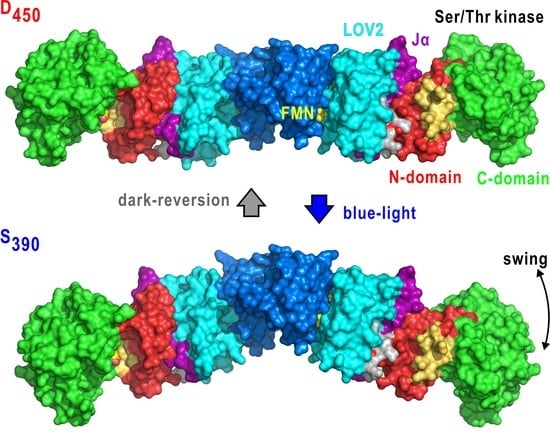
Domain Organization in Plant Blue-Light Receptor Phototropin2 of Arabidopsis thaliana Studied by Small-Angle X-ray Scattering
Abstract
:1. Introduction
2. Results
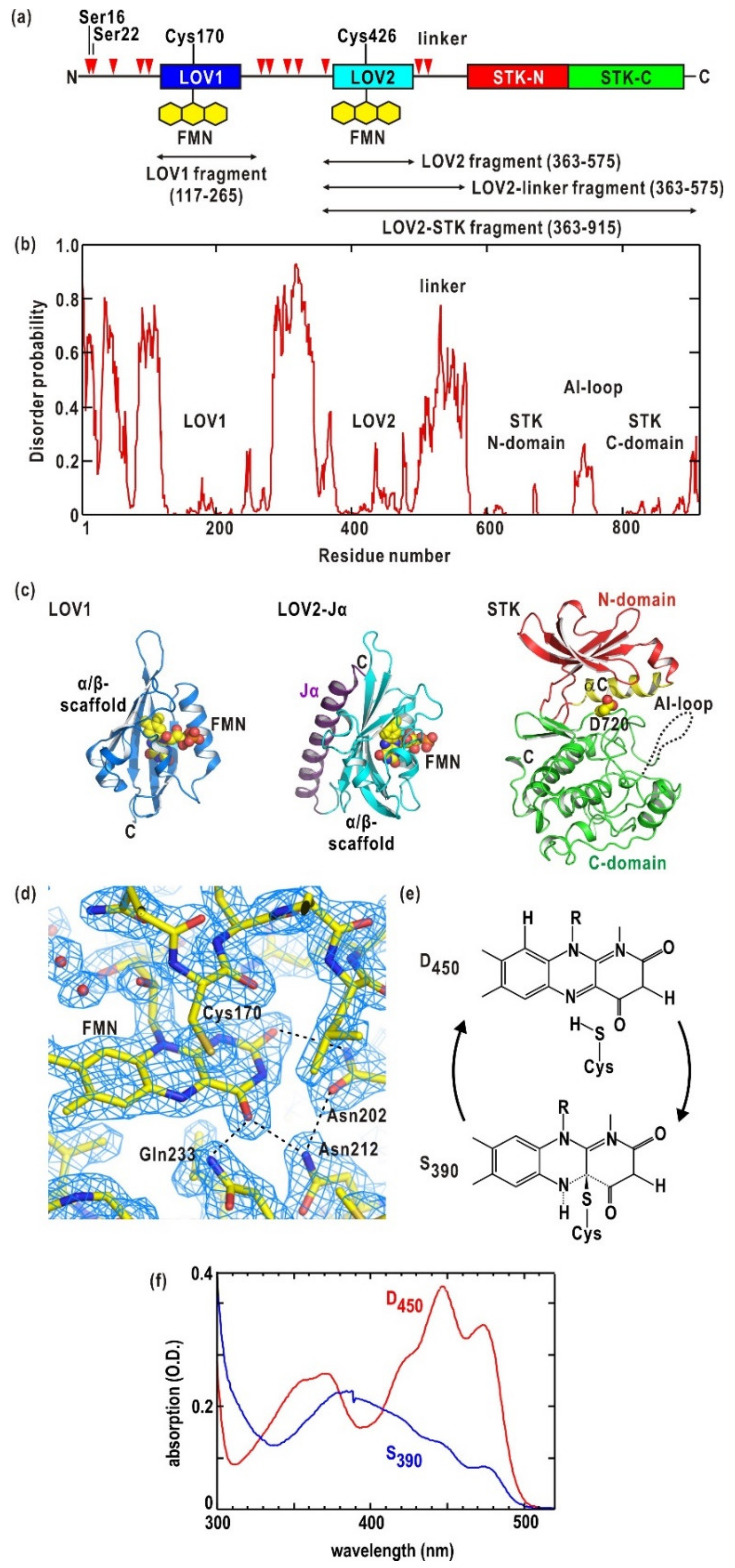
2.1. Photochemical Properties
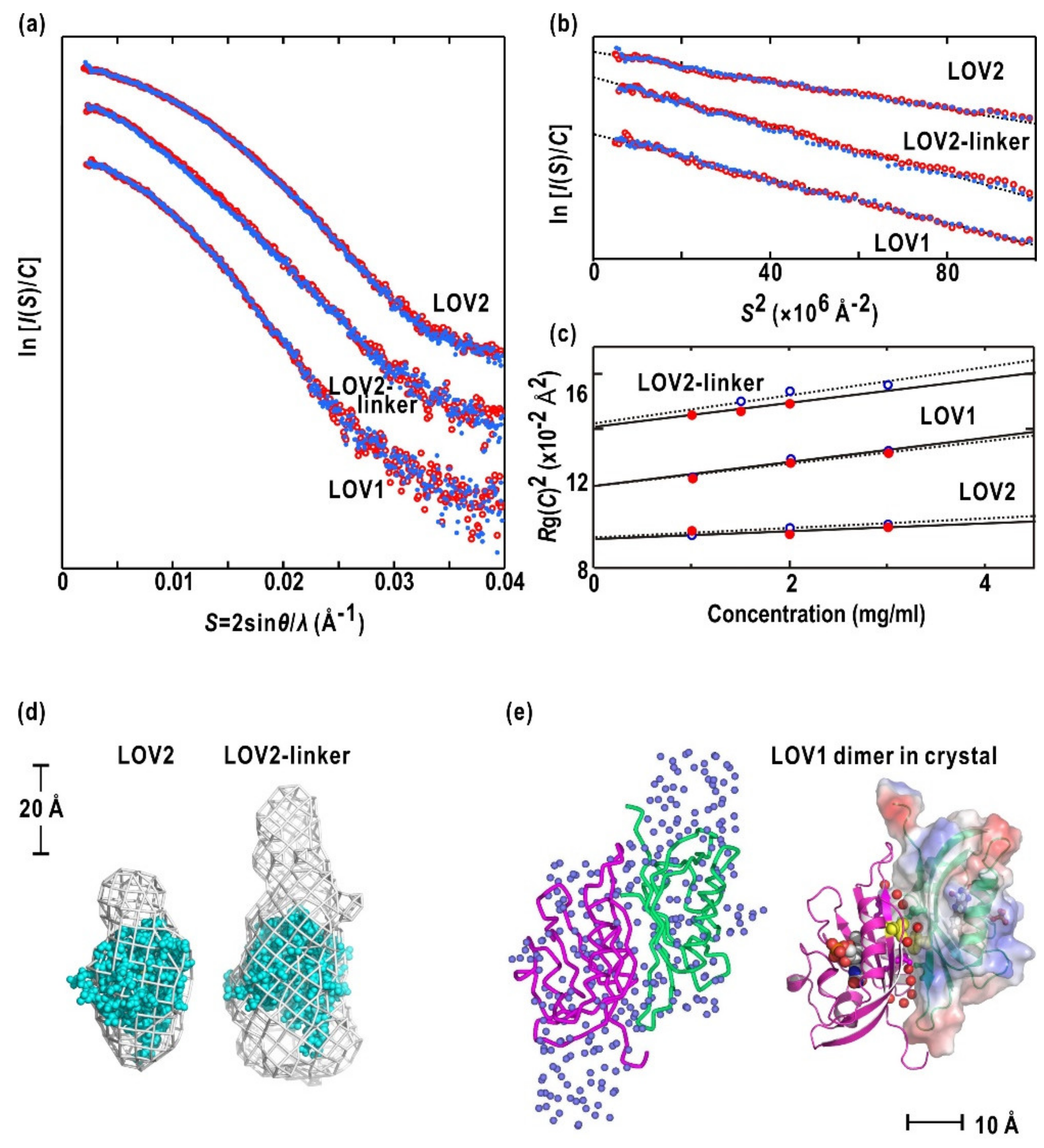
2.2. LOV1, LOV2, and LOV2-Linker Fragments
2.3. Asp720Asn-Mutated LOV2-STK
2.4. Asp720Asn-Mutated Full-Length Phot2
3. Discussion
3.1. Structures of LOV1, LOV2, and LOV2-Linker
3.2. Molecular Shape and BL-Induced Rearrangement of Domains in LOV2-STK
3.3. Organization and BL-Induced Rearrangement of Domains in Phot2
3.4. Future Structural Studies of Phot2
4. Materials and Methods
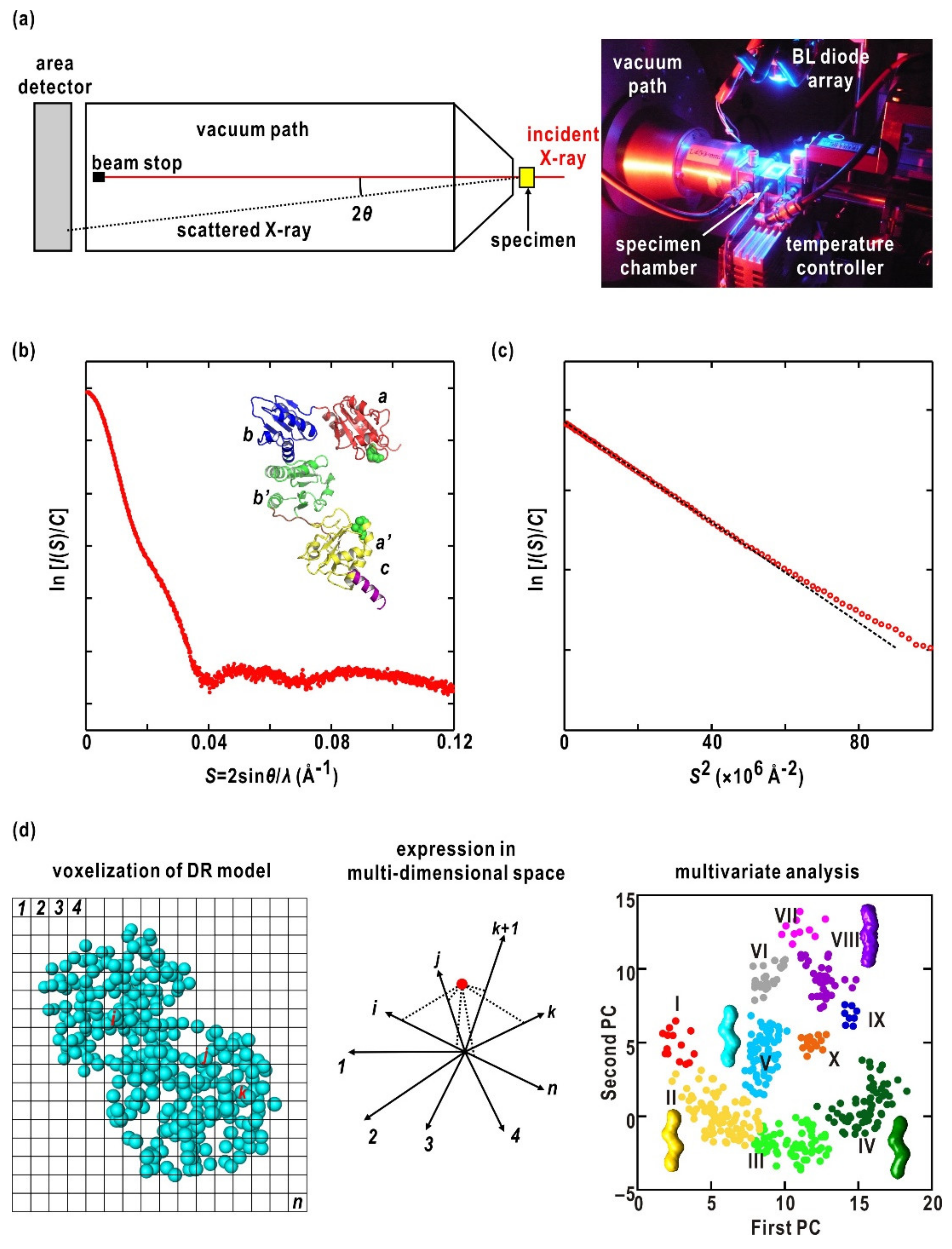
4.1. Specimen Preparation
4.2. SAXS Measurements
4.3. SAXS Analysis
4.4. Molecular Shape
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BL | Blue-light |
| D450 | Dark state |
| DR | Dummy residue (which approximates amino acid residues in ab initio modeling for small-angle X-ray scattering data) |
| FMN | Flavin mononucleotide |
| LOV | Light-oxygen-voltage sensing domain |
| PAS | Per-Arnt-Sim |
| phot | Phototropin |
| phot1 | Phototropin1 |
| phot2 | Phototropin2 |
| PKA | Protein kinase A |
| S390 | Blue-light activated adduct state |
| SAXS | Small-angle X-ray scattering |
| SH2 | Src homology 2 domain |
| STK | Serine-threonine kinase domain |
| TEM | Transmission electron microscopy |
| WHD | Winged-helix domain |
References
- Christie, J.M. Phototropin blue-light receptors. Annu. Rev. Plant Biol. 2007, 58, 21–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.W.; Rothfels, C.J.; Melkonian, M.; Villarreal, J.C.; Stevenson, D.W.; Graham, S.W.; Wong, G.K.; Mathews, S.; Pryer, K.M. The origin and evolution of phototropins. Front. Plant Sci. 2015, 6, 637. [Google Scholar] [CrossRef]
- Petroutsos, D.; Tokutsu, R.; Maruyama, S.; Flori, S.; Greiner, A.; Magneschi, L.; Cusant, L.; Kottke, T.; Mittag, M.; Hegemann, P.; et al. Blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 2016, 537, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Christie, J.M.; Reymond, P.; Powell, G.K.; Bernasconi, P.; Raibekas, A.A.; Liscum, E.; Briggs, W.R. Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 1998, 282, 1698–1701. [Google Scholar] [CrossRef]
- Sakai, T.; Kagawa, T.; Kasahara, M.; Swartz, T.E.; Christie, J.M.; Briggs, W.R.; Wada, M.; Okada, K. Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 2001, 98, 6969–6974. [Google Scholar] [CrossRef] [Green Version]
