Production of Lipopeptide Biosurfactant by a Hydrocarbon-Degrading Antarctic Rhodococcus
Abstract
1. Introduction
2. Results and Discussion
2.1. Screening for the Synthesis of Biosurfactant
2.1.1. Hydrophobicity Assay
2.1.2. Drop-Collapse Test
2.1.3. Oil Spread Test
2.1.4. Microplate Assay
2.1.5. Emulsification Index (E24)
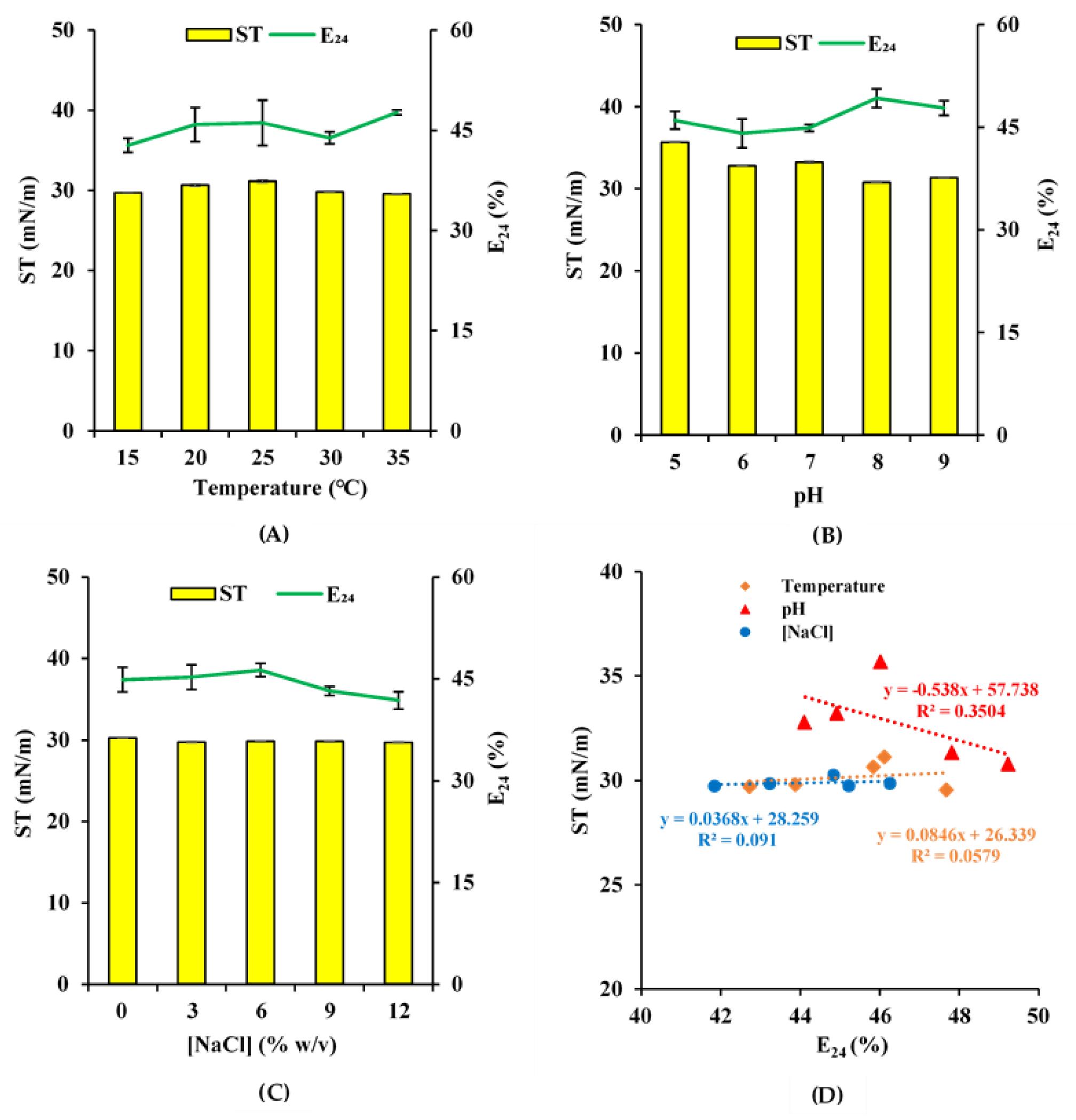
2.2. Biosurfactant Stability Test
2.3. Recovery of Crude Biosurfactant and Estimation of Carbohydrate, Protein and Lipid Content
2.4. Thin-Layer Chromatography
2.5. Fourier-Transform Infrared Spectroscopy Analysis
2.6. Genomic Analyses and Anti-SMASH Prediction
3. Materials and Methods
3.1. Chemical and Reagents
3.2. Microorganism
3.3. Media and Growth Conditions
3.4. Screening of Bacteria for Biosurfactant Production
3.4.1. Hydrophobicity Assay
3.4.2. Drop Collapse Test
3.4.3. Oil Displacement Test
3.4.4. Qualitative Microplate Analysis
3.4.5. Emulsification Capacity Assay
3.5. Determination of Surfactant Stability
3.5.1. Biosurfactant Stability Test
3.5.2. Surface Tension Measurements
3.6. Extraction of Biosurfactant
3.7. Carbohydrate, Protein and Lipid Estimation
3.8. Thin-Layer Chromatography Analysis
3.9. Infrared Analysis
3.10. DNA Extraction, Genome Sequencing and Annotation
3.11. Secondary Metabolites Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BATH | Bacterial adhesion to hydrocarbon |
| BGC | Biosynthetic gene cluster |
| BH | Bushnell Haas |
| BLAST | Basic Local Alignment Search Tool |
| BSA | Bovine serum albumin |
| FTIR | Fourier-transform infrared |
| MTBE | Methyl-tert-butyl ether |
| NRPS | Non-ribosomal peptide synthetase |
| ORF | Open reading frame |
| PBS | Phosphate-buffered saline |
| PCP | Peptidyl carrier protein |
| PKS | Polyketide synthase |
| RAST | Rapid Annotations using Subsystems Technology |
| ST | Surface tension |
| TE | Thioesterase |
| TLC | Thin-layer chromatography |
References
- Aislabie, J.M.; Balks, M.R.; Foght, J.M.; Waterhouse, E.J. Hydrocarbon spills on Antarctic soils: effects and management. Environ. Sci. Technol. 2004, 38, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Aislabie, J.; Saul, D.J.; Foght, J.M. Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles 2006, 10, 171–179. [Google Scholar] [CrossRef] [PubMed]
- McDonald, R.; Knox, O.G.G. Cold region bioremediation of hydrocarbon contaminated soils: Do we know enough? Environ. Sci. Technol. 2014, 48, 9980–9981. [Google Scholar] [CrossRef] [PubMed]
- Gesheva, V.; Stackebrandt, E.; Vasileva-Tonkova, E. Biosurfactant production by halotolerant Rhodococcus fascians from Casey Station, Wilkes Land, Antarctica. Curr. Microbiol. 2010, 61, 112–117. [Google Scholar] [CrossRef]
- Malavenda, R.; Rizzo, C.; Michaud, L.; Gerçe, B.; Bruni, V.; Syldatk, C.; Hausmann, R.; Lo Giudice, A. Biosurfactant production by Arctic and Antarctic bacteria growing on hydrocarbons. Polar Biol. 2015, 38, 1565–1574. [Google Scholar] [CrossRef]
- Desai, J.D.; Banat, I.M. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 1997, 61, 47–64. [Google Scholar] [CrossRef]
