Molecular Network and Culture Media Variation Reveal a Complex Metabolic Profile in Pantoea cf. eucrina D2 Associated with an Acidified Marine Sponge
Abstract
1. Introduction
2. Results and Discussion
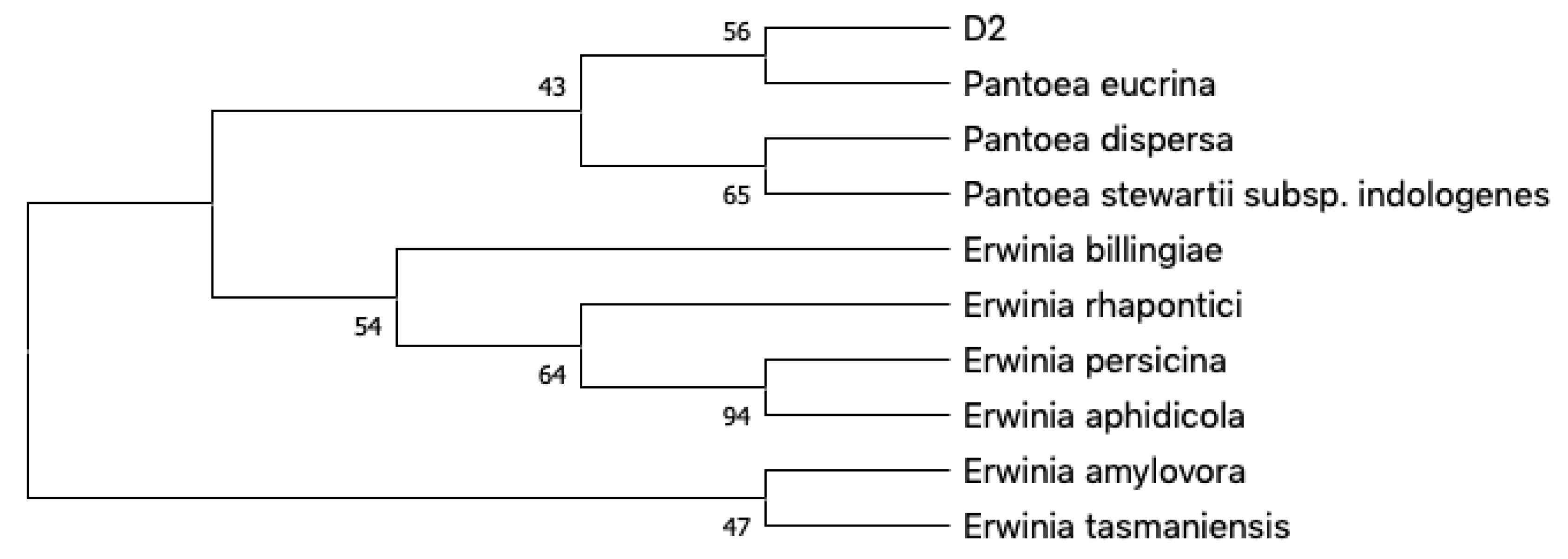
2.1. Isolation and Identification of Bacterial Strains from the Sponge Chondrosia reniformis Living at Naturally Lowered pH Conditions
