Research Progress on the Roles of Cytokinin in Plant Response to Stress
Abstract
:1. Introduction
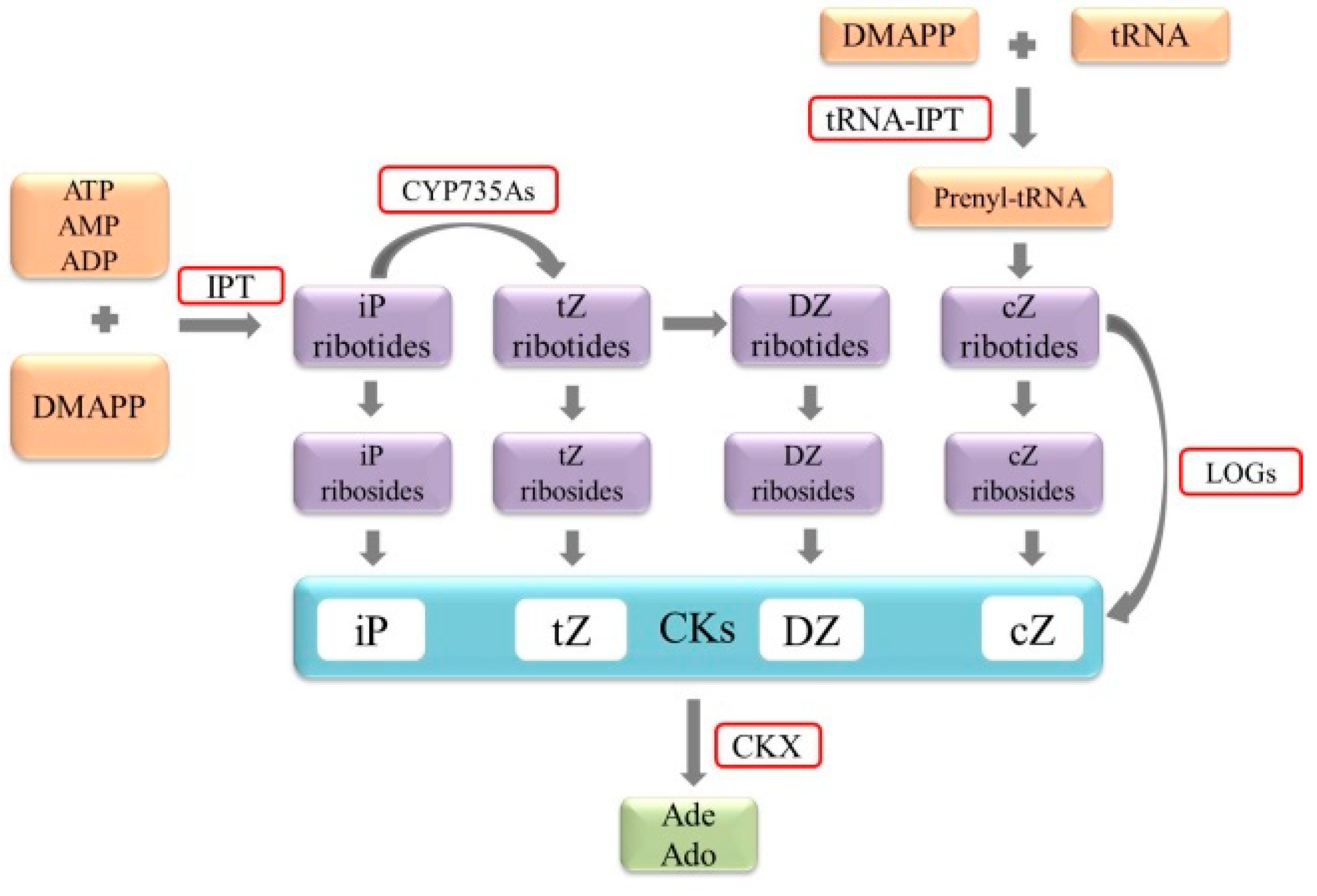
2. Synthesis and Metabolism
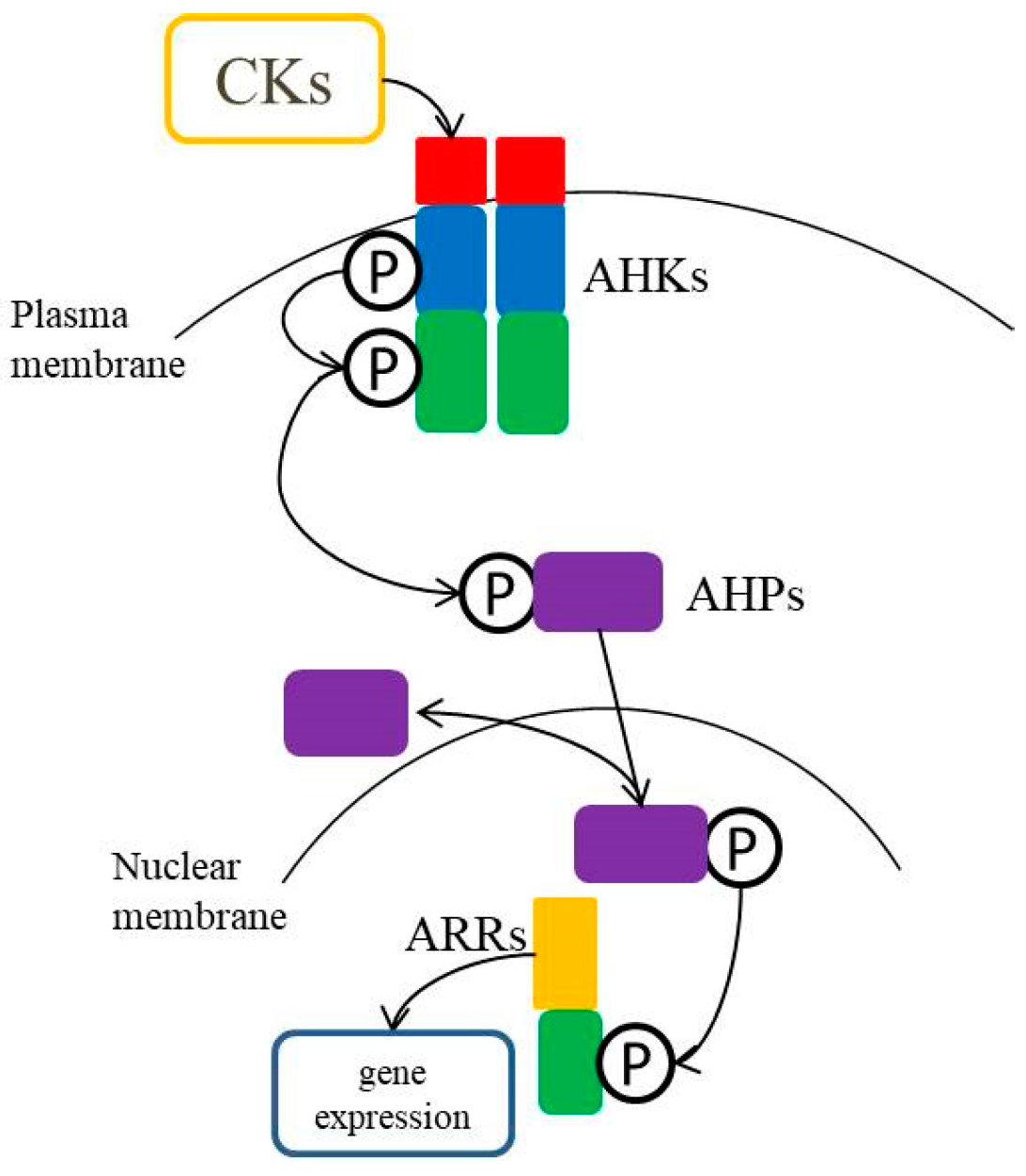
3. Cytokinin Signaling Pathway
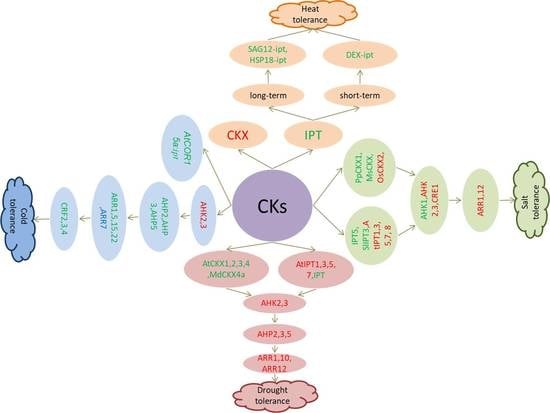
4. Role of Cytokinin in Abiotic Stress Response
4.1. Heat Stress Response
4.2. Cold Stress Response
4.3. Salt Stress Response
4.4. Drought Stress Response
5. Conclusions and Prospects
5.1. Cytokinin Response Factors (CRFs) Are Key Factors Involved in the Role of Cytokinin in Abiotic Stress
5.2. Learning from Different Stresses
5.3. Enhancing Practical Application
Author Contributions
Funding
Conflicts of Interest
References
- Honig, M.; Plihalova, L.; Husickova, A.; Nisler, J.; Doležal, K. Role of cytokinins in senescence, antioxidant defence and photosynthesis. Int. J. Mol. Sci. 2018, 19, 4045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: A coping strategy. Environ. Sci. Pollut. Res. Int. 2018, 25, 33103–33118. [Google Scholar] [CrossRef] [PubMed]
- Mi, X.; Wang, X.; Wu, H.; Gan, L.; Ding, J.; Li, Y. Characterization and expression analysis of cytokinin biosynthesis genes in Fragaria vesca. Plant Growth Regul. 2017, 82, 139–149. [Google Scholar] [CrossRef]
- Keshishian, E.A.; Hallmark, H.T.; Ramaraj, T.; Plackova, L.; Sundararajan, A.; Schilkey, F.D.; Novak, O.; Rashotte, A.M. Salt and oxidative stresses uniquely regulate tomato cytokinin levels and transcriptomic response. Plant. Direct. 2018, 2, e00071. [Google Scholar] [CrossRef] [Green Version]
- Prerostova, S.; Dobrev, P.I.; Gaudinova, A.; Knirsch, V.; Korber, N.; Pieruschka, R.; Fiorani, F.; Brzobohatý, B.; Cerný, M.; Spichal, L.; et al. Cytokinins: Their Impact on molecular and growth responses to drought stress and recovery in Arabidopsis. Front. Plant Sci. 2018, 9, 655. [Google Scholar] [CrossRef] [Green Version]
- Kiba, T.; Takei, K.; Kojima, M.; Sakakibara, H. Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell. 2013, 27, 452–461. [Google Scholar] [CrossRef] [Green Version]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant. Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [Green Version]
- Kudo, T.; Kiba, T.; Sakakibara, H. Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 2010, 52, 53–60. [Google Scholar] [CrossRef]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant. Cell. 2009, 21, 3152–3169. [Google Scholar] [CrossRef] [Green Version]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef] [Green Version]
- Zurcher, E.; Muller, B. Biology, Cytokinin synthesis, signaling, and function—Advances and new insights. Int. Rev. Cell Mol. Biol. 2016, 324, 1–38. [Google Scholar] [PubMed]
- Sakamoto, T.; Sakakibara, H.; Kojima, M.; Yamamoto, Y.; Nagasaki, H.; Inukai, Y.; Sato, Y.; Matsuoka, M. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 2006, 142, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frebort, I.; Kowalska, M.; Hluska, T.; Frebortova, J.; Galuszka, P. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef]
- Werner, T.; Kollmer, I.; Bartrina, I.; Holst, K.; Schmulling, T. New insights into the biology of cytokinin degradation. Plant Biol. 2006, 8, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Spichal, L.; Rakova, N.; Riefler, M.; Mizuno, T.; Romanov, G.A.; Strnad, M.; Schmulling, T. Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant and Cell Physiol. 2004, 45, 1299–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galuszka, P.; Popelkova, H.; Werner, T.; Frebortova, J.; Pospisilova, H.; Mik, V.; Kollmer, I.; Schmulling, T.; Frebort, I. Biochemical characterization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. J. Plant Growth Regul. 2007, 26, 255–267. [Google Scholar] [CrossRef]
- Kowalska, M.; Galuszka, P.; Frebortova, J.; Sebela, M.; Beres, T.; Hluska, T.; Smehilova, M.; Bilyeu, K.D.; Frebort, I. Vacuolar and cytosolic cytokinin dehydrogenases of Arabidopsis thaliana: Heterologous expression, purification and properties. Phytochemistry 2010, 71, 1970–1978. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.; Xin, Q. Genome-wide analysis and identification of cytokinin oxidase/dehydrogenase (CKX) gene family in foxtail millet (Setaria italica). Crop J. 2014, 2, 244–254. [Google Scholar] [CrossRef] [Green Version]