- Uzoigwe, C.; Burgess, J.G.; Ennis, C.J.; Rahman, P.K.S.M. Bioemulsifiers are not biosurfactants and require different screening approaches. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. Critical review on biosurfactant analysis, purification and characterization using rhamnolipid as a model biosurfactant. Bioresour. Technol. 2017, 232, 389–397. [Google Scholar] [CrossRef]
- Perfumo, A.; Banat, I.M.; Marchant, R. Going green and cold: Biosurfactants from low-temperature environments to biotechnology applications. Trends Biotechnol. 2018, 36, 277–289. [Google Scholar] [CrossRef]
- Habib, S.; Ahmad, S.A.; Johari, W.L.W.; Shukor, M.Y.A.; Alias, S.A.; Khalil, K.A.; Yasid, N.A. Evaluation of conventional and response surface level optimisation of n-dodecane (n-C12) mineralisation by psychrotolerant strains isolated from pristine soil at Southern Victoria Island, Antarctica. Microb. Cell Factories 2018, 17, 44. [Google Scholar] [CrossRef]
- Kuyukina, M.S.; Ivshina, I.B. Application of Rhodococcus in bioremediation of contaminated environments. In Biology of Rhodococcus; Alvarez, H.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 231–262. [Google Scholar]
- Kuyukina, M.S.; Ivshina, I.B. Rhodococcus biosurfactants: Biosynthesis, properties, and potential applications. In Biology of Rhodococcus; Alvarez, H.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 291–313. [Google Scholar]
- Kügler, J.H.; Le Roes-Hill, M.; Syldatk, C.; Hausmann, R. Surfactants tailored by the class Actinobacteria. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Lang, S.; Philp, J.C. Surface-active lipids in rhodococci. Antonie Van Leeuwenhoek 1998, 74, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Wang, Y.; Sun, F.; Liu, Z.; Lai, Q.; Shao, Z. A novel lipopeptide produced by a Pacific Ocean deep-sea bacterium, Rhodococcus sp. TW53. J. Appl. Microbiol. 2008, 105, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Yalaoui-Guellal, D.; Fella-Temzi, S.; Djafri-Dib, S.; Brahmi, F.; Banat, I.M.; Madani, K. Biodegradation potential of crude petroleum by hydrocarbonoclastic bacteria isolated from Soummam wadi sediment and chemical-biological proprieties of their biosurfactants. J. Pet. Sci. Eng. 2020, 184, 106554. [Google Scholar] [CrossRef]
- Ferrari, B.C.; Bissett, A.; Snape, I.; van Dorst, J.; Palmer, A.S.; Ji, M.; Siciliano, S.D.; Stark, J.S.; Winsley, T.; Brown, M.V. Geological connectivity drives microbial community structure and connectivity in polar, terrestrial ecosystems. Environ. Microbiol. 2016, 18, 1834–1849. [Google Scholar] [CrossRef]
- Ji, M.; Greening, C.; Vanwonterghem, I.; Carere, C.R.; Bay, S.K.; Steen, J.A.; Montgomery, K.; Lines, T.; Beardall, J.; van Dorst, J.; et al. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 2017, 552, 400–403. [Google Scholar] [CrossRef]
- De Pascale, D.; De Santi, C.; Fu, J.; Landfald, B. The microbial diversity of Polar environments is a fertile ground for bioprospecting. Mar. Genom. 2012, 8, 15–22. [Google Scholar] [CrossRef]