- Kagawa, T.; Sakai, T.; Suetsugu, N.; Oikawa, K.; Ishiguro, S.; Kato, T.; Tabata, S.; Okada, K.; Wada, M. Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 2001, 291, 2138–2141. [Google Scholar] [CrossRef] [PubMed]
- Jarillo, J.A.; Gabrys, H.; Capel, J.; Alonso, J.M.; Ecker, J.R.; Cashmore, A.R. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 2001, 410, 952–954. [Google Scholar] [CrossRef]
- Kinoshita, T.; Doi, M.; Suetsugu, N.; Kagawa, T.; Wada, M.; Shimazaki, K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 2001, 414, 656–660. [Google Scholar] [CrossRef]
- De Carbonnel, M.; Davis, P.; Roelfsema, M.R.; Inoue, S.; Schepens, I.; Lariguet, P.; Geisler, M.; Shimazaki, K.; Hangarter, R.; Fankhauser, C. The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol. 2010, 152, 1391–1405. [Google Scholar] [CrossRef] [Green Version]
- Aihara, Y.; Tabata, R.; Suzuki, T.; Shimazaki, K.; Nagatani, A. Molecular basis of the functional specificities of phototropin 1 and 2. Plant J. 2008, 56, 364–375. [Google Scholar] [CrossRef]
- Moglich, A.; Yang, X.; Ayers, R.A.; Moffat, K. Structure and function of plant photoreceptors. Annu. Rev. Plant Biol. 2010, 61, 21–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moglich, A.; Ayers, R.A.; Moffat, K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 2009, 17, 1282–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.Y.; Tseng, T.S.; Kaiserli, E.; Sullivan, S.; Christie, J.M.; Briggs, W.R. Physiological roles of the light, oxygen, or voltage domains of phototropin 1 and phototropin 2 in Arabidopsis. Plant Physiol. 2007, 143, 517–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, B.L.; Zhulin, I.B. PAS domains: Internal sensors of oxygen, redox potential and light. Microbiol. Mol. Biol. Rev. 1999, 63, 479–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crosson, S.; Moffat, K. Structure of a flavin-binding plant photoreceptor domain: Insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 2001, 98, 2995–3000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedorov, R.; Schlichting, I.; Hartmann, E.; Domratcheva, T.; Fuhrmann, M.; Hegemann, P. Crystal Structures and Molecular Mechanism of a Light-Induced Signaling Switch: The Phot-LOV1 Domain from Chlamydomonas reinhardtii. Biophys. J. 2003, 84, 2474–2482. [Google Scholar] [CrossRef] [Green Version]
- Halavaty, A.S.; Moffat, K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry 2007, 46, 14001–14009. [Google Scholar] [CrossRef]
- Nakasako, M.; Zikihara, K.; Matsuoka, D.; Katsura, H.; Tokutomi, S. Structural basis of the LOV1 dimerization of Arabidopsis phototropins 1 and 2. J. Mol. Biol. 2008, 381, 718–733. [Google Scholar] [CrossRef]