- Massonneau, A.; Houbaherin, N.; Pethe, C.; Madzak, C.; Falque, M.; Mercy, M.; Kopecny, D.; Majira, A.; Rogowsky, P.M.; Laloue, M. Maize cytokinin oxidase genes: Differential expression and cloning of two new cDNAs. J. Exp. Bot. 2004, 55, 2549–2557. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; Stock, A.M. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009, 63, 133–154. [Google Scholar] [CrossRef] [Green Version]
- Cheung, J.; Hendrickson, W.A. Sensor domains of two-component regulatory systems. Curr. Opin. Microbiol. 2010, 13, 116–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grefen, C.; Harter, K. Plant two-component systems: Principles, functions, complexity and cross talk. Planta 2004, 219, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Sheen, J.; Muller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pils, B.; Heyl, A. Unraveling the evolution of cytokinin signaling. Plant Physiol. 2009, 151, 782–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochida, K.; Yoshida, T.; Sakurai, T.; Yamaguchishinozaki, K.; Shinozaki, K.; Tran, L.P. Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 2010, 17, 303–324. [Google Scholar] [CrossRef] [Green Version]
- Schaller, G.E.; Kieber, J.J.; Shiu, S.H. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book 2008, 6, e0112. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.; Chen, H.C.; Sheen, J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002, 129, 500–515. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, C.; Ohashi, Y.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi, C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 2004, 16, 1365–1377. [Google Scholar] [CrossRef] [Green Version]
- Schaller, G.E.; Shiu, S.H.; Armitage, J.P. Two-component systems and their co-option for eukaryotic signal transduction. Curr. Biol. 2011, 21, R320–R330. [Google Scholar] [CrossRef] [Green Version]
- Kabbara, S.; Schmulling, T.; Papon, N. CHASEing cytokinin receptors in plants, bacteria, fungi, and beyond. Trends Plant Sci. 2018, 23, 179–181. [Google Scholar] [CrossRef]
- Wang, F.; Cheng, S.; Wu, Y.; Ren, B.; Qian, W. A bacterial receptor PcrK Senses the plant hormone cytokinin to promote adaptation to oxidative stress. Cell Rep. 2017, 21, 2940–2951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchison, C.E.; Li, J.; Argueso, C.T.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Sakurai, K.; Imamura, A.; Nakamura, A.; Ueguchi, C.; Mizuno, T. Compilation and characterization of histidine-containing phosphotransmitters implicated in His-to-Asp phosphorelay in plants: AHP signal transducers of Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2000, 64, 2486–2489. [Google Scholar] [CrossRef]
- Mahonen, A.P.; Bishopp, A.; Higuchi, M.; Nieminen, K.; Kinoshita, K.; Tormakangas, K.; Ikeda, Y.; Oka, A.; Kakimoto, T.; Helariutta, Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 2006, 311, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, C.; Chen, Q.; Chen, H.; Ren, B.; Li, X.; Zuo, J. S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 2013, 4, 1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, K.; Mathews, D.E.; Kim, H.J.; Street, I.H.; Wildes, S.L.; Chiang, Y.H.; Mason, M.G.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J.; et al. Functional characterization of type-B response regulators in the Arabidopsis cytokinin response. Plant Physiol. 2013, 162, 212–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argyros, R.D.; Mathews, D.E.; Chiang, Y.; Palmer, C.M.; Thibault, D.M.; Etheridge, N.; Argyros, D.A.; Mason, M.G.; Kieber, J.J.; Schaller, G.E. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 2008, 20, 2102–2116. [Google Scholar] [CrossRef] [Green Version]
- Ishida, K.; Yamashino, T.; Yokoyama, A.; Mizuno, T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Zubo, Y.O.; Schaller, G.E. Role of the cytokinin-activated type-B response regulators in hormone crosstalk. Plants 2020, 9, 166. [Google Scholar] [CrossRef] [Green Version]
- Brandstatter, I.; Kieber, J.J. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. The Plant Cell. 1998, 10, 1009–1019. [Google Scholar] [CrossRef] [Green Version]
