Post-Translational Modifications of Cytochrome c in Cell Life and Disease
Abstract
:1. Introduction
2. The Pleiotropic Role of Cytochrome c in Cell Homeostasis and Diseases
3. Post-Translational Modifications of Cytochrome c: Regulation, Functionality, and Structural Changes
3.1. Phosphorylation
3.2. Nitration and Nitrosylation
3.3. Acetylation
3.4. Glycosylation and Glycations
3.5. Deamidations
3.6. Sulfoxidation
3.7. Homocysteinylation
3.8. Carbonylation
4. Conservation and Evolution of Cytochrome c Residues
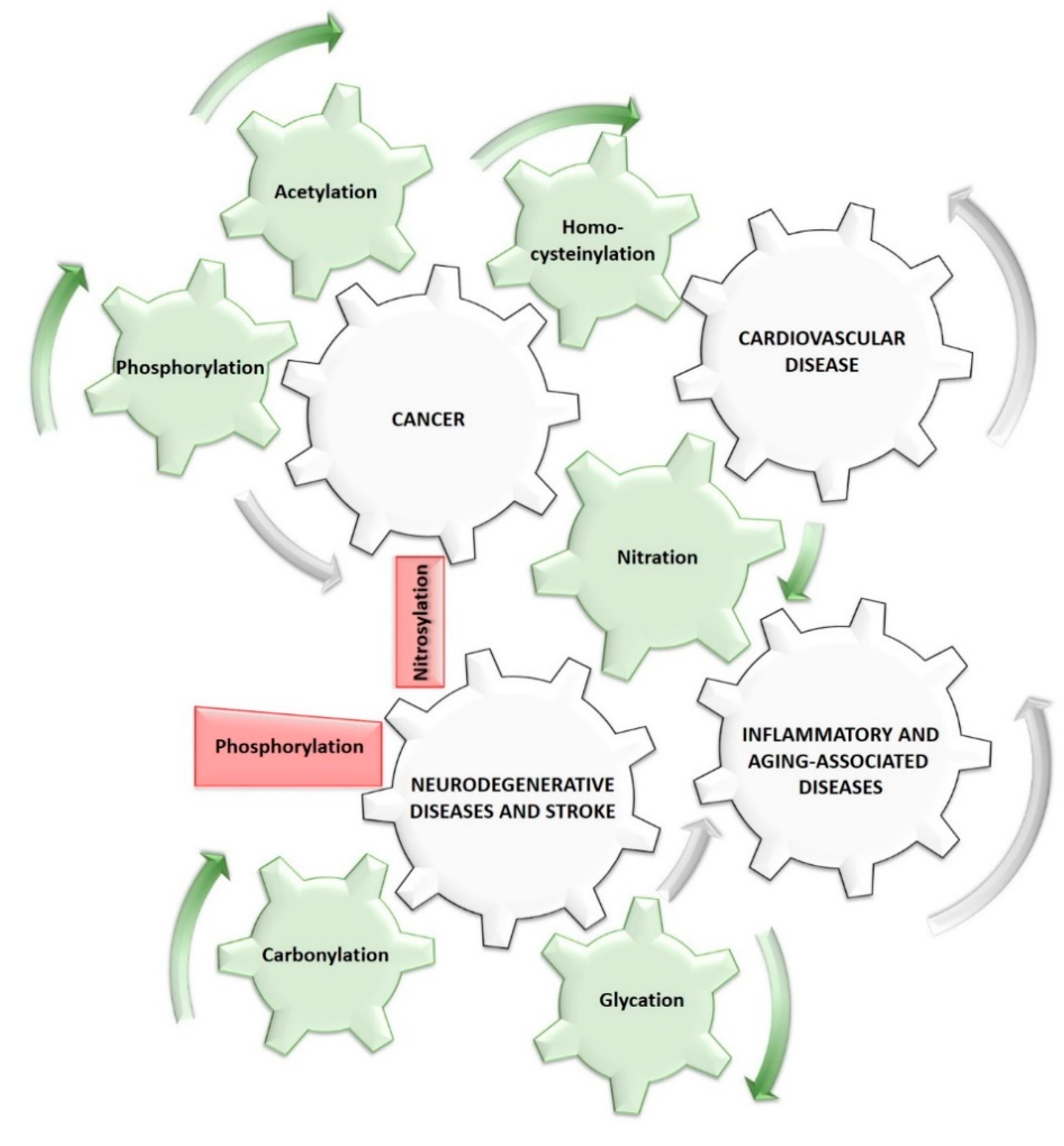
5. Clinical Relevance of Post-Translationally Modified Cytochrome c
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AcK Apaf-1 | N-ε-acetyl-L-Lys Apoptosis protease-activating factor-1 |
| CL | Cardiolipin |
| CIII | Complex III |
| CIV | Complex IV |
| Cytochrome c | Cc |
| Cytochrome c1 | Cc1 |
| CcO | Cytochrome c oxidase |
| ECT | Electron transport chain |
| IP3 | Inositol 1,4,5-trisphosphate |
| MAMs | Mitochondria-associated endoplasmic reticulum membranes |
| PCD | Programmed cell death |
| pCMF pI | p-carboxymethyl-L-phenylalanine Isoelectric point |
| PTMs | Post-translational modifications |
| RER | Rugose endoplasmic reticulum |
| RNS | Reactive nitrogen species |
| ROS | Reactive oxygen species |
| VDAC | Voltage-dependent anion channel |
References
- Pieczenik, S.R.; Neustadt, J. Mitochondrial dysfunction and molecular pathways of disease. Exp. Mol. Pathol. 2007, 83, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Moreno, I.; García-Heredia, J.M.; Díaz-Quintana, A.; De La Rosa, M.Á. Cytochrome c signalosome in mitochondria. Eur. Biophys. J. 2011, 40, 1301–1315. [Google Scholar] [CrossRef] [PubMed]
- Hüttemann, M.; Pecina, P.; Rainbolt, M.; Sanderson, T.H.; Kagan, V.E.; Samavati, L.; Doan, J.W.; Lee, I. The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: From respiration to apoptosis. Mitochondrion 2011, 11, 369–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Zhang, J.; Ying, W. Mitochondrial Electron Transport Chain Inhibition Suppresses LPS-Induced Inflammatory Responses via TREM1/STAT3 Pathway in BV2 Microglia. ROS generation and uncoupling (Review). Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef]
- Liu, X.; Kim, C.N.; Yang, J.; Jemmerson, R.; Wang, X. Induction of Apoptotic Program in Cell-Free Extracts: Requirement for dATP and Cytochrome c. Cell 1996, 86, 147–157. [Google Scholar] [CrossRef] [Green Version]
- D’Herde, K.; De Prest, B.; Mussche, S.; Schotte, P.; Beyaert, R.; Van Coster, R.; Roels, F. Ultrastructural localization of cytochrome c in apoptosis demonstrates mitochondrial heterogeneity. Cell Death Differ. 2000, 7, 331–337. [Google Scholar] [CrossRef]
- González-Arzola, K.; Velázquez-Cruz, A.; Guerra-Castellano, A.; Casado-Combreras, M.A.; Pérez-Mejías, G.; Quintana, A.D.; Díaz-Moreno, I.; De La Rosa, M.A. New moonlighting functions of mitochondrial cytochrome c in the cytoplasm and nucleus. FEBS Lett. 2019, 593, 3101–3119. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Beltrán, B.; Díaz-Quintana, A.; González-Arzola, K.; Velazquez-Campoy, A.; De La Rosa, M.Á.; Díaz-Moreno, I. Cytochrome c1 exhibits two binding sites for cytochrome c in plants. Biochim. et Biophys. Acta (BBA)-Gen. Subj. 2014, 1837, 1717–1729. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Beltrán, B.; Díaz-Moreno, I.; González-Arzola, K.; Castellano, A.G.; Velazquez-Campoy, A.; De La Rosa, M.Á.; Díaz-Quintana, A. Respiratory complexes III and IV can each bind two molecules of cytochromecat low ionic strength. FEBS Lett. 2015, 589, 476–483. [Google Scholar] [CrossRef] [Green Version]
- Lagunas, A.; Castellano, A.G.; Nin-Hill, A.; Díaz-Moreno, I.; De La Rosa, M.A.; Samitier, J.; Rovira, C.; Gorostiza, P. Long distance electron transfer through the aqueous solution between redox partner proteins. Nat. Commun. 2018, 9, 5157. [Google Scholar] [CrossRef]
- Pérez-Mejías, G.; Olloqui-Sariego, J.L.; Guerra-Castellano, A.; Díaz-Quintana, A.; Calvente, J.J.; Andreu, R.; De La Rosa, M.A.; Díaz-Moreno, I. Physical contact between cytochrome c1 and cytochrome c increases the driving force for electron transfer. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2020, 148277. [Google Scholar] [CrossRef]