- Hitomi, K.; Christie, J.M.; Arvai, A.S.; Hartfield, K.A.; Pratt, A.J.; Tainer, J.A.; Getzoff, E.D. Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability. J. Biol. Chem. 2012, 287, 22295–22304. [Google Scholar]
- Halavaty, A.S.; Moffat, K. Coiled-coil dimerization of the LOV2 domain of the blue-light photoreceptor phototropin 1 from Arabidopsis thaliana. Acta Crystallogr. Sect. F 2013, 69, 1316–1321. [Google Scholar] [CrossRef] [Green Version]
- Salomon, M.; Christie, J.M.; Knieb, E.; Lempert, U.; Briggs, W.R. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 2000, 39, 9401–9410. [Google Scholar] [CrossRef] [PubMed]
- Swartz, T.E.; Corchnoy, S.B.; Christie, J.M.; Lewis, J.W.; Szundi, I.; Briggs, W.R.; Bogomolni, R.A. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 2001, 276, 36493–36500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crosson, S.; Moffat, K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002, 14, 1067–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasahara, M.; Swartz, T.E.; Olney, M.A.; Onodera, A.; Mochizuki, N.; Fukuzawa, H.; Asamizu, E.; Tabata, S.; Kanegae, H.; Takano, M.; et al. Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 2002, 129, 762–773. [Google Scholar] [CrossRef] [Green Version]
- Bogre, L.; Okresz, L.; Henriques, R.; Anthony, R.G. Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 2003, 8, 424–431. [Google Scholar] [CrossRef]
- Akamine, P.; Madhusudan Wu, J.; Xuong, N.H.; Ten Eyck, L.F.; Taylor, S.S. Dynamic Features of cAMP-dependent protein kinase revealed by apoenzyme crystal structure. J. Mol. Biol. 2003, 327, 159–171. [Google Scholar] [CrossRef]
- Christie, J.M.; Swartz, T.E.; Bogomolni, R.A.; Briggs, W.R. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002, 32, 205–219. [Google Scholar] [CrossRef]
- Sullivan, S.; Thomson, C.E.; Lamont, D.J.; Jones, M.A.; Christie, J.M. In vivo phosphorylation site mapping and functional characterization of Arabidopsis phototropin 1. Mol. Plant 2008, 1, 178–194. [Google Scholar] [CrossRef]
- Oide, M.; Okajima, K.; Kashojiya, S.; Takayama, Y.; Oroguchi, T.; Hikima, T.; Yamamoto, M.; Nakasako, M. Blue Light-excited Light-Oxygen-Voltage-sensing Domain 2 (LOV2) Triggers a Rearrangement of the Kinase Domain to Induce Phosphorylation Activity in Arabidopsis Phototropin1. J. Biol. Chem. 2016, 291, 19975–19984. [Google Scholar] [CrossRef] [Green Version]