- Benaud, N.; Zhang, E.; van Dorst, J.; Brown, M.V.; Kalaitzis, J.A.; Neilan, B.A.; Ferrari, B.C. Harnessing long-read amplicon sequencing to uncover NRPS and Type I PKS gene sequence diversity in polar desert soils. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef]
- Donadio, S.; Monciardini, P.; Sosio, M. Polyketide synthases and nonribosomal peptide synthetases: The emerging view from bacterial genomics. Nat. Prod. Rep. 2007, 24, 1073–1109. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Distribution and phylogeny of nonribosomal peptide and polyketide biosynthetic pathways in eukaryotes. Proc. Natl. Acad. Sci. USA 2014, 111, E3947. [Google Scholar] [CrossRef]
- Rosenberg, M.; Gutnick, D.; Rosenberg, E. Adherence of bacteria to hydrocarbons: A simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 1980, 9, 29–33. [Google Scholar] [CrossRef]
- Satpute, S.K.; Banpurkar, A.G.; Dhakephalkar, P.K.; Banat, I.M.; Chopade, B.A. Methods for investigating biosurfactants and bioemulsifiers: A review. Crit. Rev. Biotechnol. 2010, 30, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.K.; Collins-Thompson, D.L.; Lee, H.; Trevors, J.T. A drop-collapsing test for screening surfactant-producing microorganisms. J. Microbiol. Methods 1991, 13, 271–279. [Google Scholar] [CrossRef]
- Bodour, A.A.; Miller-Maier, R.M. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J. Microbiol. Methods 1998, 32, 273–280. [Google Scholar] [CrossRef]
- Youssef, N.H.; Duncan, K.E.; Nagle, D.P.; Savage, K.N.; Knapp, R.M.; McInerney, M.J. Comparison of methods to detect biosurfactant production by diverse microorganisms. J. Microbiol. Methods 2004, 56, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Vaux, D.J.; Cottingham, M. Method and apparatus for measuring surface configuration. U.S. Patent 722,447,0B2, 29 May 2007. [Google Scholar]
- Chen, C.-Y.; Baker, S.C.; Darton, R.C. The application of a high throughput analysis method for the screening of potential biosurfactants from natural sources. J. Microbiol. Methods 2007, 70, 503–510. [Google Scholar] [CrossRef]
- Du Noüy, P.L. An interfacial tensiometer for universal use. J. Gen. Physiol. 1925, 7, 625–631. [Google Scholar] [CrossRef]
- White, D.A.; Hird, L.C.; Ali, S.T. Production and characterization of a trehalolipid biosurfactant produced by the novel marine bacterium Rhodococcus sp., strain PML026. J. Appl. Microbiol. 2013, 115, 744–755. [Google Scholar] [CrossRef]
- Kazemi, K.; Zhang, B.; Lye, L.M. Production of biosurfactant by Rhodococcus erythropolis sp. cultivated in a novel fish waste compost extract substrate. In Proceedings of the Solid Waste Management Conference, London, ON, Canada, 1–4 June 2016; p. 10. [Google Scholar]
- Israelachvili, J.N.; Mitchell, D.J.; Ninham, B.W. Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J. Chem. Soc. Faraday Trans. 1976, 72, 1525–1568. [Google Scholar] [CrossRef]
- Champion, J.T.; Gilkey, J.C.; Lamparski, H.; Retterer, J.; Miller, R.M. Electron microscopy of rhamnolipid (biosurfactant) morphology: Effects of pH, cadmium, and octadecane. J. Colloid Interface Sci. 1995, 170, 569–574. [Google Scholar] [CrossRef]