- Pasdois, P.; Parker, J.E.; Griffiths, E.J.; Halestrap, A.P. The role of oxidized cytochrome c in regulating mitochondrial reactive oxygen species production and its perturbation in ischaemia. Biochem. J. 2011, 436, 493–505. [Google Scholar] [CrossRef] [Green Version]
- Allen, S.; Balabanidou, V.; Sideris, D.P.; Lisowsky, T.; Tokatlidis, K. Erv1 Mediates the Mia40-dependent Protein Import Pathway and Provides a Functional Link to the Respiratory Chain by Shuttling Electrons to Cytochrome c. J. Mol. Biol. 2005, 353, 937–944. [Google Scholar] [CrossRef]
- Dabir, D.V.; Leverich, E.P.; Kim, S.-K.; Tsai, F.D.; Hirasawa, M.; Knaff, D.B.; Koehler, C.M. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007, 26, 4801–4811. [Google Scholar] [CrossRef] [Green Version]
- Soltys, B.J.; Andrews, D.W.; Jemmerson, R.; Gupta, R.S. Cytochrome-c localizes in secretory granules in pancreas and anterior pituitary. Cell Biol. Int. 2001, 25, 331–338. [Google Scholar] [CrossRef]
- Ow, Y.-L.P.; Green, D.R.; Hao, Z.; Mak, T.W. Cytochrome c: Functions beyond respiration. Nat. Rev. Mol. Cell Biol. 2008, 9, 532–542. [Google Scholar] [CrossRef]
- Gilchrist, D.G. PROGRAMMED CELL DEATH IN PLANT DISEASE: The Purpose and Promise of Cellular Suicide. Annu. Rev. Phytopathol. 1998, 36, 393–414. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, Y.; Steller, H. Programmed cell death in animal development and disease. Cell 2011, 147, 742–758. [Google Scholar] [CrossRef] [Green Version]
- Dorstyn, L.; Read, S.; Cakouros, D.; Huh, J.R.; Hay, B.A.; Kumar, S. The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J. Cell Biol. 2002, 156, 1089–1098. [Google Scholar] [CrossRef] [Green Version]
- Garrido, C.; Galluzzi, L.; Brunet, M.; E Puig, P.; Didelot, C.; Kroemer, G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006, 13, 1423–1433. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Fábregas, J.; Díaz-Moreno, I.; González-Arzola, K.; Janocha, S.; Navarro, J.A.; Hervás, M.; Bernhardt, R.; Díaz-Quintana, A.; De La Rosa, M.Á. NewArabidopsis thalianaCytochromecPartners: A Look Into the Elusive Role of Cytochromecin Programmed Cell Death in Plants. Mol. Cell. Proteom. 2013, 12, 3666–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Fábregas, J.; Díaz-Moreno, I.; González-Arzola, K.; Díaz-Quintana, A.; A De La Rosa, M. A common signalosome for programmed cell death in humans and plants. Cell Death Dis. 2014, 5, e1314. [Google Scholar] [CrossRef] [PubMed]
- González-Arzola, K.; Díaz-Moreno, I.; Cano-González, A.; Díaz-Quintana, A.; Velázquez-Campoy, A.; Moreno-Beltrán, B.; López-Rivas, A.; De la Rosa, M.A. Structural basis for inhibition of the histone chaperone activity of SET/TAF-Iβ by cytochrome c. Proc. Natl. Acad. Sci. USA 2015, 112, 9908–9913. [Google Scholar]
- González-Arzola, K.; Díaz-Quintana, A.; Rivero-Rodríguez, F.; Velázquez-Campoy, A.; De La Rosa, M.A.; Díaz-Moreno, I. Histone chaperone activity of Arabidopsis thaliana NRP1 is blocked by cytochrome c. Nucleic Acids Res. 2017, 45, 2150–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elena-Real, C.A.; Díaz-Quintana, A.; González-Arzola, K.; Velázquez-Campoy, A.; Orzáez, M.; López-Rivas, A.; Gil-Caballero, S.; De la Rosa, M.Á.; Díaz-Moreno, I. Cytochrome c speeds up caspase cascade activation by blocking 14-3-3ε-dependent Apaf-1 inhibition. Cell Death Dis. 2018, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Quintana, A.; Pérez-Mejías, G.; Guerra-Castellano, A.; De La Rosa, M.A.; Díaz-Moreno, I. Wheel and Deal in the Mitochondrial Inner Membranes: The Tale of Cytochrome c and Cardiolipin. Oxidative Med. Cell. Longev. 2020, 2020, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everse, J.; Liu, C.-J.J.; Coates, P.W. Physical and catalytic properties of a peroxidase derived from cytochrome c. Biochim. et Biophys. Acta (BBA)-Mol. Basis Dis. 2011, 1812, 1138–1145. [Google Scholar] [CrossRef]
- Ascenzi, P.; Coletta, M.; Wilson, M.T.; Fiorucci, L.; Marino, M.; Polticelli, F.; Sinibaldi, F.; Santucci, R. Cardiolipin-cytochromeccomplex: Switching cytochromecfrom an electron-transfer shuttle to a myoglobin- and a peroxidase-like heme-protein. IUBMB Life 2015, 67, 98–109. [Google Scholar] [CrossRef]
- Jemmerson, R.; Liu, J.; Hausauer, D.; Lam, K.-P.; Mondino, A.A.; Nelson, R.D. A Conformational Change in Cytochromecof Apoptotic and Necrotic Cells Is Detected by Monoclonal Antibody Binding and Mimicked by Association of the Native Antigen with Synthetic Phospholipid Vesicles†. Biochemistry 1999, 38, 3599–3609. [Google Scholar] [CrossRef]