- Okajima, K.; Aihara, Y.; Takayama, Y.; Nakajima, M.; Kashojiya, S.; Hikima, T.; Oroguchi, T.; Kobayashi, A.; Sekiguchi, Y.; Yamamoto, M.; et al. Light-induced conformational changes of LOV1 (light oxygen voltage-sensing domain 1) and LOV2 relative to the kinase domain and regulation of kinase activity in Chlamydomonas phototropin. J. Biol. Chem. 2014, 289, 413–422. [Google Scholar] [CrossRef] [Green Version]
- Harper, S.M.; Neil, L.C.; Gardner, K.H. Structural basis of a phototropin light switch. Science 2003, 301, 1541–1544. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.M.; Neil, L.C.; Day, I.J.; Hore, P.J.; Gardner, K.H. Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. J. Am. Chem. Soc. 2004, 126, 3390–3391. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Swartz, T.E.; Bogomolni, R.A.; Kliger, D.S. LOV story: The signaling state of the phot1 LOV2 photocycle involves chromophore-triggered protein structure relaxation, as probed by far-ultra-violet time-resolved optical rotatory dispersion spectroscopy. Biochemistry 2007, 46, 4619–4624. [Google Scholar] [CrossRef] [PubMed]
- Kashojiya, S.; Yoshihara, S.; Okajima, K.; Tokutomi, S. The linker between LOV2-Jalpha and STK plays an essential role in the kinase activation by blue light in Arabidopsis phototropin1, a plant blue light receptor. FEBS Lett. 2016, 590, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Christie, J.M.; Yang, H.; Richter, G.L.; Sullivan, S.; Thomson, C.E.; Lin, J.; Titapiwatanakun, B.; Ennis, M.; Kaiserli, E.; Lee, O.R.; et al. phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 2011, 9, e1001076. [Google Scholar] [CrossRef] [PubMed]
- Demarsy, E.; Schepens, I.; Okajima, K.; Hersch, M.; Bergmann, S.; Christie, J.; Shimazaki, K.; Tokutomi, S.; Fankhauser, C. Phytochrome Kinase Substrate 4 is phosphorylated by the phototropin 1 photoreceptor. EMBO J. 2012, 31, 3457–3467. [Google Scholar] [CrossRef]
- Takemiya, A.; Sugiyama, N.; Fujimoto, H.; Tsutsumi, T.; Yamauchi, S.; Hiyama, A.; Tada, Y.; Christie, J.M.; Shimazaki, K. Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nat. Commun. 2013, 4, 2094. [Google Scholar] [CrossRef] [Green Version]
- Nakasako, M.; Iwata, T.; Matsuoka, D.; Tokutomi, S. Light-induced structural changes of LOV domain-containing polypeptides from Arabidopsis phototropin 1 and 2 studied by small-angle X-ray scattering. Biochemistry 2004, 43, 14881–14890. [Google Scholar] [CrossRef]
- Nakasako, M.; Hirata, M.; Shimizu, N.; Hosokawa, S.; Matsuoka, D.; Oka, T.; Yamamoto, M.; Tokutomi, S. Crystallization and preliminary X-ray diffraction experiments of LOV1 domains of phototropin 1 and 2 from Arabidopsis thaliana. Acta Crystallogr. Sect. F 2008, 64, 617–621. [Google Scholar] [CrossRef] [Green Version]