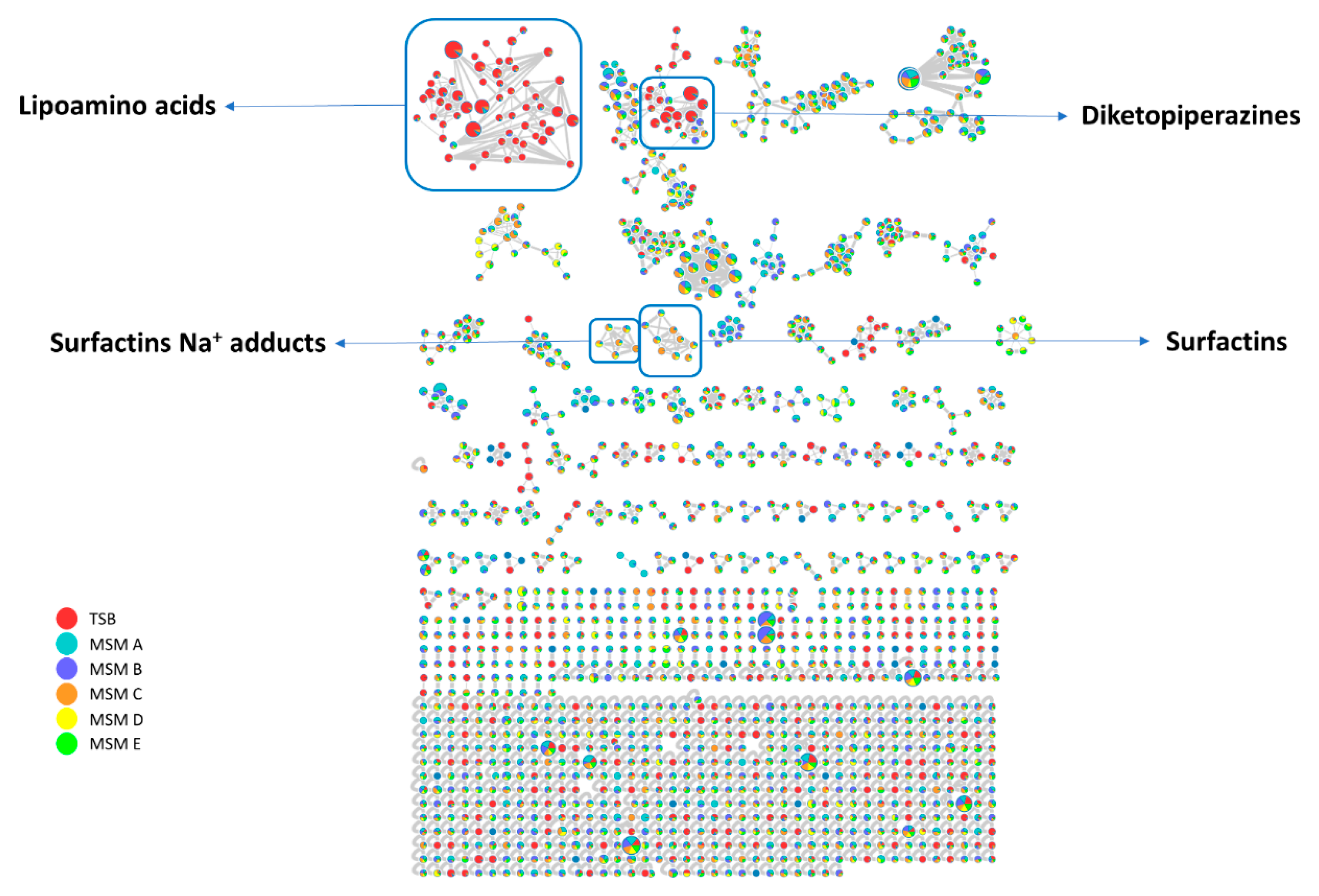
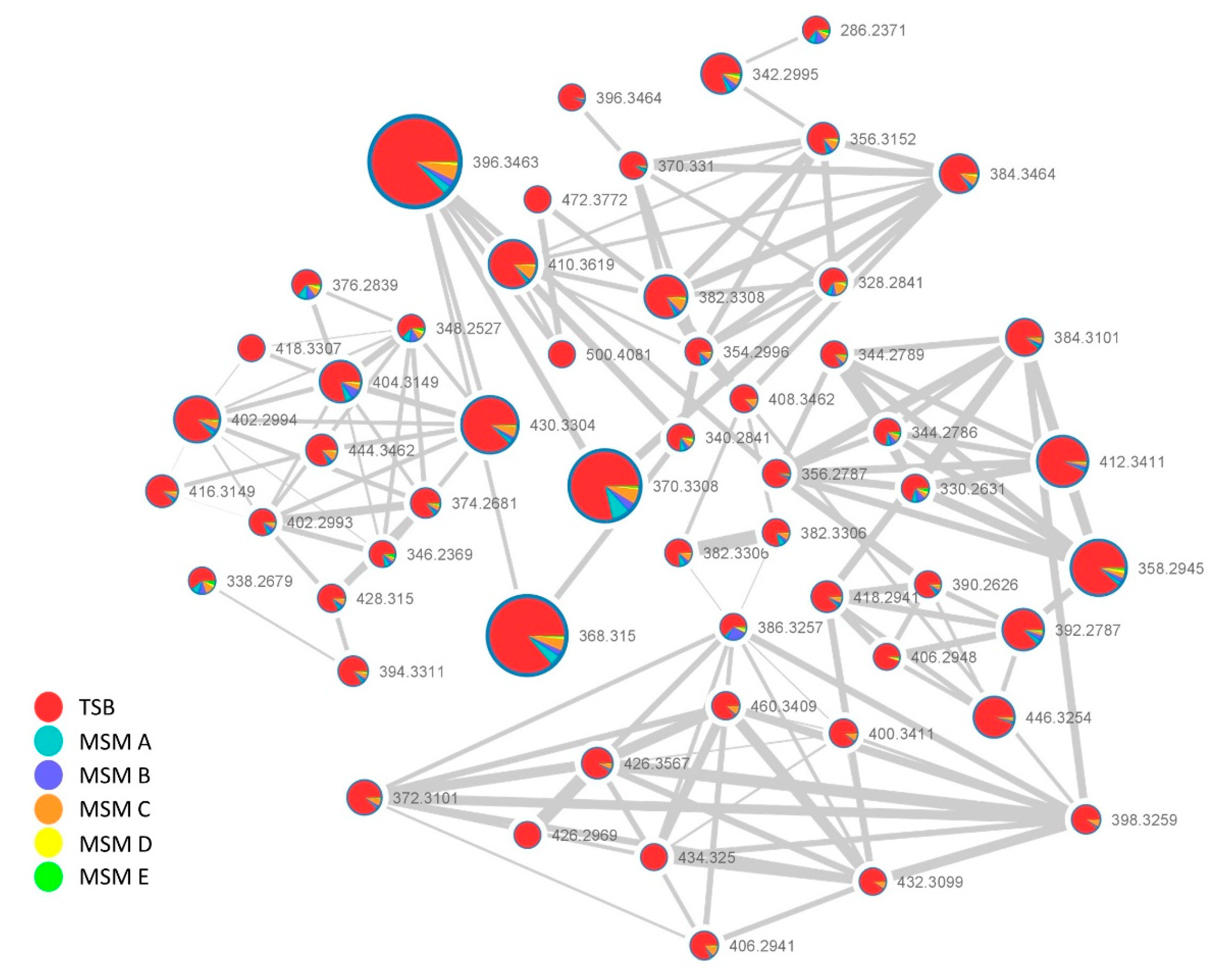
2.2. MS/MS-Based Molecular Networking Analysis of Pantoea eucrina D2 Metabolome Grown under Different Culture Conditions
2.2.1. Surfactins Molecular Cluster
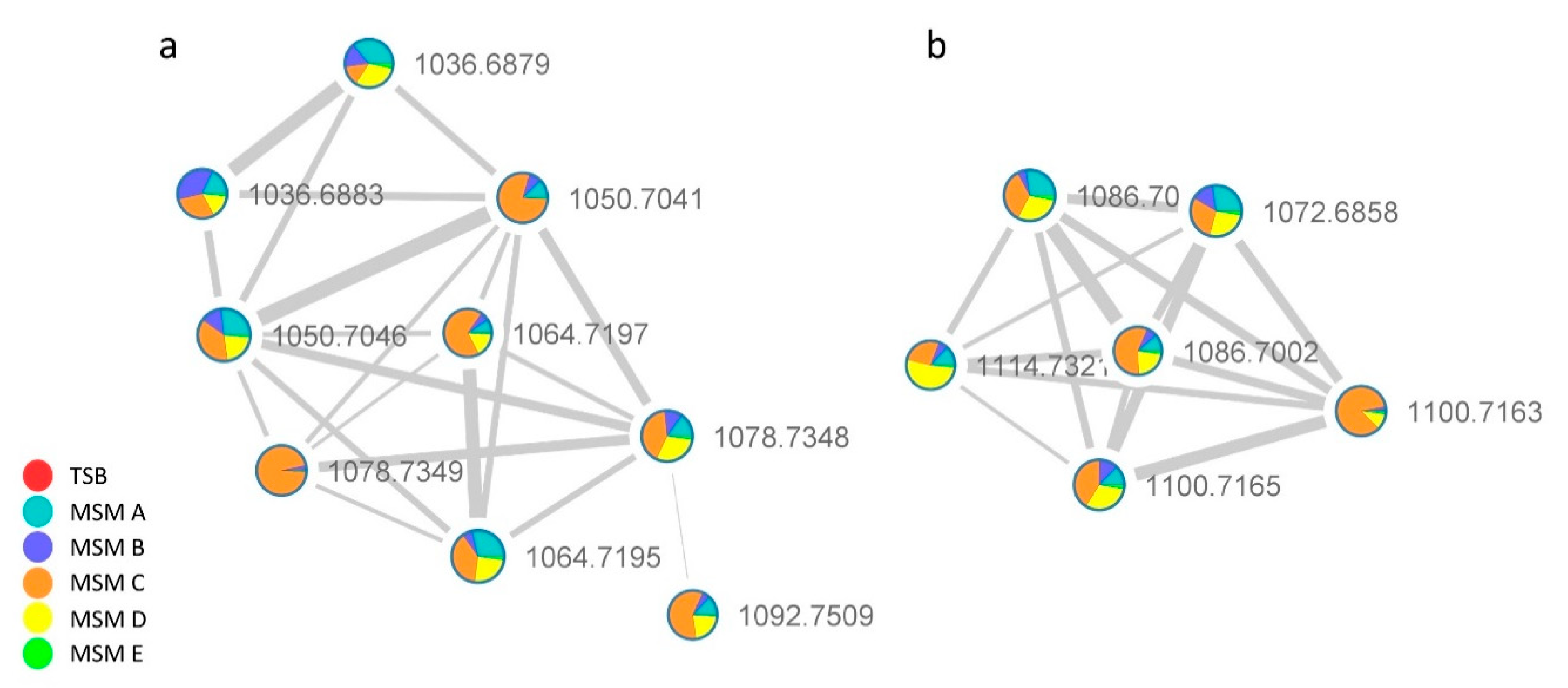
2.2.2. Lipoamino Acids Molecular Cluster
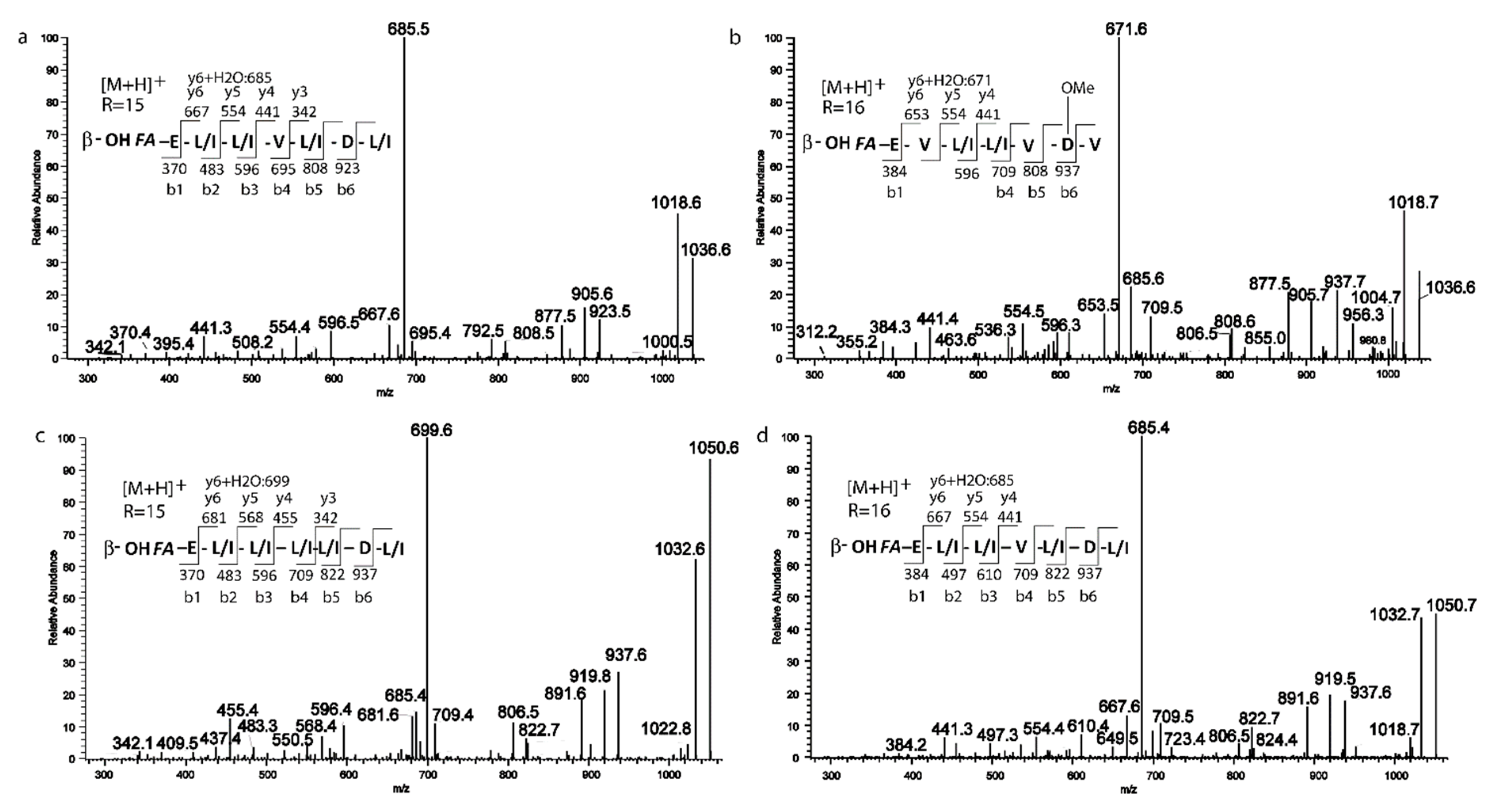
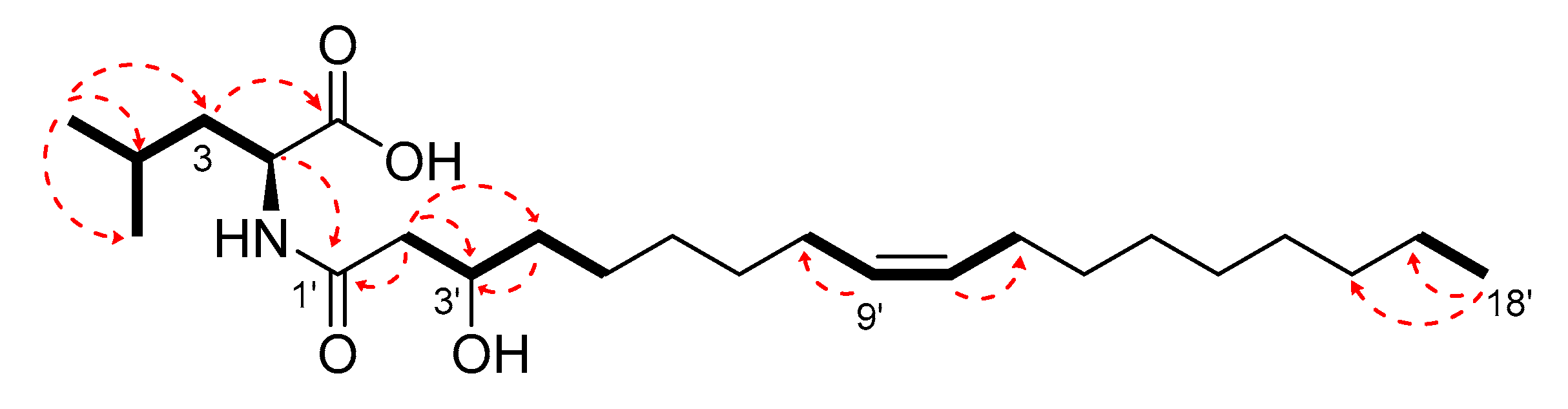
2.3. Isolation and Structure Elucidation of Pure Compounds
2.4. Minimum Inhibition Concentration (MIC)
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Media and Buffers
3.3. Sponge Collection and Bacterial Isolation at Different pH
3.4. 16S Sequencing
3.5. Bacterial Cultivation, OSMAC Cultures and Extraction Methodologies
3.6. Mass Spectrometry Analysis
3.7. Molecular Networking Building
3.8. Medium-Scale Cultivation and Isolation of Pure Lipoamino Acids
3.9. Hydrolysis and Advanced Marfey’s Analysis.
3.10. Minimum Inhibition Concentration (MIC) Assessment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Walterson, A.M.; Stavrinides, J. Pantoea:insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015, 39, 968–984. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. 2016, 23, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.M.H.; Soares, H.M.; Soares, H.M. Promising bacterial genera for agricultural practices: An insight on plant growth-promoting properties and microbial safety aspects. Sci. Total Environ. 2019, 682, 779–799. [Google Scholar] [CrossRef] [PubMed]
- Smits, T.; Duffy, B.; Blom, J.; Ishimaru, C.A.; Stockwell, V.O. Pantocin A, a peptide-derived antibiotic involved in biological control by plant-associated Pantoea species. Arch. Microbiol. 2019, 201, 713–722. [Google Scholar] [CrossRef]