- Kagan, V.E.; Borisenko, G.G.; Tyurina, Y.Y.; Tyurin, V.A.; Jiang, J.; Potapovich, A.I.; Kini, V.; Amoscato, A.A.; Fujii, Y. Oxidative lipidomics of apoptosis: Redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine. Free. Radic. Biol. Med. 2004, 37, 1963–1985. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Zalk, R.; Gincel, D.; Vardi, N. Subcellular localization of VDAC in mitochondria and ER in the cerebellum. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2004, 1657, 105–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, S.Z.A.; Zhao, D.; Khan, S.H.; Yang, L. Regulatory Mechanisms of Endoplasmic Reticulum Resident IP3 Receptors. J. Mol. Neurosci. 2015, 56, 938–948. [Google Scholar] [CrossRef] [PubMed]
- Camara, A.K.S.; Zhou, Y.; Wen, P.-C.; Tajkhorshid, E.; Kwok, W.-M. Mitochondrial VDAC1: A Key Gatekeeper as Potential Therapeutic Target. Front. Physiol. 2017, 8, 460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patergnani, S.; Suski, J.M.; Agnoletto, C.; Bononi, A.; Bonora, M.; De Marchi, E.; Giorgi, C.; Retta, S.F.; Missiroli, S.; Poletti, F.; et al. Calcium signaling around Mitochondria Associated Membranes (MAMs). Cell Commun. Signal. 2011, 9, 19. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Ma, Q.; Wang, Q.; Sun, X.; Zhang, Z.; Ji, L.; Huang, Q. The relationship between mitochondria Ca2+ intake mediated by mitochondria-associated endoplasmic reticulum membranes and tumor genesis. J. Cell Signal 2019, 4, 199. [Google Scholar]
- Vance, J.E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990, 265, 7248–7256. [Google Scholar]
- Theurey, P.; Rieusset, J. Mitochondria-Associated Membranes Response to Nutrient Availability and Role in Metabolic Diseases. Trends Endocrinol. Metab. 2017, 28, 32–45. [Google Scholar] [CrossRef] [Green Version]
- Boehning, D.; Patterson, R.L.; Sedaghat, L.; Glebova, N.O.; Kurosaki, T.; Snyder, S.H. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat. Cell Biol. 2003, 5, 1051–1061. [Google Scholar] [CrossRef]
- Zalk, R.; Israelson, A.; Garty, E.S.; Azoulay-Zohar, H.; Shoshan-Barmatz, V. Oligomeric states of the voltage-dependent anion channel and cytochrome c release from mitochondria. Biochem. J. 2005, 386, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Szabadkai, G.; Bianchi, K.; Várnai, P.; De Stefani, D.; Wieckowski, M.R.; Cavagna, D.; Nagy, A.I.; Balla, T.; Rizzuto, R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 2006, 175, 901–911. [Google Scholar] [CrossRef] [Green Version]
- Shoshan-Barmatz, V.; Keinan, N.; Abu-Hamad, S.; Tyomkin, D.; Aram, L. Apoptosis is regulated by the VDAC1 N-terminal region and by VDAC oligomerization: Release of cytochrome c, AIF and Smac/Diablo. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2010, 1797, 1281–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoshan-Barmatz, V.; Maldonado, E.N.; Krelin, Y. VDAC1 at the crossroads of cell metabolism, apoptosis and cell stress. Cell Stress 2017, 1, 11–36. [Google Scholar] [CrossRef] [PubMed]
- Ameisen, J.C. On the origin, evolution, and nature of programmed cell death: A timeline of four billion years. Cell Death Differ. 2002, 9, 367–393. [Google Scholar] [CrossRef] [PubMed]
- Paine, M.J.I.; Garner, A.P.; Powell, D.; Sibbald, J.; Sales, M.; Pratt, N.; Smith, T.; Tew, D.G.; Wolf, C.R. Cloning and characterization of a novel human dual flavin reductase. J. Biol. Chem. 2000, 275, 1471–1478. [Google Scholar] [CrossRef] [Green Version]
- Walsh, C. Posttranslational Modification of Proteins: Expanding Nature’s Inventory; Roberts and Company Publishers: Greenwood Village, CO, USA, 2006; p. 490. [Google Scholar]
- Karve, T.M.; Cheema, A.K. Small Changes Huge Impact: The Role of Protein Posttranslational Modifications in Cellular Homeostasis and Disease. J. Amino Acids 2011, 2011, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.M. Post-Translational Modifications of Protein Backbones: Unique Functions, Mechanisms, and Challenges. Biochemistry 2018, 57, 177–185. [Google Scholar] [CrossRef]
- Kalpage, H.A.; Bazylianska, V.; Recanati, M.A.; Fite, A.; Liu, J.; Wan, J.; Mantena, N.; Malek, M.H.; Podgorski, I.; Heath, E.I.; et al. Tissue-specific regulation of cytochrome c by post-translational modifications: Respiration, the mitochondrial membrane potential, ROS, and apoptosis. FASEB J. 2018, 33, 1540–1553. [Google Scholar] [CrossRef]