- Oide, M.; Okajima, K.; Nakagami, H.; Kato, T.; Sekiguchi, Y.; Oroguchi, T.; Hikima, T.; Yamamoto, M.; Nakasako, M. Blue-light excited LOV1 and LOV2 domains cooperatively regulate the kinase activity of full-length phototropin2 from Arabidopsis. J. Biol. Chem. 2018, 293, 963–972. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Nakasako, M.; Okajima, K.; Iwata, A.; Kashojiya, S.; Matsui, Y.; Tokutomi, S. Light-induced movement of the LOV2 domain in an Asp720Asn mutant LOV2-kinase fragment of Arabidopsis phototropin 2. Biochemistry 2011, 50, 1174–118314. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Mokecular Graphyics System, DeLano Scientific, Palo Alto, CA, USA. 2020. Available online: http://www.pymol.org (accessed on 7 September 2020).
- Nakasako, M.; Maeno, A.; Kurimoto, E.; Harada, T.; Yamaguchi, Y.; Oka, T.; Takayama, Y.; Iwata, A.; Kato, K. Redox-dependent domain rearrangement of protein disulfide isomerase from a thermophilic fungus. Biochemistry 2010, 49, 6953–6962. [Google Scholar] [CrossRef] [PubMed]
- Svergun, D.I.; Petoukhov, M.V.; Koch, M.H.J. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001, 80, 2946–2953. [Google Scholar] [CrossRef] [Green Version]
- Pilz, I. Chapter 8: Proteins. In Small-Angle X-Ray Scattering; Glatter, O., Kratky, O., Eds.; Academic Press: New York, NY, USA, 1982. [Google Scholar]
- Svergun, D.I.; Koch, M.H.J.; Timmins, P.A.; May, R.P. Small angle X-ray and neutron scattering from solutions of biological macromolecules; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Oroguchi, T.; Hashimoto, H.; Shimizu, T.; Sato, M.; Ikeguchi, M. Intrinsic dynamics of restriction endonuclease EcoO109I studied by molecular dynamics simulations and X-ray scattering data analysis. Biophys. J. 2009, 96, 2808–2822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakasako, M.; Iwata, T.; Inoue, K.; Tokutomi, S. Light-induced global structural changes in phytochrome A regulating photomorphogenesis in plants. FEBS J. 2005, 272, 603–612. [Google Scholar] [CrossRef]
- Watanabe, Y.; Inoko, Y. Size-exclusion chromatography combined with small-angle X-ray scattering optics. J. Chromatogr. A 2009, 1216, 7461–7465. [Google Scholar] [CrossRef]
- Graewert, M.A.; Franke, D.; Jeffries, C.M.; Blanchet, C.E.; Ruskule, D.; Kuhle, K.; Flieger, A.; Schäfer, B.; Tartsch, B.; Meijers, R.; et al. Automated pipeline for purification, biophysical and x-ray analysis of biomacromolecular solutions. Sci. Rep. 2015, 5, 10734. [Google Scholar] [CrossRef]