- To, J.P.; Haberer, G.; Ferreira, F.; Deruere, J.; Mason, M.G.; Schaller, G.E.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 2004, 16, 658–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Zhang, X.; Gong, Z.; Yang, S.; Shi, Y. ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J. 2017, 89, 354–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Dai, X.; Li, J.; Liu, N.; Liu, X.; Li, S. The type-B cytokinin response regulator ARR1 inhibits shoot regeneration in an ARR12-dependent manner in Arabidopsis. Plant Cell. 2020, 32, 2271–2291. [Google Scholar] [CrossRef]
- To, J.P.; Deruere, J.; Maxwell, B.B.; Morris, V.F.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 2007, 19, 3901–3914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horak, J.; Grefen, C.; Berendzen, K.W.; Hahn, A.; Stierhof, Y.; Stadelhofer, B.; Stahl, M.; Koncz, C.; Harter, K. The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant. Biol. 2008, 8, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, S.; Kiba, T.; Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Genes Encoding Pseudo-Response Regulators: Insight into His-to-Asp Phosphorelay and Circadian Rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 791–803. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.C.; Weir, N.R.; Hill, K.; Zhang, W.; Kim, H.J.; Shiu, S.; Schaller, G.E.; Kieber, J.J. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012, 158, 1666–1684. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Shi, Y.; Zhang, X.; Du, H.; Xu, B.; Huang, B. Melatonin suppression of heat-induced leaf senescence involves changes in abscisic acid and cytokinin biosynthesis and signaling pathways in perennial ryegrass (Lolium perenne L.). Environ. Exp. Bot. 2017, 138, 36–45. [Google Scholar] [CrossRef]
- Hu, S.; Ding, Y.; Zhu, C. Sensitivity and responses of chloroplasts to heat stress in plants. Front. Plant. Sci. 2020, 11, 375. [Google Scholar] [CrossRef] [Green Version]
- Dobra, J.; Cerný, M.; Storchova, H.; Dobrev, P.I.; Skalak, J.; Jedelský, P.; Luksanova, H.; Gaudinova, A.; Pesek, B.; Malbeck, J.; et al. The impact of heat stress targeting on the hormonal and transcriptomic response in Arabidopsis. Plant Sci. 2015, 231, 52–61. [Google Scholar] [CrossRef]
- Skalak, J.; Cerný, M.; Jedelský, P.; Dobra, J.; Ge, E.; Novak, J.; Hronkova, M.; Dobrev, P.I.; Vankova, R.; Brzobohatý, B. Stimulation of ipt overexpression as a tool to elucidate the role of cytokinins in high temperature responses of Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 2861–2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Huang, B. Effects of foliar-applied ethylene inhibitor and synthetic cytokinin on creeping bentgrass to enhance heat tolerance. Crop Sci. 2009, 49, 1876–1884. [Google Scholar] [CrossRef]
- Cerny, M.; Jedelský, P.; Novak, J.; Schlosser, A.; Brzobohatý, B. Cytokinin modulates proteomic, transcriptomic and growth responses to temperature shocks in Arabidopsis. Plant Cell Environ. 2014, 37, 1641–1655. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Gianfagna, T.; Huang, B. Proteomic changes associated with expression of a gene (ipt) controlling cytokinin synthesis for improving heat tolerance in a perennial grass species. J. Exp. Bot. 2010, 61, 3273–3289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Cui, K.; Wang, W.; Li, Q.; Fahad, S.; Hu, Q.; Huang, J.; Nie, L.; Mohapatra, P.K.; Peng, S. Heat-induced cytokinin transportation and degradation are associated with reduced panicle cytokinin expression and fewer spikelets per panicle in rice. Front. Plant Sci. 2017, 8, 371. [Google Scholar] [CrossRef] [Green Version]
- Zatloukal, M.; Gemrotova, M.; Doležal, K.; Havlicek, L.; Spichal, L.; Strnad, M. Novel potent inhibitors of A. thaliana cytokinin oxidase/dehydrogenase. Bioorg. Med. Chem 2008, 16, 9268–9275. [Google Scholar] [CrossRef]
- Prerostova, S.; Dobrev, P.I.; Kramna, B.; Alena, G.; Knirsch, V.; Spichal, L.; Zatloukal, M.; Vankova, R. Heat acclimation and inhibition of cytokinin degradation positively affect heat stress tolerance of Arabidopsis. Front. Plant Sci. 2020, 11, 87. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Gupta, S.; Rashotte, A.M. Characterization of two tomato AP2/ERF genes, SlCRF1 and SlCRF2 in hormone and stress responses. Plant Cell Reports. 2014, 33, 35–45. [Google Scholar] [CrossRef]