- Kalpage, H.A.; Wan, J.; Morse, P.T.; Zurek, M.P.; Turner, A.A.; Khobeir, A.; Yazdi, N.; Hakim, L.; Liu, J.; Vaishnav, A.; et al. Cytochrome c phosphorylation: Control of mitochondrial electron transport chain flux and apoptosis. Int. J. Biochem. Cell Biol. 2020, 121, 105704. [Google Scholar] [CrossRef]
- Lee, I.; Salomon, A.R.; Yu, K.; Doan, J.W.; I Grossman, L.; Hüttemann, M. New Prospects for an Old Enzyme: Mammalian CytochromecIs Tyrosine-Phosphorylated in Vivo. Biochemistry 2006, 45, 9121–9128. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Lee, I.; Salomon, A.R.; Yu, K.; Hüttemann, M. Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochim. Biophys. Acta Bioenerg. 2008, 1777, 1066–1071. [Google Scholar] [CrossRef] [Green Version]
- Sanderson, T.H.; Mahapatra, G.; Pecina, P.; Ji, Q.; Yu, K.; Sinkler, C.; Varughese, A.; Kumar, R.; Bukowski, M.J.; Tousignant, R.N.; et al. Cytochrome c Is Tyrosine 97 Phosphorylated by Neuroprotective Insulin Treatment. PLOS ONE 2013, 8, e78627. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, G.; Varughese, A.; Ji, Q.; Lee, I.; Liu, J.; Vaishnav, A.; Sinkler, C.; Kapralov, A.A.; Moraes, C.T.; Sanderson, T.H.; et al. Phosphorylation of cytochrome c threonine 28 regulates electron transport chain activity in kidney: Implications for amp kinase. J. Biol. Chem. 2017, 292, 64–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, J.; Kalpage, H.A.; Vaishnav, A.; Liu, J.; Lee, I.; Mahapatra, G.; Turner, A.A.; Zurek, M.P.; Ji, Q.; Moraes, C.T.; et al. Regulation of Respiration and Apoptosis by Cytochrome c Threonine 58 Phosphorylation. Sci. Rep. 2019, 9, 15–16. [Google Scholar] [CrossRef] [PubMed]
- Kalpage, H.A.; Vaishnav, A.; Liu, J.; Varughese, A.; Wan, J.; Turner, A.A.; Ji, Q.; Zurek, M.P.; Kapralov, A.A.; Kagan, V.E.; et al. Serine-47 phosphorylation of cytochrome c in the mammalian brain regulates cytochrome c oxidase and caspase-3 activity. FASEB J. 2019, 33, 13503–13514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.R.; Magliery, T.J.; Pastrnak, M.; Schultz, P.G. Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. Proc. Natl. Acad. Sci. USA 1997, 94, 10092–10097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewals, K.; Ovaa, H. Unnatural amino acid incorporation in E. coli: Current and future applications in the design of therapeutic proteins. Front. Chem. 2014, 2, 15. [Google Scholar] [CrossRef]
- Leisle, L.; I Valiyaveetil, F.; Mehl, R.A.; Ahern, C.A. Incorporation of Non-Canonical Amino Acids. Adv. Exp. Med. Biol. 2015, 869, 119–151. [Google Scholar] [CrossRef]
- Meineke, B.; Heimgärtner, J.; Eirich, J.; Landreh, M.; Elsässer, S.J. Site-Specific Incorporation of Two ncAAs for Two-Color Bioorthogonal Labeling and Crosslinking of Proteins on Live Mammalian Cells. Cell Rep. 2020, 31, 107811. [Google Scholar] [CrossRef]
- Kawahata, N.; Yang, M.G.; Luke, G.P.; Shakespeare, W.C.; Sundaramoorthi, R.; Wang, Y.; Johnson, D.; Merry, T.; Violette, S.; Guan, W.; et al. A novel phosphotyrosine mimetic 4′-carboxymethyloxy-3′-phosphonophenylalanine (cpp): Exploitation in the design of nonpeptide inhibitors of pp60Src SH2 domain. Bioorganic Med. Chem. Lett. 2001, 11, 2319–2323. [Google Scholar] [CrossRef]
- A Erlanson, D.; McDowell, R.S.; He, M.M.; Randal, M.; Simmons, R.L.; Kung, J.; Waight, A.; Hansen, S.K. Discovery of a New Phosphotyrosine Mimetic for PTP1B Using Breakaway Tethering. J. Am. Chem. Soc. 2003, 125, 5602–5603. [Google Scholar] [CrossRef]
- Rothman, D.M.; Petersson, E.J.; Vázquez, M.E.; Brandt, G.S.; Dougherty, D.A.; Imperiali, B. Caged Phosphoproteins. J. Am. Chem. Soc. 2005, 127, 846–847. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Schultz, P.G. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat. Biotechnol. 2006, 24, 1436–1440. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Supekova, L.; Schultz, P.G. A Genetically Encoded Metabolically Stable Analogue of Phosphotyrosine in Escherichia coli. ACS Chem. Biol. 2007, 2, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Serwa, R.; Wilkening, I.; Del Signore, G.; Mühlberg, M.; Claussnitzer, I.; Weise, C.; Gerrits, M.; Hackenberger, C.P.R. Chemoselective Staudinger-Phosphite Reaction of Azides for the Phosphorylation of Proteins. Angew. Chem. Int. Ed. 2009, 48, 8234–8239. [Google Scholar] [CrossRef]