- Oide, M.; Hikima, T.; Oroguchi, T.; Kato, T.; Yamaguchi, Y.; Yoshihara, S.; Yamamoto, M.; Nakasako, M.; Okajima, K. Molecular shape under far-red light and red light-induced association of Arabidopsis phytochrome B. FEBS J. 2020, 287, 1612–1625. [Google Scholar] [CrossRef]
- Chacón, P.; Morán, F.; Díaz, J.F.; Pantos, E.; Andreu, J.M. Low-resolution structures of proteins in solution retrieved from X-ray scattering with a genetic algorithm. Biophys. J. 1998, 74, 2760–2775. [Google Scholar] [CrossRef] [Green Version]
- Svergun, D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999, 76, 2879–2886. [Google Scholar] [CrossRef] [Green Version]
- Franke, D.; Svergun, D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009, 42, 342–346. [Google Scholar] [CrossRef] [Green Version]
- Jeffries, C.M.; Svergun, D.I. High-Throughput Studies of Protein Shapes and Interactions by Synchrotron Small-Angle X-Ray Scattering. Methods Mol. Biol. 2015, 1261, 277–301. [Google Scholar] [PubMed]
- Volkov, V.V.; Svergun, D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003, 36, 860–864. [Google Scholar] [CrossRef] [Green Version]
- Tuukkanen, A.T.; Kleywegt, G.J.; Svergun, D.I. Resolution of ab initio shapes determined from small-angle scattering. IUCrJ 2016, 3, 440–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oide, M.; Sekiguchi, Y.; Fukuda, A.; Okajima, K.; Oroguchi, T.; Nakasako, M. Classification of ab initio models of proteins restored from small-angle X-ray scattering. J. Synchrotron Rad. 2018, 25, 1379–1388. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Kofler, M.; Hantschel, O.; Gish, G.D.; Grebien, F.; Salah, E.; Neudecker, P.; Kay, L.E.; Turk, B.E.; Superti-Furga, G.; et al. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell 2008, 134, 793–803. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, A.; Takayama, Y.; Sakai, H.; Chong, K.T.; Takeuchi, S.; Nakagawa, A.; Nada, S.; Okada, M.; Tsukihara, T. Structure of the carboxyl-terminal Src kinase, Csk. J. Biol. Chem. 2002, 277, 14351–14354. [Google Scholar] [CrossRef] [Green Version]
- Nagar, B.; Hantschel, O.; Seeliger, M.; Davies, J.M.; Weis, W.I.; Superti-Furga, G.; Kuriyan, J. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol. Cell 2006, 21, 787–798. [Google Scholar] [CrossRef]
- LaRonde-LeBlanc, N.; Wlodawer, A. Crystal structure of A. Fulgidus Rio2 defines a new family of serine protein kinases. Structure 2004, 12, 1585–1594. [Google Scholar] [CrossRef] [Green Version]
