):5980. https://doi.org/10.3390/ijms21175980
Chicago/Turabian StyleHamzelou, Sara, Karthik Shantharam Kamath, Farhad Masoomi-Aladizgeh, Matthew M. Johnsen, Brian J. Atwell, and Paul A. Haynes. 2020. "Wild and Cultivated Species of Rice Have Distinctive Proteomic Responses to Drought" International Journal of Molecular Sciences 21, no. 17: 5980. https://doi.org/10.3390/ijms21175980
APA StyleHamzelou, S., Kamath, K. S., Masoomi-Aladizgeh, F., Johnsen, M. M., Atwell, B. J., & Haynes, P. A. (2020). Wild and Cultivated Species of Rice Have Distinctive Proteomic Responses to Drought. International Journal of Molecular Sciences, 21(17), 5980. https://doi.org/10.3390/ijms21175980