- Fan, C.; Ip, K.; Söll, D. Expanding the genetic code of Escherichia coli with phosphotyrosine. FEBS Lett. 2016, 590, 3040–3047. [Google Scholar] [CrossRef] [Green Version]
- Hoppmann, C.; Wong, A.; Yang, B.; Li, S.; Hunter, T.; Shokat, K.M.; Wang, L. Site-specific incorporation of phosphotyrosine using an expanded genetic code. Nat. Chem. Biol. 2017, 13, 842–844. [Google Scholar] [CrossRef]
- Luo, X.; Fu, G.; Wang, R.E.; Zhu, X.; Zambaldo, C.; Liu, R.; Liu, T.; Lyu, X.; Du, J.; Xuan, W.; et al. Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria. Nat. Chem. Biol. 2017, 13, 845–849. [Google Scholar] [CrossRef] [Green Version]
- Chin, J.W.; Santoro, S.W.; Martin, A.B.; King, D.S.; Wang, L.; Schultz, P.G. Addition of p-azido-L-phenylaianine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 2002, 124, 9026–9027. [Google Scholar] [CrossRef]
- Zhang, M.S.; Brunner, S.F.; Huguenin-Dezot, N.; Liang, A.D.; Schmied, W.H.; Rogerson, D.T.; Chin, J.W. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing. Nat. Methods 2017, 14, 729–736. [Google Scholar] [CrossRef]
- Rogerson, D.T.; Sachdeva, A.; Wang, K.; Haq, T.; Kazlauskaite, A.; Hancock, S.M.; Huguenin-Dezot, N.; Muqit, M.M.K.; Fry, A.M.; Bayliss, R.; et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat. Chem. Biol. 2015, 11, 496–503. [Google Scholar] [CrossRef] [Green Version]
- Sokolovsky, M.; Riordan, J.F.; Vallee, B.L. Conversion of 3-nitrotyrosine to 3-aminotyrosine in peptides and proteins. Biochem. Biophys. Res. Commun. 1967, 27, 20–25. [Google Scholar] [CrossRef]
- Neumann, H.; Peak-Chew, S.Y.; Chin, J.W. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008, 4, 232–234. [Google Scholar] [CrossRef] [PubMed]
- Hancock, S.M.; Uprety, R.; Deiters, A.; Chin, J.W. Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair. J. Am. Chem. Soc. 2010, 132, 14819–14824. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wan, W.; Russell, W.K.; Pai, P.-J.; Wang, Z.; Russell, D.H.; Liu, W.R. Genetic incorporation of an aliphatic keto-containing amino acid into proteins for their site-specific modifications. Bioorganic Med. Chem. Lett. 2010, 20, 878–880. [Google Scholar] [CrossRef]
- Pérez-Mejías, G.; Velázquez-Cruz, A.; Guerra-Castellano, A.; Baños-Jaime, B.; Díaz-Quintana, A.; González-Arzola, K.; De La Rosa, M.Á.; Díaz-Moreno, I. Exploring protein phosphorylation by combining computational approaches and biochemical methods. Comput. Struct. Biotechnol. J. 2020, 18, 1852–1863. [Google Scholar] [CrossRef]
- Castellano, A.G.; Díaz-Quintana, A.; Moreno-Beltrán, B.; López-Prados, J.; Nieto, P.M.; Meister, W.; Staffa, J.; Teixeira, M.; Hildebrandt, P.; De La Rosa, M.Á.; et al. Mimicking Tyrosine Phosphorylation in Human Cytochrome cby the Evolved tRNA Synthetase Technique. Chem.-A Eur. J. 2015, 21, 15004–15012. [Google Scholar] [CrossRef]
- Castellano, A.G.; Díaz-Moreno, I.; Velazquez-Campoy, A.; De La Rosa, M.Á.; Díaz-Quintana, A. Structural and functional characterization of phosphomimetic mutants of cytochrome c at threonine 28 and serine 47. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2016, 1857, 387–395. [Google Scholar] [CrossRef] [Green Version]
- Pecina, P.; Borisenko, G.G.; Belikova, N.A.; Tyurina, Y.Y.; Pecinova, A.; Lee, I.; Samhan-Arias, A.K.; Przyklenk, K.; Kagan, V.E.; Hüttemann, M. Phosphomimetic substitution of cytochrome c tyrosine 48 decreases respiration and binding to cardiolipin and abolishes ability to trigger downstream caspase activation. Biochemistry 2010, 49, 6705–6714. [Google Scholar] [CrossRef]
- García-Heredia, J.M.; Díaz-Quintana, A.; Salzano, M.; Orzáez, M.; Pérez-Payá, E.; Teixeira, M.; De La Rosa, M.A.; Díaz-Moreno, I. Tyrosine phosphorylation turns alkaline transition into a biologically relevant process and makes human cytochrome c behave as an anti-apoptotic switch. JBIC J. Biol. Inorg. Chem. 2011, 16, 1155–1168. [Google Scholar] [CrossRef]
- Moreno-Beltrán, B.; Guerra-Castellano, A.; Díaz-Quintana, A.; Del Conte, R.; García-Mauriño, S.M.; Díaz-Moreno, S.; González-Arzola, K.; Santos-Ocaña, C.; Velázquez-Campoy, A.; De La Rosa, M.A.; et al. Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48. Proc. Natl. Acad. Sci. USA 2017, 114, E3041–E3050. [Google Scholar] [CrossRef] [Green Version]