- Okajima, K.; Kashojiya, S.; Tokutomi, S. Photosensitivity of kinase activation by blue light involves the lifetime of a cysteinyl-flavin adduct intermediate, S390, in the photoreaction cycle of the LOV2 domain in phototropin, a plant blue light receptor. J. Biol. Chem. 2012, 287, 40972–40981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakasako, M.; Matusoka, D.; Zikihara, K.; Tokutomi, S. Quaternary structure of LOV-domain containing polypeptides of Arabidopsis FKF1 protein. FEBS Lett. 2005, 579, 1067–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somers, D.E.; Schultz, T.F.; Milnamow, M.; Kay, S.A. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 2000, 101, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Schultz, T.F.; Kiyosue, T.; Yanovsky, M.; Wada, M.; Kay, S.A. A role of LKP2 in the circadian clock of Arabidopsis. Plant Cell 2001, 13, 2659–2670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Gross, S.; Roussos, C.; Meurer, S.; Muller-Esterl, W.; Papapetropoulos, A. Structural and functional characterization of the dimerization region of soluble guanylyl cyclise. J. Biol. Chem. 2004, 279, 24935–24943. [Google Scholar] [CrossRef] [Green Version]
- Zoltowski, B.D.; Schwerdtfeger, C.; Widom, J.; Loros, J.J.; Bilwes, A.M.; Dunlap, J.C.; Crane, B.R. Conformational switching in fungal light sensor Vivid. Science 2007, 316, 1054–1057. [Google Scholar] [CrossRef] [Green Version]
- Key, J.; Hefti, M.; Purcell, E.B.; Moffat, K. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: Signaling, dimerization, and mechanism. Biochemistry 2007, 46, 3614–3623. [Google Scholar] [CrossRef]
- Kashojiya, S.; Okajima, K.; Shimada, T.; Tokutomi, S. Essential role of the A’ alpha/AbetaA’alpha/Abeta gap in the N-terminal upstream of LOV2 for the blue light signaling from LOV2 to kinase in Arabidopsis photototropin1, a plant blue light receptor. PLoS ONE 2015, 10, e0124284. [Google Scholar] [CrossRef]
- Hantschel, O.; Nagar, B.; Guettler, S.; Kretzschmar, J.; Dorey, K.; Kuriyan, J.; Superti-Furga, G. A Myristoyl/Phosphotyrosine Switch Regulates c-Abl. Cell 2003, 112, 845–857. [Google Scholar] [CrossRef]
- LaRonde-LeBlanc, N.; Guszczynski, T.; Copeland, T.; Wlodawer, A. Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide–metal ion complexes. FEBS J. 2005, 272, 2800–2810. [Google Scholar] [CrossRef]
- Pfeifer, A.; Mathes, T.; Lu, Y.; Hegemann, P.; Kottke, T. Blue light induces global and localized conformational changes in the kinase domain of full-length phototropin. Biochemistry 2010, 49, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Tokutomi, S.; Matsuoka, D.; Zikihara, K. Molecular structure and regulation of phototropin kinase by blue light. Biochim. Biophys. Acta 2008, 1784, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Okajima, K. Molecular mechanism of phototropin light signaling. J. Plant Res. 2016, 129, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, N.; Kong, S.G.; Kasahara, M.; Wada, M. Both LOV1 and LOV2 domains of phototropin2 function as the photosensory domain for hypocotyl phototropic responses in Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 2013, 100, 60–69. [Google Scholar] [CrossRef]