- Veerasamy, M.; He, Y.; Huang, B. Leaf senescence and protein metabolism in creeping bentgrass exposed to heat stress and treated with cytokinins. J. Am. Soc. Hortic. Sci. 2007, 132, 467–472. [Google Scholar] [CrossRef] [Green Version]
- Sobol, S.; Chayut, N.; Nave, N.; Kafle, D.; Hegele, M.; Kaminetsky, R.; Wunsche, J.N.; Samach, A. Genetic variation in yield under hot ambient temperatures spotlights a role for cytokinin in protection of developing floral primordia. Plant Cell Environ. 2014, 37, 643–657. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Cui, K.; Wang, W.; Li, Q.; Fahad, S.; Hu, Q.; Huang, J.; Nie, L.; Peng, S. Heat-induced phytohormone changes are associated with disrupted early reproductive development and reduced yield in rice. Sci. Rep. 2016, 6, 34978. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.X.; Fu, C.; Yang, W.W.; Zhang, Q.; Fan, H.; Liu, J. The involvement of TsFtsH8 in Thellungiella salsuginea tolerance to cold and high light stresses. Acta Physiol. Plant 2016, 38, 3. [Google Scholar] [CrossRef]
- Sui, N. Photoinhibition of Suaeda salsa to chilling stress is related to energy dissipation and water-water cycle. Photosynthetica 2015, 53, 207–212. [Google Scholar] [CrossRef]
- Feng, Z.; Deng, Y.; Fan, H.; Sun, Q.J.; Sui, N.; Wang, B.S. Effects of NaCl stress on the growth and photosynthetic characteristics of Ulmus pumila L. seedlings in sand culture. Photosynthetica. 2014, 52, 313–320. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Q.; Cui, F.; Hou, L.; Zhao, S.; Xia, H.; Qiu, J.; Li, T.; Zhang, Y.; Wang, X. Genome-wide analysis of gene expression provides new insights into cold responses in Thellungiella salsuginea. Front. Plant Sci. 2017, 8, 713. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.C.; Li, M.; Xie, X.; Han, G.; Sui, N.; Wang, B. Deficiency of phytochrome B alleviates chilling-induced photoinhibition in rice. Am. J. Bot. 2013, 100, 1860–1870. [Google Scholar] [CrossRef]
- Cheng, S.; Yang, Z.; Wang, M.; Song, J.; Sui, N.; Fan, H. Salinity improves chilling resistance in Suaeda salsa. Acta Physiol. Plant. 2014, 36, 1823–1830. [Google Scholar] [CrossRef]
- Liu, W.; Ji, S.; Fang, X.; Wang, Q.; Li, Z.; Yao, F.; Hou, L.; Dai, S. Protein kinase LTRPK1 influences cold adaptation and microtubule stability in rice. J. Plant Growth Regul. 2013, 32, 483–490. [Google Scholar] [CrossRef]
- Koc, I.; Yuksel, I.; Caetano-Anolles, G. Metabolite-centric reporter pathway and tripartite network analysis of Arabidopsis under cold stress. Front. Bioeng. Biotechnol. 2018, 6, 121. [Google Scholar] [CrossRef]
- Li, S.; Yang, Y.; Zhang, Q.; Liu, N.; Xu, Q.; Hu, L. Differential physiological and metabolic response to low temperature in two zoysiagrass genotypes native to high and low latitude. PLoS ONE 2018, 13, e0198885. [Google Scholar] [CrossRef]
- Fenollosa, E.; Gamez, A.; Munne-Bosch, S. Plasticity in the hormonal response to cold stress in the invasive plant Carpobrotus edulis. J. Plant Physiol. 2018, 231, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Zhao, H.; Liu, W.; Li, L.; He, Y. Role of cytokinin and salicylic acid in plant growth at low temperatures. Plant Growth Regul. 2008, 57, 211–221. [Google Scholar] [CrossRef]
- Khan, T.A.; Fariduddin, Q.; Yusuf, M. Low-temperature stress: Is phytohormones application a remedy? Environ. Sci. Pollut. Res. Int. 2017, 24, 21574–21590. [Google Scholar] [CrossRef] [PubMed]
- Belintani, N.G.; Guerzoni, J.T.; Moreira, R.M.; Vieira, L.G. Improving low-temperature tolerance in sugarcane by expressing the ipt gene under a cold inducible promoter. Biologia Plantarum. 2011, 56, 71–77. [Google Scholar] [CrossRef]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef] [Green Version]
- Kang, N.Y.; Cho, C.; Kim, J. Inducible expression of Arabidopsis response regulator 22 (ARR22), a type-C ARR, in transgenic Arabidopsis enhances drought and freezing tolerance. PLoS ONE 2013, 8, e79248. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, J. Arabidopsis response Regulator1 and Arabidopsis histidine phosphotransfer Protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 2013, 161, 408–424. [Google Scholar] [CrossRef] [Green Version]