- Teng, T.; Li, X.; Zhang, L.; Li, Y. Identification and Characterization of pantocin wh-1, a Novel Cyclic Polypeptide Produced by Pantoea dispersa W18. Molecules 2020, 25, 485. [Google Scholar] [CrossRef]
- De Maayer, P.; Chan, W.Y.; Blom, J.; Venter, S.; Duffy, B.; Smits, T.H.; Coutinho, T.A. The large universal Pantoea plasmid LPP-1 plays a major role in biological and ecological diversification. BMC Genom. 2012, 13, 625. [Google Scholar] [CrossRef]
- Rezzonico, F.; Smits, T.; Duffy, B. Misidentification slanders Pantoea agglomerans as a serial killer. J. Hosp. Infect. 2012, 81, 137–139. [Google Scholar] [CrossRef]
- Touré, S.; Desrat, S.; Péllissier, L.; Allard, P.-M.; Wolfender, J.-L.; Dusfour, I.; Stien, D.; Eparvier, V. Characterization, Diversity, and Structure-Activity Relationship Study of Lipoamino Acids from Pantoea sp. and Synthetic Analogues. Int. J. Mol. Sci. 2019, 20, 1083. [Google Scholar] [CrossRef]
- Gambi, M.; Gaglioti, M.; Teixido, N. I sistemi di emissione di CO2 dell’isola d’Ischia. Mem. Della Carta Geol. D’italia 2019, 105, 55–64. [Google Scholar]
- Foo, S.A.; Byrne, M.; Ricevuto, E.; Gambi, M.C. The Carbon Dioxide Vents of Ischia, Italy, A Natural System to Assess Impacts of Ocean Acidification on Marine Ecosystems: An Overview of Research and Comparisons with Other Vent Systems. Oceanogr. Mar. Biol. 2018, 237–310. [Google Scholar] [CrossRef]
- Hall-Spencer, J.M.; Rodolfo-Metalpa, R.; Martin, S.; Ransome, E.; Fine, M.; Turner, S.M.; Rowley, S.J.; Tedesco, D.; Buia, M.C. Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nature 2008, 454, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Erwin, P.M.; Coma, R.; López-Sendino, P.; Serrano, E.; Ribes, M. Stable symbionts across the HMA-LMA dichotomy: Low seasonal and interannual variation in sponge-associated bacteria from taxonomically diverse hosts. FEMS Microbiol. Ecol. 2015, 91, 91. [Google Scholar] [CrossRef] [PubMed]