- Marqués, A.M.; Pinazo, A.; Farfan, M.; Aranda, F.J.; Teruel, J.A.; Ortiz, A.; Manresa, A.; Espuny, M.J. The physicochemical properties and chemical composition of trehalose lipids produced by Rhodococcus erythropolis 51T7. Chem. Phys. Lipids 2009, 158, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Mouafo, T.H.; Mbawala, A.; Ndjouenkeu, R. Effect of different carbon sources on biosurfactants’ production by three strains of Lactobacillus spp. BioMed Res. Int. 2018, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.; Upasani, V.N. Evaluation of rhamnolipid production by a halotolerant novel strain of Pseudomonas aeruginosa. Bioresour. Technol. 2019, 288, 121577. [Google Scholar] [CrossRef] [PubMed]
- Willumsen, P.A.; Karlson, U. Screening of bacteria, isolated from PAH-contaminated soils, for production of biosurfactants and bioemulsifiers. Biodegradation 1997, 7, 415–423. [Google Scholar] [CrossRef]
- Plaza, G.A.; Zjawiony, I.; Banat, I.M. Use of different methods for detection of thermophilic biosurfactant-producing bacteria from hydrocarbon-contaminated and bioremediated soils. J. Pet. Sci. Eng. 2006, 50, 71–77. [Google Scholar] [CrossRef]
- Cirigliano, M.C.; Carman, G.M. Isolation of a bioemulsifier from Candida lipolytica. Appl. Environ. Microbiol. 1984, 48, 747–750. [Google Scholar] [CrossRef]
- Rosenberg, E.; Ron, E.Z. High- and low-molecular-mass microbial surfactants. Appl. Microbiol. Biotechnol. 1999, 52, 154–162. [Google Scholar] [CrossRef]
- Cooper, D.G.; Macdonald, C.R.; Duff, S.J.B.; Kosaric, N. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation additions. Appl. Environ. Microbiol. 1981, 42, 408–412. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Chaudhary, D.K.; Jeong, S.-W.; Kim, J. Oil-degrading properties of a psychrotolerant bacterial strain, Rhodococcus sp. Y2-2, in liquid and soil media. World J. Microbiol. Biotechnol. 2018, 34. [Google Scholar] [CrossRef]
- Balan, S.S.; Kumar, C.G.; Jayalakshmi, S. Aneurinifactin, a new lipopeptide biosurfactant produced by a marine Aneurinibacillus aneurinilyticus SBP-11 isolated from Gulf of Mannar: Purification, characterization and its biological evaluation. Microbiol. Res. 2017, 194, 1–9. [Google Scholar] [CrossRef]
- Kiran, G.S.; Priyadharsini, S.; Sajayan, A.; Priyadharsini, G.B.; Poulose, N.; Selvin, J. Production of lipopeptide biosurfactant by a marine Nesterenkonia sp. and its application in food industry. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Boland, J.A.; Meijer, W.G. The pathogenic actinobacterium Rhodococcus equi: What’s in a name? Mol. Microbiol. 2019, 112, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.M.; Ponamoreva, O.N.; Nechaeva, I.A.; Petrikov, K.V.; Delegan, Y.A.; Surin, A.K.; Linklater, D.; Filonov, A.E. Characterization of biosurfactants produced by the oil-degrading bacterium Rhodococcus erythropolis S67 at low temperature. World J. Microbiol. Biotechnol. 2018, 34, 20. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, G.J.; Ciapina, E.M.P.; Gomes de Barros, E.; Junior, N.P. Biosurfactant production by Rhodococcus erythropolis and its application to oil removal. Braz. J. Microbiol 2010, 41, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Sekine, M.; Tanikawa, S.; Omata, S.; Saito, M.; Fujisawa, T.; Tsukatani, N.; Tajima, T.; Sekigawa, T.; Kosugi, H.; Matsuo, Y.; et al. Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ. Microbiol. 2006, 8, 334–346. [Google Scholar] [CrossRef]
- Yarza, P.; Spröer, C.; Swiderski, J.; Mrotzek, N.; Spring, S.; Tindall, B.J.; Gronow, S.; Pukall, R.; Klenk, H.-P.; Lang, E.; et al. Sequencing orphan species initiative (SOS): Filling the gaps in the 16S rRNA gene sequence database for all species with validly published names. Syst. Appl. Microbiol. 2013, 36, 69–73. [Google Scholar] [CrossRef]
- Strnad, H.; Patek, M.; Fousek, J.; Szokol, J.; Ulbrich, P.; Nesvera, J.; Paces, V.; Vlcek, C. Genome sequence of Rhodococcus erythropolis strain CCM2595, a phenol derivative-degrading bacterium. Genome Announc. 2014, 2. [Google Scholar] [CrossRef]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Wieland Brown, L.C.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef]
- Ceniceros, A.; Dijkhuizen, L.; Petrusma, M.; Medema, M.H. Genome-based exploration of the specialized metabolic capacities of the genus Rhodococcus. BMC Genom. 2017, 18, 593. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Metcalf, W.W. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genom. 2013, 14, 611. [Google Scholar] [CrossRef]
- Martínez-Núñez, M.A.; y López, V.E.L. Nonribosomal peptides synthetases and their applications in industry. Sustain. Chem. Process. 2016, 4, 1–8. [Google Scholar] [CrossRef]
- Rausch, C.; Hoof, I.; Weber, T.; Wohlleben, W.; Huson, D.H. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol. Biol. 2007, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Roongsawang, N.; Washio, K.; Morikawa, M. Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int. J. Mol. Sci. 2011, 12, 141–172. [Google Scholar] [CrossRef] [PubMed]
- Roongsawang, N.; Lim, S.P.; Washio, K.; Takano, K.; Kanaya, S.; Morikawa, M. Phylogenetic analysis of condensation domains in the nonribosomal peptide synthetases. FEMS Microbiol. Lett. 2005, 252, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, J.; Marahiel, M.A. Chapter 21—Nonribosomal peptide synthesis. In Handbook of Biologically Active Peptides, 2nd ed.; Kastin, A.J., Ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 138–149. [Google Scholar]
- Konz, D.; Doekel, S.; Marahiel, M.A. Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J. Bacteriol. 1999, 181, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-Y.; Chen, C.-L.; Lee, Y.-H.; Cheng, Y.-C.; Wu, Y.-C.; Shu, H.-Y.; Götz, F.; Liu, S.-T. Nonribosomal synthesis of fengycin on an enzyme complex formed by fengycin synthetases. J. Biol. Chem. 2007, 282, 5608–5616. [Google Scholar] [CrossRef]
- Cronan, J.E.; Waldrop, G.L. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 2002, 41, 407–435. [Google Scholar] [CrossRef]