- Zwack, P.J.; Compton, M.A.; Adams, C.I.; Rashotte, A.M. Cytokinin response factor 4 (CRF4) is induced by cold and involved in freezing tolerance. Plant Cell Rep. 2016, 35, 573–584. [Google Scholar] [CrossRef]
- Jeon, J.; Cho, C.; Lee, M.R.; Binh, N.V.; Kim, J. Cytokinin response factor2 (CRF2) and CRF3 regulate lateral root development in response to cold stress in Arabidopsis. Plant Cell 2016, 28, 1828–1843. [Google Scholar] [CrossRef] [Green Version]
- Jeon, J.; Kim, N.Y.; Kim, S.; Kang, N.Y.; Novak, O.; Ku, S.; Cho, C.; Lee, D.J.; Lee, E.; Strnad, M.A.; et al. subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 2010, 285, 23371–23386. [Google Scholar] [CrossRef] [Green Version]
- Veselova, S.V.; Farhutdinov, R.; Veselov, S.Y.; Kudoyarova, G.R.; Veselov, D.S.; Hartung, W. The effect of root cooling on hormone content, leaf conductance and root hydraulic conductivity of durum wheat seedlings (Triticum durum L.). J. Plant Physiol. 2005, 162, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Sebolt, A.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Song, J.; Wang, B.-S. NaCl increases the activity of the plasma membrane H+-ATPase in C3 halophyte Suaeda salsa callus. Acta Physiol. Plant. 2009, 32, 27–36. [Google Scholar] [CrossRef]
- Song, J.; Wang, B. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 2015, 115, 541–553. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Li, Y.; Han, G.; Song, J.; Wang, B. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct. Plant Biol. 2018, 45, 350–361. [Google Scholar] [CrossRef]
- Feng, Z.T.; Deng, Y.Q.; Zhang, S.C.; Liang, X.; Yuan, F.; Hao, J.L.; Zhang, J.C.; Sun, S.F.; Wang, B.S. K(+) accumulation in the cytoplasm and nucleus of the salt gland cells of Limonium bicolor accompanies increased rates of salt secretion under NaCl treatment using NanoSIMS. Plant Sci. 2015, 238, 286–296. [Google Scholar] [CrossRef]
- Liu, S.; Wang, W.; Li, M.; Wan, S.; Sui, N. Antioxidants and unsaturated fatty acids are involved in salt tolerance in peanut. Acta Physiol. Plant. 2017, 39, 207. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, R.; Ma, Y.; Song, J. Physiological and molecular evidence for Na+ and Cl− exclusion in the roots of two Suaeda salsa populations. Aquat. Bot. 2018, 146, 1–7. [Google Scholar] [CrossRef]
- Sui, N.; Han, G. Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. Acta Physiol. Plant. 2014, 36, 983–992. [Google Scholar] [CrossRef]
- Wang, F.; Xu, Y.; Wang, S.; Shi, W.; Liu, R.; Feng, G.; Song, J. Salinity affects production and salt tolerance of dimorphic seeds of Suaeda salsa. Plant Physiol. Biochem. 2015, 95, 41–48. [Google Scholar] [CrossRef]
- Lin, J.; Li, J.P.; Yuan, F.; Yang, Z.; Wang, B.S.; Chen, M. Transcriptome profiling of genes involved in photosynthesis in Elaeagnus angustifolia L. under salt stress. Photosynthetica 2018, 56, 998–1009. [Google Scholar] [CrossRef]
- Han, N.; Lan, W.; He, X.; Shao, Q. Expression of a Suaeda salsa vacuolar H+/Ca2+ transporter gene in Arabidopsis contributes to physiological changes in salinity. Plant Mol. Biol. Report. 2011, 30, 470–477. [Google Scholar] [CrossRef]
- Han, N.; Shao, Q.; Bao, H.; Wang, B. Cloning and characterization of a Ca2+/H+ antiporter from halophyte Suaeda salsa L. Plant Mol. Biol. Rep. 2010, 29, 449–457. [Google Scholar] [CrossRef]
- Li, K.; Pang, C.H.; Ding, F.; Sui, N.; Feng, Z.T.; Wang, B.S. Overexpression of Suaeda salsa stroma ascorbate peroxidase in Arabidopsis chloroplasts enhances salt tolerance of plants. South Afr. J. Bot. 2012, 78, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.C.; Liu, W.; Qiu, L.; Zhang, S.M.; Ma, L.; Zhang, H. Overexpression of glutathione S-transferase gene increases salt tolerance of Arabidopsis. Russ. J. Plant Physiol. 2010, 57, 233–240. [Google Scholar] [CrossRef]
- Vankova, R.; Gaudinova, A.; Dobrev, P.I.; Malbeck, J.; Haisel, D.; Motyka, V. Comparison of salinity and drought stress effects on abscisic acid metabolites activity of cytokinin oxidase/dehydrogenase and chlorophyll levels in radish audtabacco. Ecol. Quest. 2010, 14, 99–100. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Liu, J.; Zhai, L.; Gan, Z.; Zhang, G.; Yang, S.; Wang, Y.; Wu, T.; Zhang, X.; Xu, X.; et al. Natural variation in cytokinin maintenance improves salt tolerance in apple rootstocks. Plant Cell Environ. 2019, 42, 424–436. [Google Scholar] [CrossRef]