- Björk, J.R.; O’Hara, R.B.; Ribes, M.; Coma, R.; Montoya, J.M. The dynamic core microbiome: Structure, stability and resistance. bioRxiv 2017, 137885. [Google Scholar] [CrossRef]
- Goodwin, C.E.; Rodolfo-Metalpa, R.; Picton, B.; Hall-Spencer, J.M. Effects of ocean acidification on sponge communities. Mar. Ecol. 2013, 35, 41–49. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Brady, C.L.; Cleenwerck, I.; Venter, S.; Vancanneyt, M.; Swings, J.; Coutinho, T.A. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 2008, 31, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.L.; Cleenwerck, I.; Venter, S.N.; Engelbeen, K.; De Vos, P.; Coutinho, T.A. Emended description of the genus Pantoea, description of four species from human clinical samples, Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov. and Pantoea conspicua sp. nov., and transfer of Pectobacterium cypripedii (Hori 1911) Brenner et al. 1973 emend. Hauben et al. 1998 to the genus as Pantoea cypripedii comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 2430–2440. [Google Scholar] [CrossRef]
- Tindall, B.J. The combination Enterobacter agglomerans is to be cited as Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 and the combination Pantoea agglomerans is to be cited as Pantoea agglomerans (Beijerinck 1888) Gavini et al. 1989. Opinion 90. Judicial Commission of the International Committee on Systematics of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 3582–3583. [Google Scholar] [CrossRef]
- Brady, S.F.; Wright, S.A.; Lee, J.C.; Sutton, A.E.; Zumoff, C.H.; Wodzinski, R.S.; Beer, S.V.; Clardy, J. Pantocin B, an antibiotic from Erwinia herbicola discovered by heterologous expression of cloned genes. J. Am. Chem. Soc. 1999, 121, 11912–11913. [Google Scholar] [CrossRef]