- Lea, W.; Abbas, A.S.; Sprecher, H.; Vockley, J.; Schulz, H. Long-chain acyl-CoA dehydrogenase is a key enzyme in the mitochondrial beta-oxidation of unsaturated fatty acids. Biochim. Biophys. Acta 2000, 1485, 121–128. [Google Scholar] [CrossRef]
- Hu, F.; Liu, Y.; Li, S. Rational strain improvement for surfactin production: Enhancing the yield and generating novel structures. Microb. Cell Fact. 2019, 18, 42. [Google Scholar] [CrossRef]
- Kumar, C.S.; Anuradha, C.M.; Rao, K.V.; Swamy, K.V. In silico characterization of fatty acid synthase of Mycobacterium tuberculosis H37Rv. Internet J. Genom. Proteom. 2005, 2. [Google Scholar]
- Chutoam, P.; Charoensawan, V.; Wongtrakoongate, P.; Kum-arth, A.; Buphamalai, P.; Tungpradabkul, S. RpoS and oxidative stress conditions regulate succinyl-CoA: 3-ketoacid-coenzyme A transferase (SCOT) expression in Burkholderia pseudomallei. Microbiol. Immunol. 2013, 57, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, K.; Akiyama, T.; Shoda, M. Cloning, Sequencing, and Characterization of the Iturin A Operon. J. Bacteriol. 2001, 183, 6265–6273. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Y.; Zhou, K.; Huang, W.T.; Zhou, P.; Yang, S.; Zhao, X.; Xie, J.; Xia, L.; Ding, X. A comprehensive genomic and growth proteomic analysis of antitumor lipopeptide bacillomycin Lb biosynthesis in Bacillus amyloliquefaciens X030. Appl. Microbiol. Biotechnol. 2019, 103, 7647–7662. [Google Scholar] [CrossRef] [PubMed]
- Röttig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2—A web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011, 39, W362–W367. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Haynes, S.W.; Ames, B.D.; Wang, P.; Vien, L.P.; Walsh, C.T.; Tang, Y. Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat. Chem. Biol. 2012, 8, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Keating, T.A.; Ehmann, D.E.; Kohli, R.M.; Marshall, C.G.; Trauger, J.W.; Walsh, C.T. Chain termination steps in nonribosomal peptide synthetase assembly lines: Directed acyl-S-enzyme breakdown in antibiotic and siderophore biosynthesis. ChemBioChem 2001, 2, 99–107. [Google Scholar] [CrossRef]
- Yalaoui-Guellal, D.; Brahmi, F.; Touati, A.; Champs, C.D.; Banat, I.M.; Madani, K. Production of biosurfactants by hydrocarbons degrading bacteria isolated from Soummam watershed sediments of Bejaia in Algeria. Environ. Prog. Sustain. Energy 2018, 37, 189–195. [Google Scholar] [CrossRef]
- Kennicutt, M.C.; Chown, S.L.; Cassano, J.J.; Liggett, D.; Peck, L.S.; Massom, R.; Rintoul, S.R.; Storey, J.; Vaughan, D.G.; Wilson, T.J.; et al. A roadmap for Antarctic and Southern Ocean science for the next two decades and beyond. Antarct. Sci. 2015, 27, 3–18. [Google Scholar] [CrossRef]
- Antarctic Treaty System. Protocol on Environmental Protection to the Antarctic Treaty; Final Report of the XIth Antarctic Special Consultative Meeting; Secretariat of the Antarctic Treaty: Madrid, Spain, 1991; Available online: https://www.ats.aq/e/protocol.html (accessed on 23 June 2020).