- Guerra-Castellano, A.; Díaz-Quintana, A.; Pérez-Mejías, G.; Elena-Real, C.A.; González-Arzola, K.; García-Mauriño, S.M.; De La Rosa, M.A.; Díaz-Moreno, I. Oxidative stress is tightly regulated by cytochrome c phosphorylation and respirasome factors in mitochondria. Proc. Natl. Acad. Sci. USA 2018, 115, 7955–7960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Moreno, I.; García-Heredia, J.M.; Díaz-Quintana, A.; Teixeira, M.; De La Rosa, M.Á. Nitration of tyrosines 46 and 48 induces the specific degradation of cytochrome c upon change of the heme iron state to high-spin. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2011, 1807, 1616–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, H.; Ohshima, Y.; Takusagawa, M.; Ikota, N.; Takahashi, Y.; Shimizu, S.; Ozawa, T. Functional Modification of Cytochrome c by Peroxynitrite in an Electron Transfer Reaction. Chem. Pharm. Bull. 2001, 49, 1547–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Moreno, I.; Nieto, P.M.; Del Conte, R.; Gairí, M.; García-Heredia, J.M.; De La Rosa, M.Á.; Díaz-Quintana, A. A Non-damaging Method to Analyze the Configuration and Dynamics of Nitrotyrosines in Proteins. Chem.-A Eur. J. 2012, 18, 3872–3878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Heredia, J.M.; Díaz-Moreno, I.; Nieto, P.M.; Orzáez, M.; Kocanis, S.; Teixeira, M.; Pérez-Payá, E.; Díaz-Quintana, A.; De La Rosa, M.Á. Nitration of tyrosine 74 prevents human cytochrome c to play a key role in apoptosis signaling by blocking caspase-9 activation. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2010, 1797, 981–993. [Google Scholar] [CrossRef] [PubMed]
- García-Heredia, J.M.; Díaz-Moreno, I.; Díaz-Quintana, A.; Orzáez, M.; Navarro, J.A.; Hervás, M.; De La Rosa, M.Á. Specific nitration of tyrosines 46 and 48 makes cytochromecassemble a non-functional apoptosome. FEBS Lett. 2011, 586, 154–158. [Google Scholar] [CrossRef] [Green Version]
- Vlasova, I.I.; Tyurin, V.A.; Kapralov, A.A.; Kurnikov, I.V.; Osipov, A.N.; Potapovich, M.V.; Stoyanovsky, D.A.; Kagan, V.E. Nitric oxide inhibits peroxidase activity of cytochrome c.cardiolipin complex and blocks cardiolipin oxidation. J. Biol. Chem. 2006, 281, 14554–14562. [Google Scholar] [CrossRef] [Green Version]
- Kruglik, S.G.; Yoo, B.-K.; Lambry, J.C.; Martin, J.-L.; Negrerie, M. Structural changes and picosecond to second dynamics of cytochrome c in interaction with nitric oxide in ferrous and ferric redox states. Phys. Chem. Chem. Phys. 2017, 19, 21317–21334. [Google Scholar] [CrossRef] [Green Version]
- Azzi, A.; Montecucco, C.; Richter, C. The use of acetylated ferricytochrome C for the detection of superoxide radicals produced in biological membranes. Biochem. Biophys. Res. Commun. 1975, 65, 597–603. [Google Scholar] [CrossRef]
- Minakami, S.; Titani, K.; Ishikura, H. The structure of cytochrome c. II. Properties of acetylated cytochrome c. J. Biochem. 1958, 45, 341–348. [Google Scholar] [CrossRef]
- Wada, K.; Okunuki, K. Studies on Chemically Modified Cytochrome cI. The Acetylated Cytochrome c. J. Biochem. 1968, 64, 667–687. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, Y.; Tan, Y.; Goto, Y. Comparison of the Conformational Stability of the Molten Globule and Native States of Horse Cytochrome c. J. Mol. Biol. 1994, 237, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Takemori, S.; Wada, K.; Sekuzu, I.; Okunuki, K.; Takemori, K.W.S. Reaction of Cytochrome a with Chemically Modified Cytochrome c and Basic Proteins. Nat. Cell Biol. 1962, 195, 456–457. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, S.S.; Krasnikov, B.F.; O Pereverzev, M.; Skulachev, V.P. The antioxidant functions of cytochrome c. FEBS Lett. 1999, 462, 192–198. [Google Scholar] [CrossRef] [Green Version]
- Bazylianska, V. The Effect of Acetylation of Cytochrome c on Its Functions in Prostate Cancer; Wayne State University: Detroit, MI, USA, 2017. [Google Scholar]
- Méndez, J.; Cruz, M.M.; Delgado, Y.; Figueroa, C.M.; Orellano, E.A.; Morales, M.; Monteagudo, A.; Griebenow, K. Delivery of Chemically Glycosylated Cytochrome c Immobilized in Mesoporous Silica Nanoparticles Induces Apoptosis in HeLa Cancer Cells. Mol. Pharm. 2014, 11, 102–111. [Google Scholar] [CrossRef] [Green Version]