- Silvi, S.; Barghini, P.; Aquilanti, A.; Juárez-Jiménez, B.; Fenice, M. Physiologic and metabolic characterization of a new marine isolate (BM39) of Pantoea sp. producing high levels of exopolysaccharide. Microb. Cell Factories 2013, 12, 10. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 1–4. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Pathak, K.V.; Bose, A.; Keharia, H. Characterization of Novel Lipopeptides Produced by Bacillus tequilensis P15 Using Liquid Chromatography Coupled Electron Spray Ionization Tandem Mass Spectrometry (LC–ESI–MS/MS). Int. J. Pept. Res. Ther. 2013, 20, 133–143. [Google Scholar] [CrossRef]
- Bartal, A.; Vigneshwari, A.; Bóka, B.; Vörös, M.; Takacs, I.; Kredics, L.; Manczinger, L.; Varga, M.; Vágvölgyi, C.; Szekeres, A. Effects of Different Cultivation Parameters on the Production of Surfactin Variants by a Bacillus subtilis Strain. Molecules 2018, 23, 2675. [Google Scholar] [CrossRef]
- Kecskeméti, A.; Bartal, A.; Bóka, B.; Kredics, L.; Manczinger, L.; Kadaikunnan, S.; Alharby, N.S.; Khaled, J.M.; Varga, M.; Vágvölgyi, C.; et al. High-Frequency Occurrence of Surfactin Monomethyl Isoforms in the Ferment Broth of a Bacillus subtilis Strain Revealed by Ion Trap Mass Spectrometry. Molecules 2018, 23, 2224. [Google Scholar] [CrossRef]
- Peela, S.; Raju, K.B.; Ramana, T. Studies on a new marine streptomycete BT-408 producing polyketide antibiotic SBR-22 effective against methicillin resistant Staphylococcus aureus. Microbiol. Res. 2005, 160, 119–126. [Google Scholar] [CrossRef]
- Preetha, R.; Jayaprakash, N.; Philip, R.; Singh, I.S.B. Optimization of carbon and nitrogen sources and growth factors for the production of an aquaculture probiotic (Pseudomonas MCCB 103) using response surface methodology. J. Appl. Microbiol. 2006, 102, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Poblete-Castro, I.; Wittmann, C.; Nikel, P.I. Biochemistry, genetics and biotechnology of glycerol utilization in Pseudomonas species. Microb. Biotechnol. 2020, 13, 32–53. [Google Scholar] [CrossRef]