- Janek, T.; Krasowska, A.; Czyżnikowska, Z.; Lukaszewicz, M. Trehalose lipid biosurfactant reduces adhesion of microbial pathogens to polystyrene and silicone surfaces: An experimental and computational approach. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Giuliano, L.; Bruni, V.; Scarfi, S.; Golyshin, P.N. Characterization of Antarctic hydrocarbon-degrading bacteria capable of producing bioemulsifiers. New Microbiol. 1999, 22, 249–256. [Google Scholar]
- Smykla, J.; Porazinska, D.L.; Iakovenko, N.S.; Devetter, M.; Drewnik, M.; Hii, Y.S.; Emslie, S.D. Geochemical and biotic factors influencing the diversity and distribution of soil microfauna across ice-free coastal habitats in Victoria Land, Antarctica. Soil Biol. Biochem. 2018, 116, 265–276. [Google Scholar] [CrossRef]
- Cooper, D.G.; Goldenberg, B.G. Surface-active agents from two Bacillus species. Appl. Environ. Microbiol. 1987, 53, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.G.; Zajic, J.E.; Gerson, D.F. Production of surface-active lipids by Corynebacterium lepus. Appl. Environ. Microbiol. 1979, 37, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Kuyukina, M.S.; Ivshina, I.B.; Philp, J.C.; Christofi, N.; Dunbar, S.A.; Ritchkova, M.I. Recovery of Rhodococcus biosurfactants using methyl tertiary-butyl ether extraction. J. Microbiol. Methods 2001, 46, 149–156. [Google Scholar] [CrossRef]
- Du Bois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]

| Strain | Rhodococcus sp. ADL36 | R. erythropolis PR4 [49] | R. erythropolis CCM2925 [51] |
|---|---|---|---|
| Genomic organization | |||
| Genomic size (Mb) | 8.44 | 6.89 | 6.37 |
| G + C (%) 1 | 63.28 | 62.29 | 62.50 |
| Number of CDS 2 | 8462 | 6293 | 5743 |
| tRNAs | 82 | 54 | 53 |
| rRNA gene clusters | 6 | 15 | 12 |
| Assembly and annotation level | Contigs (draft) | Complete (whole) | Complete (whole) |
| Isolate origin | Antarctic soil | Pacific Ocean | Unspecified soil |
| Region | Cluster Type | Gene Cluster(s) | Description | Similarity (%) |
|---|---|---|---|---|
| 1.1 | NRPS | Heterobactin A/S2 | NRP 1 | 100 |
| 2.1 | Bacteriocin | Branched-chain fatty acids | Other | 75 |
| 2.2 | Linear azol(in)e-containing peptides | Diisonitrile antibiotic SF2768 | NRP | 11 |
| 2.3 | T1PKS 2 | — | — | — |
| 3.1 | NRPS | Erythrochelin | NRP | 57 |
| 4.1 | NRPS | — | — | — |
| 4.2 | NRPS | Monensin | Polyketide | 5 |
| 4.3 | Terpene | Isorenieratene | Terpene | 42 |
| 4.4 | Ectoine | Ectoine | Other | 75 |
| 6.1 | NRPS | Bacillomycin D, iturin, mycosubtilin | Polyketide + NRP: lipopeptide | 20, 22, 20 |
| 8.1 | NRPS | Coelichelin | NRP | 27 |
| 8.2 | NRPS, terpene | SF2575 | Polyketide: type II + saccharide: hybrid/tailoring | 6 |
| 9.1 | Lanthipeptide | — | — | — |
| 10.1 | Butyrolactone | — | — | — |
| 10.2 | Aminoglycoside/aminocyclitol cluster | Acarbose | Saccharide | 7 |
| 13.1 | NRPS | Rifamorpholine A–E, rifamycin | Polyketide | 4, 4 |
| 67.1 | NRPS | — | — | — |