- Oide, M.; Kato, T.; Oroguchi, T.; Nakasako, M. Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy. FEBS J. 2020, 287, 3472–3493. [Google Scholar] [CrossRef]
- Guinier, A.; Fournet, G. Small-Angle Scattering of X-Rays; Wiley: New York, NY, USA, 1955. [Google Scholar]
- Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Ibel, K.; Stuhrmann, H.B. Comparison of neutron and X-ray scattering of dilute myoglobin solutions. J. Mol. Biol. 1975, 93, 255–265. [Google Scholar] [CrossRef]
- Petoukhov, M.V.; Svergun, D.I. Ambiguity assessment of small-angle scattering curves from monodisperse systems. Acta Crystallogr. Sect. D 2015, 71, 1051–1058. [Google Scholar] [CrossRef]
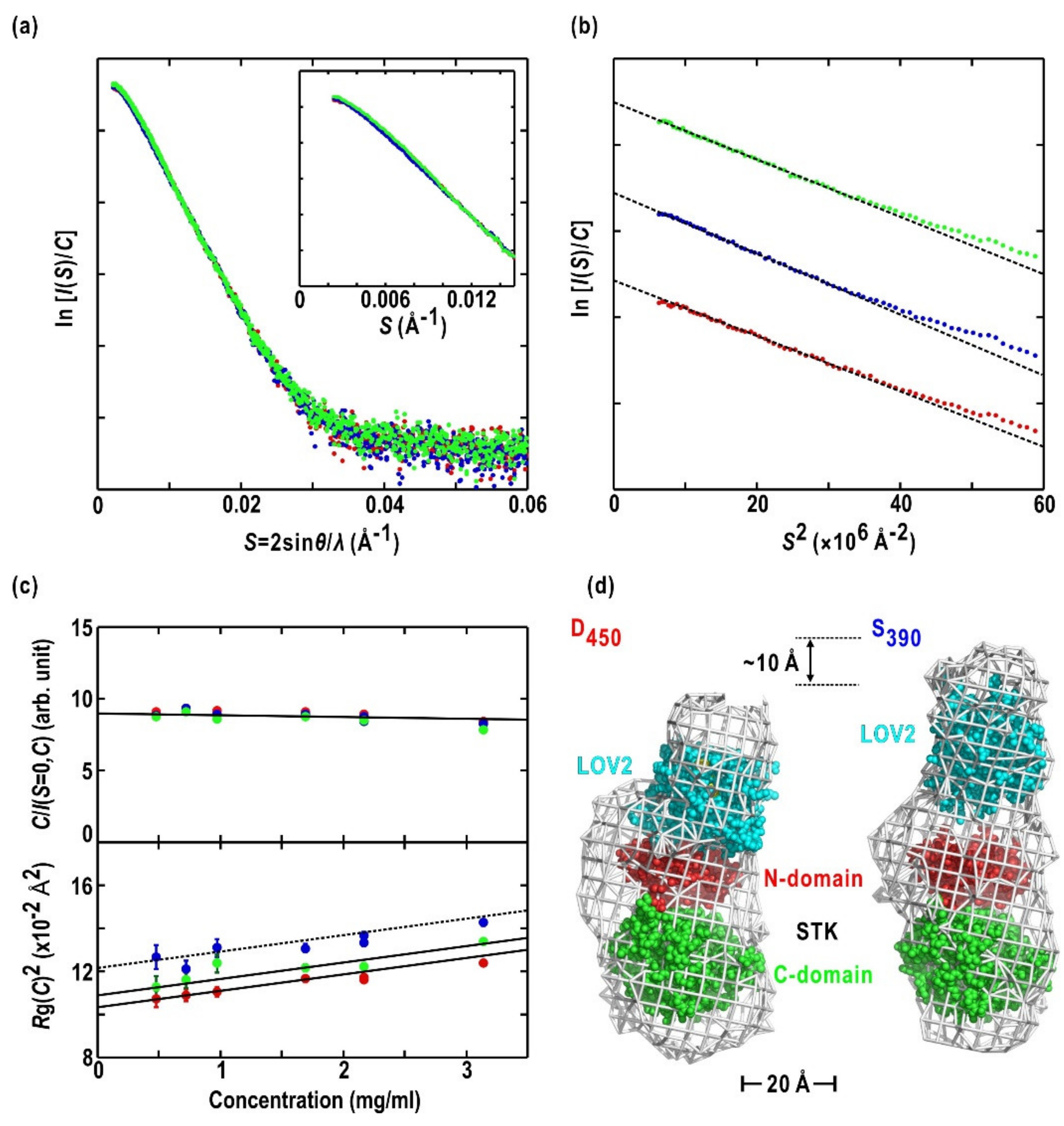

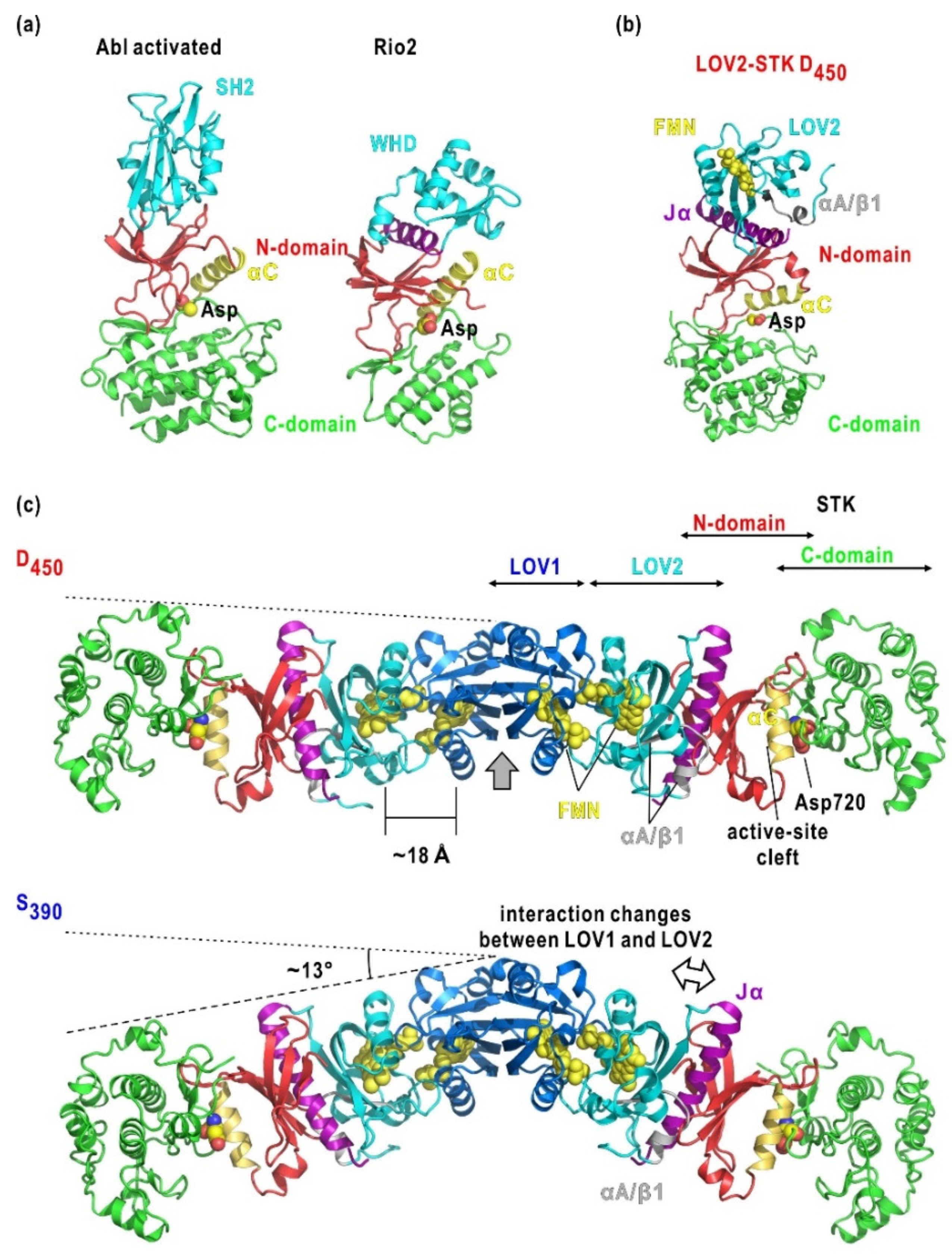
| Fragment (Mutation) | Residue Start/eEd | Oligomeric State | Rg Dark/Light (Å) | Dmax Dark/Light (Å) |
|---|---|---|---|---|
| LOV1 [18,41] | 117/265 | dimer | 22.2 ± 0.3/22.6 ± 0.3 | 94 ± 3/94 ± 3 |
| LOV2 [41] | 363/500 | monomer | 20.2 ± 0.2/20.1 ± 0.3 | 72 ± 2/72 ± 2 |
| LOV2-linker [41] | 363/575 | monomer | 24.3 ± 0.3/25.0 ± 0.4 | 107 ± 3/110 ± 3 |
| LOV2-STK [22] (Asp720Asn) | 363/915 | monomer | 32.4 ± 0.4/34.8 ± 0.7 | 133 ± 2/140 ± 2 |
| Full-length [43] (Asp720Asn) | 1/915 | dimer | 44.3 ± 1.4/43.7 ± 1.4 | 188 ± 2/186 ± 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakasako, M.; Oide, M.; Takayama, Y.; Oroguchi, T.; Okajima, K. Domain Organization in Plant Blue-Light Receptor Phototropin2 of Arabidopsis thaliana Studied by Small-Angle X-ray Scattering. Int. J. Mol. Sci. 2020, 21, 6638. https://doi.org/10.3390/ijms21186638
Nakasako M, Oide M, Takayama Y, Oroguchi T, Okajima K. Domain Organization in Plant Blue-Light Receptor Phototropin2 of Arabidopsis thaliana Studied by Small-Angle X-ray Scattering. International Journal of Molecular Sciences. 2020; 21(18):6638. https://doi.org/10.3390/ijms21186638
Chicago/Turabian StyleNakasako, Masayoshi, Mao Oide, Yuki Takayama, Tomotaka Oroguchi, and Koji Okajima. 2020. "Domain Organization in Plant Blue-Light Receptor Phototropin2 of Arabidopsis thaliana Studied by Small-Angle X-ray Scattering" International Journal of Molecular Sciences 21, no. 18: 6638. https://doi.org/10.3390/ijms21186638