- Zhou, D.; Hu, F.; Lin, J.; Wang, W.; Li, S. Genome and transcriptome analysis of Bacillus velezensis BS-37, an efficient surfactin producer from glycerol, in response to d-/l-leucine. MicrobiologyOpen 2019, 8, e00794. [Google Scholar] [CrossRef]
- Bus, J.; Sies, I.; Jie, M.S.L.K. 13C-NMR of methyl, methylene and carbonyl carbon atoms of methyl alkenoates and alkynoates. Chem. Phys. Lipids 1976, 17, 501–518. [Google Scholar] [CrossRef]
- Guo, H.; Rischer, M.; Sperfeld, M.; Weigel, C.; Menzel, K.D.; Clardy, J.; Beemelmanns, C. Natural products and morphogenic activity of γ-Proteobacteria associated with the marine hydroid polyp Hydractinia echinata. Bioorganic Med. Chem. 2017, 25, 6088–6097. [Google Scholar] [CrossRef] [PubMed]
- Effenberger, K.A.; James, R.C.; Urabe, V.K.; Dickey, B.J.; Linington, R.G.; Jurica, M. The Natural Product N-Palmitoyl-l-leucine Selectively Inhibits Late Assembly of Human Spliceosomes. J. Biol. Chem. 2015, 290, 27524–27531. [Google Scholar] [CrossRef]
- McKellar, R.; Paquet, A.; Ma, C.-Y. Antimicrobial activity of fatty N-acylamino acids against Gram-positive foodborne pathogens. Food Microbiol. 1992, 9, 67–76. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 10th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; Volume 35. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Connolly, P.; Bloomfield, S.F.; Denyer, S. The use of impedance for preservative efficacy testing of pharmaceuticals and cosmetic products. J. Appl. Bacteriol. 1994, 76, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Werbrouck, H.; Vermeulen, A.; Van Coillie, E.; Messens, W.; Herman, L.; Devlieghere, F.; Uyttendaele, M. Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. Int. J. Food Microbiol. 2009, 134, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.D.; Bisno, A.L.; Parisi, J.T.; McLaughlin, B.; Hester, M.G.; Luther, R.W. Nosocomial Septicemia Due to Multiply Antibiotic-Resistant Staphylococcus epidermidis. Ann. Intern. Med. 1982, 96, 1. [Google Scholar] [CrossRef]
- Corvec, S.; Poirel, L.; Naas, T.; Drugeon, H.; Nordmann, P. Genetics and Expression of the Carbapenem-Hydrolyzing Oxacillinase Gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 1530–1533. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.-Z.; Poole, K. Fluoroquinolone susceptibilities of efflux-mediated multidrug-resistant Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Burkholderia cepacia. J. Antimicrob. Chemother. 2001, 48, 549–552. [Google Scholar] [CrossRef]
- Brady, S.F.; Clardy, J. Long-ChainN-Acyl Amino Acid Antibiotics Isolated from Heterologously Expressed Environmental DNA. J. Am. Chem. Soc. 2000, 122, 12903–12904. [Google Scholar] [CrossRef]
- Pohnert, G.; Jung, V.; Haukioja, E.; Lempa, K.; Boland, W. New fatty acid amides from regurgitant of Lepidopteran (Noctuidae, Geometridae) caterpillars. Tetrahedron 1999, 55, 11275–11280. [Google Scholar] [CrossRef]


| Strain | Identity |
|---|---|
| G | Pseudoalteromonas sp. |
| H | Shewanella sp. |
| S1 | Pseudomonas sp. |
| U1 | Sphingobium sp. |
| Z1 | Sphingobium sp. |
| X1 | Stenotrophomonas sp. |
| J1 | Acinetobacter sp. |
| D2 | Pantoea sp. |
| MIC (μg/mL) | |||||
|---|---|---|---|---|---|
| Compound | S. aureus 6538P | L. monocyogenes MB677 | S. epidermidis ATCC 35984 | A. baumannii 13 | S. maltophilia ATCC 13,637 |
| 1 | 10 | 8.0 | 13 | – | – |
| 2 | 13 | 8.0 | 13 | – | – |
| 3 | 10 | 4.0 | 10 | – | – |