| 181.1 | Lasso peptide | — | — | — |
| Strain(s) | Origin | Enrichment Substrate | ST (mN/m) 1 | E24 (%) 2 | Surfactant Content (%) | Reference(s) |
|---|---|---|---|---|---|---|
| Rhodococcus sp. TW53 | Deep-sea sediment, west Pacific Ocean | n-hexadecane | 34.4 (S) 3, 30.7 (P) 4 | n/a 5 | n.d 6 | [15] |
| R. ruber MP4 | Soummam sediments, Bridge Skala of Bejaia, Algeria | Petroleum + glucose | n.d | 78.50 (S), 87.77 (P) | Protein: 10.5; lipid: 64.2 | [16,72] |
| Rhodococcus sp. ADL36 | Fellfield soils, coastal ice-free environments, Southern Victoria Island, Antarctica | Diesel | 29.7 | 45.33 | Protein: 25.0; lipid: 64.0; sugar: < 3.0 | This study |
| Strain(s) | Origin | Substrate | Biosurfactant Produced | ST (mN/m) 1 | E24 (%) 2 | T (°C) 3 | Reference(s) |
|---|---|---|---|---|---|---|---|
| Rhodococcus spp. | [5] | ||||||
| 176; 179; 181; 187 | South Shetlands Islands (Antarctica) and Svalbard Islands (Norwegian Arctic) | Tetradecane | Glycolipid | 28.3; 27.3; 28.6; 28.2 | 58; 37; 33; 57 | 4; 15 | |
| R. fascians A-3 | Casey Station, Wilkes Island, Antarctica | Kerosene | Rhamnose-containing lipid | 27 | n.d 4 | 28 | [4] |
| R. fascians BD8 | Arctic Archipelago of Svalbard | n-hexadecane | Trehalose lipid | 34–36 | n.d | 28 | [75] |
| Rhodococcus sp. | Terra Nova Bay (Ross Sea, Antarctica) | Aliphatic/aromatic hydrocarbons | Trehalose lipid | 32 | n.d | n/a 5 | [76] |
| Rhodococcus sp. ADL36 | Fellfield soils, coastal ice-free environments, Southern Victoria Island, Antarctica | Diesel | Lipopeptide | 29.7 | 45.3 | 20 | This study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habib, S.; Ahmad, S.A.; Wan Johari, W.L.; Abd Shukor, M.Y.; Alias, S.A.; Smykla, J.; Saruni, N.H.; Abdul Razak, N.S.; Yasid, N.A. Production of Lipopeptide Biosurfactant by a Hydrocarbon-Degrading Antarctic Rhodococcus. Int. J. Mol. Sci. 2020, 21, 6138. https://doi.org/10.3390/ijms21176138
Habib S, Ahmad SA, Wan Johari WL, Abd Shukor MY, Alias SA, Smykla J, Saruni NH, Abdul Razak NS, Yasid NA. Production of Lipopeptide Biosurfactant by a Hydrocarbon-Degrading Antarctic Rhodococcus. International Journal of Molecular Sciences. 2020; 21(17):6138. https://doi.org/10.3390/ijms21176138
Chicago/Turabian StyleHabib, Syahir, Siti Aqlima Ahmad, Wan Lutfi Wan Johari, Mohd Yunus Abd Shukor, Siti Aisyah Alias, Jerzy Smykla, Nurul Hani Saruni, Nur Syafiqah Abdul Razak, and Nur Adeela Yasid. 2020. "Production of Lipopeptide Biosurfactant by a Hydrocarbon-Degrading Antarctic Rhodococcus" International Journal of Molecular Sciences 21, no. 17: 6138. https://doi.org/10.3390/ijms21176138
APA StyleHabib, S., Ahmad, S. A., Wan Johari, W. L., Abd Shukor, M. Y., Alias, S. A., Smykla, J., Saruni, N. H., Abdul Razak, N. S., & Yasid, N. A. (2020). Production of Lipopeptide Biosurfactant by a Hydrocarbon-Degrading Antarctic Rhodococcus. International Journal of Molecular Sciences, 21(17), 6138. https://doi.org/10.3390/ijms21176138