- Delgado, Y.; Morales-Cruz, M.; Hernández-Román, J.; Martínez, Y.; Griebenow, K. Chemical glycosylation of cytochrome c improves physical and chemical protein stability. BMC Biochem. 2014, 15, 16. [Google Scholar] [CrossRef] [Green Version]
- Mercado-Uribe, H.; Andrade-Medina, M.; Espinoza-Rodríguez, J.H.; Carrillo-Tripp, M.; Scheckhuber, C.Q. Analyzing structural alterations of mitochondrial intermembrane space superoxide scavengers cytochrome-c and SOD1 after methylglyoxal treatment. PLoS ONE 2020, 15, e0232408. [Google Scholar] [CrossRef]
- A Oliveira, L.M.; A Gomes, R.; Yang, D.; Dennison, S.R.; Família, C.; Lages, A.; Coelho, A.; Murphy, R.M.; Phoenix, D.A.; Quintas, A. Insights into the molecular mechanism of protein native-like aggregation upon glycation. Biochim. et Biophys. Acta (BBA)-Proteins Proteom. 2013, 1834, 1010–1022. [Google Scholar] [CrossRef]
- Hildick-Smith, G.J.; Downey, M.C.; Gretebeck, L.M.; Gersten, R.A.; Sandwick, R.K. Ribose 5-Phosphate Glycation Reduces CytochromecRespiratory Activity and Membrane Affinity. Biochemistry 2011, 50, 11047–11057. [Google Scholar] [CrossRef] [Green Version]
- Sharma, G.S.; Warepam, M.; Bhattacharya, R.; Singh, L.R. Covalent Modification by Glyoxals Converts Cytochrome c Into its Apoptotically Competent State. Sci. Rep. 2019, 9, 4781. [Google Scholar] [CrossRef]
- Flatmark, T. On the heterogeneity of beef heart cytochrome c III.A kinetic study of the non-enzymic deamidation of the main sub-fractions. Acta Chem Scand 1966, 20, 1487–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flatmark, T. Multiple molecular forms of bovine heart cytochrome c. A comparative study of their physiochemical properties and their reactions in biological systems. J. Biol. Chem. 1967, 242, 2454–2459. [Google Scholar] [PubMed]
- Aluri, H.S.; Simpson, D.C.; Allegood, J.C.; Hu, Y.; Szczepanek, K.; Gronert, S.; Chen, Q.; Lesnefsky, E.J. Electron flow into cytochrome c coupled with reactive oxygen species from the electron transport chain converts cytochrome c to a cardiolipin peroxidase: Role during ischemia–reperfusion. Biochim. et Biophys. Acta (BBA)-Gen. Subj. 2014, 1840, 3199–3207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capdevila, D.A.; Marmisollé, W.A.; Tomasina, F.; Demicheli, V.; Portela, M.; Radi, R.; Murgida, D.H. Specific methionine oxidation of cytochrome c in complexes with zwitterionic lipids by hydrogen peroxide: Potential implications for apoptosis. Chem. Sci. 2015, 6, 705–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A Capdevila, D.; Rouco, S.O.; Tomasina, F.; Tortora, V.; Demicheli, V.; Radi, R.; Murgida, D.H. Active Site Structure and Peroxidase Activity of Oxidatively Modified Cytochrome c Species in Complexes with Cardiolipin. Biochemistry 2015, 54, 7491–7504. [Google Scholar] [CrossRef]
- Wang, Z.; Ando, Y.; Nugraheni, A.D.; Ren, C.; Nagao, S.; Hirota, S. Self-oxidation of cytochrome c at methionine80 with molecular oxygen induced by cleavage of the Met–heme iron bond. Mol. BioSyst. 2014, 10, 3130–3137. [Google Scholar] [CrossRef] [Green Version]
- Ivanetich, K.M.; Bradshaw, J.J.; Kaminsky, L.S. Methionine sulfoxide cytochrome c. Biochemistry 1976, 15, 1144–1153. [Google Scholar] [CrossRef]
- Chen, Y.-R.; Deterding, L.J.; Sturgeon, B.E.; Tomer, K.B.; Mason, R.P. Protein Oxidation of Cytochrome c by Reactive Halogen Species Enhances Its Peroxidase Activity. J. Biol. Chem. 2002, 277, 29781–29791. [Google Scholar] [CrossRef] [Green Version]
- Gates, A.T.; Moore, J.L.; Sylvain, M.R.; Jones, C.M.; Lowry, M.; El-Zahab, B.; Robinson, J.W.; Strongin, R.M.; Warner, I.M. Mechanistic Investigation ofN-Homocysteinylation-Mediated Protein−Gold Nanoconjugate Assembly. Langmuir 2009, 25, 9346–9351. [Google Scholar] [CrossRef] [Green Version]
- Perła-Kaján, J.; Marczak, Ł.; Kaján, L.; Skowronek, P.; Twardowski, T.; Jakubowski, H. Modification by Homocysteine Thiolactone Affects Redox Status of Cytochromec. Biochemistry 2007, 46, 6225–6231. [Google Scholar] [CrossRef]
- Dopner, S.; Hildebrandt, P.; Rosell, F.I.; Mauk, A.G.; Von Walter, M.; Buse, G.; Soulimane, T. The structural and functional role of lysine residues in the binding domain of cytochrome c in the electron transfer to cytochrome c oxidase. JBIC J. Biol. Inorg. Chem. 1999, 261, 379–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, G.S.; Singh, L.R. Conformational status of cytochrome c upon N-homocysteinylation: Implications to cytochrome c release. Arch. Biochem. Biophys. 2017, 614, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H. Protein homocysteinylation: Possible mechanism underlying pathological consequences of elevated homocysteine levels. FASEB J. 1999, 13, 2277–2283. [Google Scholar] [CrossRef] [PubMed]