| 4 | 10 | 6.6 | 10 | – | – |
| 5 | 128 | 6.6 | 128 | – | – |
| 6 | – | 4.0 | – | – | – |
| 7 | 64 | 16 | 128 | – | – |
| 8 | – | 26 | – | – | – |
| Positive control a | 2 | 1 | 1.7 | 4 | 3.3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitale, G.A.; Sciarretta, M.; Cassiano, C.; Buonocore, C.; Festa, C.; Mazzella, V.; Núñez Pons, L.; D’Auria, M.V.; de Pascale, D. Molecular Network and Culture Media Variation Reveal a Complex Metabolic Profile in Pantoea cf. eucrina D2 Associated with an Acidified Marine Sponge. Int. J. Mol. Sci. 2020, 21, 6307. https://doi.org/10.3390/ijms21176307
Vitale GA, Sciarretta M, Cassiano C, Buonocore C, Festa C, Mazzella V, Núñez Pons L, D’Auria MV, de Pascale D. Molecular Network and Culture Media Variation Reveal a Complex Metabolic Profile in Pantoea cf. eucrina D2 Associated with an Acidified Marine Sponge. International Journal of Molecular Sciences. 2020; 21(17):6307. https://doi.org/10.3390/ijms21176307
Chicago/Turabian StyleVitale, Giovanni Andrea, Martina Sciarretta, Chiara Cassiano, Carmine Buonocore, Carmen Festa, Valerio Mazzella, Laura Núñez Pons, Maria Valeria D’Auria, and Donatella de Pascale. 2020. "Molecular Network and Culture Media Variation Reveal a Complex Metabolic Profile in Pantoea cf. eucrina D2 Associated with an Acidified Marine Sponge" International Journal of Molecular Sciences 21, no. 17: 6307. https://doi.org/10.3390/ijms21176307
APA StyleVitale, G. A., Sciarretta, M., Cassiano, C., Buonocore, C., Festa, C., Mazzella, V., Núñez Pons, L., D’Auria, M. V., & de Pascale, D. (2020). Molecular Network and Culture Media Variation Reveal a Complex Metabolic Profile in Pantoea cf. eucrina D2 Associated with an Acidified Marine Sponge. International Journal of Molecular Sciences, 21(17), 6307. https://doi.org/10.3390/ijms21176307