- Prerostova, S.; Dobrev, P.I.; Gaudinova, A.; Hosek, P.; Soudek, P.; Knirsch, V.; Vankova, R. Hormonal dynamics during salt stress responses of salt-sensitive Arabidopsis thaliana and salt-tolerant Thellungiella salsuginea. Plant Sci. 2017, 264, 188–198. [Google Scholar] [CrossRef]
- Joshi, R.; Sahoo, K.K.; Tripathi, A.K.; Kumar, R.; Gupta, B.K.; Pareek, A.; Singlapareek, S.L. Knockdown of an inflorescence meristem-specific cytokinin oxidase—OsCKX2 in rice reduces yield penalty under salinity stress condition. Plant Cell Environ. 2018, 41, 936–946. [Google Scholar] [CrossRef]
- Aremu, A.O.; Masondo, N.A.; Sunmonu, T.O.; Kulkarni, M.G.; Zatloukal, M.; Spichal, L.; Doležal, K.; Staden, J.V. A novel inhibitor of cytokinin degradation (INCYDE) influences the biochemical parameters and photosynthetic apparatus in NaCl-stressed tomato plants. Planta 2014, 240, 877–889. [Google Scholar] [CrossRef]
- Avalbaev, A.; Yuldashev, R.A.; Fedorova, K.A.; Somov, K.; Vysotskaya, L.B.; Allagulova, C.; Shakirova, F.M. Exogenous methyl jasmonate regulates cytokinin content by modulating cytokinin oxidase activity in wheat seedlings under salinity. J. Plant Physiol. 2016, 191, 101–110. [Google Scholar] [CrossRef]
- Ghanem, M.E.; Albacete, A.; Smigocki, A.C.; Frebort, I.; Pospisilova, H.; Martinezandujar, C.; Acosta, M.; Sanchezbravo, J.; Lutts, S.; Dodd, I.C.; et al. Root-synthesized cytokinins improve shoot growth and fruit yield in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 2011, 62, 125–140. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, W.; Chan, Z.; Wu, Y. Endogenous cytokinin overproduction modulates ROS homeostasis and decreases salt stress resistance in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1004. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Wang, J.; Xu, L.; Wang, A.; Huang, L.; Du, H.; Qiu, L.; Oelmuller, R. Drought stress responses in maize are diminished by piriformospora indica. Plant Signal. Behav. 2018, 13, e1414121. [Google Scholar] [CrossRef] [Green Version]
- Ghanem, M.E.; Albacete, A.; Martinezandujar, C.; Acosta, M.; Romeroaranda, M.R.; Dodd, I.C.; Lutts, S.; Perezalfocea, F. Hormonal changes during salinity-induced leaf senescence in tomato (Solanum lycopersicum L.). J. Exp. Bot. 2008, 59, 3039–3050. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, R.; Le, D.T.; Watanabe, Y.; Matsui, A.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.P. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS ONE 2012, 7, e32124. [Google Scholar] [CrossRef] [Green Version]
- Hyoung, S.; Cho, S.H.; Chung, J.H.; So, W.M.; Cui, M.H.; Shin, J.S. Cytokinin oxidase PpCKX1 plays regulatory roles in development and enhances dehydration and salt tolerance in Physcomitrella patens. Plant Cell Rep. 2019, 39, 419–430. [Google Scholar] [CrossRef]
- Li, S.; An, Y.; Hailati, S.; Zhang, J.; Cao, Y.; Liu, Y.; Geng, J.; Hu, T.; Yang, P. Overexpression of the cytokinin oxidase/dehydrogenase (CKX) from Medicago sativa enhanced salt stress tolerance of Arabidopsis. J. Plant Biol. 2019, 62, 374–386. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Onckelen, H.V.; Schmulling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [Green Version]
- Tran, L.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchishinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [Green Version]
- Mason, M.G.; Jha, D.; Salt, D.E.; Tester, M.; Hill, K.; Kieber, J.J.; Schaller, G.E. Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J. 2010, 64, 753–763. [Google Scholar] [CrossRef]
- Yin, W.; Xiao, Y.; Niu, M.; Meng, W.; Tong, H. ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell 2020, 32, 2292–2306. [Google Scholar] [CrossRef]
- Hallmark, H.T.; Rashotte, A.M. Review—Cytokinin response factors: Responding to more than cytokinin. Plant Sci. 2019, 289, 110251. [Google Scholar] [CrossRef]
- Keshishian, E.A. CRF2 and Its Role in Cytokinin Response and Abiotic Stress. Ph.D. Thesis, Auburn University, Auburn, AL, USA, 2018. Available online: https://etd.auburn.edu/handle/10415/6369 (accessed on 24 July 2018).
- Qin, L.; Wang, L.; Guo, Y.; Li, Y.; Ümüt, H.; Wang, Y. An ERF transcription factor from Tamarix hispida, ThCRF1, can adjust osmotic potential and reactive oxygen species scavenging capability to improve salt tolerance. Plant Int. J. Exp. Plant Biol. 2017, 154–166. [Google Scholar] [CrossRef]
- Kirkham, M.B.; Gardner, W.R.; Gerloff, G.C. Internal water status of kinetin-treated, salt-stressed plants. Plant Physiol. 1974, 53, 241–243. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, M.; Ashraf, M.; Jamil, A. Seed enhancement with cytokinins: Changes in growth and grain yield in salt stressed wheat plants. Plant Growth Regul. 2006, 50, 29–39. [Google Scholar] [CrossRef]
- Javid, M.G.; Sorooshzadeh, A.; Sanavy, S.A.; Allahdadi, I.; Moradi, F. Effects of the exogenous application of auxin and cytokinin on carbohydrate accumulation in grains of rice under salt stress. Plant Growth Regul. 2011, 65, 305–313. [Google Scholar] [CrossRef]
- Wu, X.; He, J.; Chen, J.; Yang, S.; Zha, D. Alleviation of exogenous 6-benzyladenine on two genotypes of eggplant (Solanum melongena Mill.) growth under salt stress. Protoplasma 2014, 251, 169–176. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, J.; Huang, B. Cytokinin-mitigation of salt-induced leaf senescence in perennial ryegrass involving the activation of antioxidant systems and ionic balance. Environ. Exp. Bot. 2016, 125, 1–11. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, F.; Zhou, J.; Chen, F.; Wang, B.; Xie, X. Phytochrome B control of total leaf area and stomatal density affects drought tolerance in rice. Plant Mol. Biol. 2012, 78, 289–300. [Google Scholar] [CrossRef]
- Zheng, Y.; Liao, C.; Zhao, S.; Wang, C.; Guo, Y. The glycosyltransferase QUA1 regulates chloroplast-associated calcium signaling during salt and drought stress in Arabidopsis. Plant Cell Physiol. 2017, 58, 329–341. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Ren, F.; Zhong, H.; Jiang, W.; Li, X. Identification and expression analysis of genes in response to high-salinity and drought stresses in Brassica napus. Acta Biochim. Biophys. Sin. 2010, 42, 154–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veselov, D.S.; Kudoyarova, G.R.; Kudryakova, N.V.; Kusnetsov, V.V. Role of cytokinins in stress resistance of plants. Russ. J. Plant. Physiol. 2017, 64, 15–27. [Google Scholar] [CrossRef]
- Werner, T.; Nehnevajova, E.; Kollmer, I.; Novak, O.; Strnad, M.; Kramer, U.; Schmulling, T. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 2010, 22, 3905–3920. [Google Scholar] [CrossRef] [Green Version]
- Calvo-Polanco, M.; Armada, E.; Zamarreno, A.M.; Garciamina, J.M.; Aroca, R. Local root ABA/cytokinin status and aquaporins regulate poplar responses to mild drought stress independently of the ectomycorrhizal fungus Laccaria bicolor. J. Exp. Bot. 2019, 70, 6437–6446. [Google Scholar] [CrossRef]
- Naidoo, G.; Naidoo, K.K. Drought stress effects on gas exchange and water relations of the invasive weed Chromolaena odorata. Flora 2018, 248, 1–9. [Google Scholar] [CrossRef]
- Rivero, R.M.; Shulaev, V.; Blumwald, E. Cytokinin-dependent photorespiration and the protection of photosynthesis during water deficit. Plant Physiol. 2009, 150, 1530–1540. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Liu, R.; Sui, N.; Shi, W.; Wang, L.; Tian, C.; Song, J. Changes in endogenous hormones and seed-coat phenolics during seed storage of two Suaeda salsa populations. Aust. J. Bot. 2016, 64, 325–332. [Google Scholar] [CrossRef]
- Shen, X.; Wang, Z.; Song, X.; Xu, J.; Jiang, C.; Zhao, Y.; Ma, C.; Zhang, H. Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 2014, 86, 303–317. [Google Scholar] [CrossRef]
- Pospisilova, H.; Jiskrova, E.; Vojta, P.; Mrizova, K.; Kokas, F.; Cudejkova, M.M.; Bergougnoux, V.; Plihal, O.; Klimesova, J.; Novak, O.; et al. Transgenic barley overexpressing a cytokinin dehydrogenase gene shows greater tolerance to drought stress. N. Biotechnol. 2016, 33, 692–705. [Google Scholar] [CrossRef]
- Ramireddy, E.; Hosseini, S.A.; Eggert, K.; Gillandt, S.; Gnad, H.; Wiren, N.V.; Schmulling, T. Root engineering in Barley: Increasing cytokinin degradation produces a larger root system, mineral enrichment in the shoot and improved drought tolerance. Plant Physiol. 2018, 177, 1078–1095. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kakimoto, T. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 2011, 23, 2169–2183. [Google Scholar] [CrossRef] [Green Version]
- Khandal, H.; Gupta, S.K.; Dwivedi, V.; Mandal, D.; Sharma, N.K.; Vishwakarma, N.K.; Pal, L.; Choudhary, M.; Francis, A.; Malakar, P.; et al. Root-specific expression of chickpea cytokinin oxidase/dehydrogenase 6 leads to enhanced root growth, drought tolerance and yield without compromising nodulation. Plant Biotechnol. J. 2020. [Google Scholar] [CrossRef] [Green Version]
- Mackova, H.; Hronkova, M.; Dobra, J.; Tureckova, V.; Novak, O.; Lubovska, Z.; Motyka, V.; Haisel, D.; Hajek, T.; Prasil, I.T.; et al. Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J. Exp. Bot. 2013, 64, 2805–2815. [Google Scholar] [CrossRef]
- Lubovska, Z.; Dobra, J.; Storchova, H.; Wilhelmova, N.; Vankova, R. Cytokinin oxidase/dehydrogenase overexpression modifies antioxidant defense against heat, drought and their combination in Nicotiana tabacum plants. J. Plant Physiol. 2014, 171, 1625–1633. [Google Scholar] [CrossRef]
- Kang, N.Y.; Cho, C.; Kim, N.Y.; Kim, J. Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis thaliana. J. Plant Physiol. 2012, 169, 1382–1391. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Ha, C.V.; Nishiyama, R.; Watanabe, Y.; Leyvagonzalez, M.A.; Fujita, Y.; Tran, U.T.; Li, W.; Tanaka, M.; Seki, M.; et al. Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. USA 2016, 113, 3090–3095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Rashotte, A.M. Expression patterns and regulation of SlCRF3 and SlCRF5 in response to cytokinin and abiotic stresses in tomato (Solanum lycopersicum). J. Plant Physiol. 2014, 171, 349–358. [Google Scholar] [CrossRef]
- Zaheer, M.S.; Raza, M.A.; Saleem, M.F.; Erinle, K.O.; Iqbal, R.; Ahmad, S. Effect of rhizobacteria and cytokinins application on wheat growth and yield under normal vs drought conditions. Commun. Soil Sci. Plant Anal. 2019, 50, 2521–2533. [Google Scholar] [CrossRef]
- Merewitz, E.; Du, H.; Yu, W.; Liu, Y.; Gianfagna, T.J.; Huang, B. Elevated cytokinin content in ipt transgenic creeping bentgrass promotes drought tolerance through regulating metabolite accumulation. J. Exp. Bot. 2012, 63, 1315–1328. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Burgess, P.; Zhang, X.; Huang, B. Enhancing cytokinin synthesis by overexpressing ipt alleviated drought inhibition of root growth through activating ROS-scavenging systems in Agrostis stolonifera. J. Exp. Bot. 2016, 67, 1979–1992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivero, R.M.; Gimeno, J.; Van Deynze, A.; Walia, H.; Blumwald, E. Enhanced cytokinin synthesis in tobacco plants expressing PSARK::IPT prevents the degradation of photosynthetic protein complexes during drought. Plant Cell Physiol. 2010, 51, 1929–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, H.; Gu, Q.; Zhang, J.; Sun, L.; Kuppu, S.; Zhang, Y.; Burow, M.D.; Payton, P.; Blumwald, E.; Zhang, H. Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiol. 2011, 52, 1904–1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuppu, S.; Mishra, N.; Hu, R.; Sun, L.; Zhu, X.; Shen, G.; Blumwald, E.; Payton, P.; Zhang, H. Water-deficit inducible expression of a cytokinin biosynthetic gene IPT improves drought tolerance in cotton. PLoS ONE 2013, 8, e64190. [Google Scholar] [CrossRef] [Green Version]
- Selvakumar, G.; Bindu, G.H.; Bhatt, R.M.; Upreti, K.K.; Paul, A.M.; Asha, A.; Shweta, K.; Sharma, M. Osmotolerant cytokinin producing microbes enhance tomato growth in deficit irrigation conditions. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2016, 88, 459–465. [Google Scholar] [CrossRef]
- Jorge, G.L.; Kisiala, A.; Morrison, E.N.; Aoki, M.; Nogueira, A.P.; Emery, R.J. Endosymbiotic Methylobacterium oryzae mitigates the impact of limited water availability in lentil (Lens culinaris Medik.) by increasing plant cytokinin levels. Environ. Exp. Bot. 2019, 162, 525–540. [Google Scholar] [CrossRef]
- Akter, N.; Islam, M.R.; Karim, M.A.; Hossain, T. Alleviation of drought stress in maize by exogenous application of gibberellic acid and cytokinin. J. Crop Sci. Biotechnol. 2014, 17, 41–48. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhang, M.; Meng, Z.; Wang, B.; Chen, M. Research Progress on the Roles of Cytokinin in Plant Response to Stress. Int. J. Mol. Sci. 2020, 21, 6574. https://doi.org/10.3390/ijms21186574
Liu Y, Zhang M, Meng Z, Wang B, Chen M. Research Progress on the Roles of Cytokinin in Plant Response to Stress. International Journal of Molecular Sciences. 2020; 21(18):6574. https://doi.org/10.3390/ijms21186574
Chicago/Turabian StyleLiu, Yun, Mingjing Zhang, Zhe Meng, Baoshan Wang, and Min Chen. 2020. "Research Progress on the Roles of Cytokinin in Plant Response to Stress" International Journal of Molecular Sciences 21, no. 18: 6574. https://doi.org/10.3390/ijms21186574