- Yin, V.; Shaw, G.S.; Konermann, L. Cytochrome c as a Peroxidase: Activation of the Precatalytic Native State by H2O2-Induced Covalent Modifications. J. Am. Chem. Soc. 2017, 139, 15701–15709. [Google Scholar] [CrossRef] [PubMed]
- Yin, V.; Mian, S.H.; Konermann, L. Lysine carbonylation is a previously unrecognized contributor to peroxidase activation of cytochrome c by chloramine-T. Chem. Sci. 2019, 10, 2349–2359. [Google Scholar] [CrossRef] [Green Version]
- Barayeu, U.; Lange, M.; Méndez, L.; Arnhold, J.; Shadyro, O.I.; Fedorova, M.; Flemmig, J. Cytochrome c autocatalyzed carbonylation in the presence of hydrogen peroxide and cardiolipins. J. Biol. Chem. 2018, 294, 1816–1830. [Google Scholar] [CrossRef] [Green Version]
- Cassina, A.M.; Hodara, R.; Souza, J.M.; Thomson, L.; Castro, L.; Ischiropoulos, H.; Freeman, B.A.; Radi, R. Cytochrome c nitration by peroxynitrite. J. Biol. Chem. 2000, 275, 21409–21415. [Google Scholar] [CrossRef] [Green Version]
- Abriata, L.A.; Cassina, A.; Tórtora, V.; Marín, M.; Souza, J.M.; Castro, L.; Vila, A.J.; Radi, R. Nitration of Solvent-exposed Tyrosine 74 on CytochromecTriggers Heme Iron-Methionine 80 Bond Disruption. J. Biol. Chem. 2008, 284, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Groves, J.T. Mechanisms of Peroxynitrite Interactions with Heme Proteins. Inorg. Chem. 2010, 49, 6317–6329. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Moreno, I.; García-Heredia, J.M.; González-Arzola, K.; Díaz-Quintana, A.; De La Rosa, M.Á. Recent Methodological Advances in the Analysis of Protein Tyrosine Nitration. ChemPhysChem 2013, 14, 3095–3102. [Google Scholar] [CrossRef]
- Corpas, F.J.; Del Río, L.A.; Barroso, J.B. Post-translational modifications mediated by reactive nitrogen species. Plant Signal. Behav. 2008, 3, 301–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schonhoff, C.M.; Gaston, B.; Mannick, J.B. Nitrosylation of Cytochromecduring Apoptosis. J. Biol. Chem. 2003, 278, 18265–18270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radi, R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. USA 2004, 101, 4003–4008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacher, P.; Beckman, J.S.; Liaudet, L. Nitric Oxide and Peroxynitrite in Health and Disease. Physiol. Rev. 2007, 87, 315–424. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Roldán, V.; García-Heredia, J.M.; Navarro, J.A.; De La Rosa, M.A.; Hervás, M. Effect of Nitration on the Physicochemical and Kinetic Features of Wild-Type and Monotyrosine Mutants of Human Respiratory Cytochromec†. Biochemistry 2008, 47, 12371–12379. [Google Scholar] [CrossRef]
- Ly, H.K.; Utesch, T.; Díaz-Moreno, I.; García-Heredia, J.M.; De La Rosa, M.Á.; Hildebrandt, P. Perturbation of the Redox Site Structure of Cytochrome c Variants upon Tyrosine Nitration. J. Phys. Chem. B 2012, 116, 5694–5702. [Google Scholar] [CrossRef]
- Souza, J.M.; Castro, L.; Cassina, A.; Batthyány, C.; Radi, R. Nitrocytochrome c: Synthesis, Purification, and Functional Studies. Methods in Enzymology 2008, 441, 197–215. [Google Scholar] [CrossRef]
- Hosp, F.; Lassowskat, I.; Santoro, V.; De Vleesschauwer, D.; Fliegner, D.; Redestig, H.; Mann, M.; Christian, S.; Hannah, M.A.; Finkemeier, I. Lysine acetylation in mitochondria: From inventory to function. Mitochondrion 2017, 33, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Sprung, R.; Chen, Y.; Xu, Y.; Ball, H.; Pei, J.; Cheng, T.; Kho, Y.; Xiao, H.; Xiao, L.; et al. Substrate and Functional Diversity of Lysine Acetylation Revealed by a Proteomics Survey. Mol. Cell 2006, 23, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Romero-Garcia, S.; Lopez-Gonzalez, J.S.; B´ez-Viveros, J.L.; Aguilar-Cazares, D.; Prado-Garcia, H. Tumor cell metabolism. Cancer Biol. Ther. 2011, 12, 939–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tak, I.-U.-R.; Ali, F.; Dar, J.S.; Magray, A.R.; Ganai, B.A.; Chishti, M. Posttranslational Modifications of Proteins and Their Role in Biological Processes and Associated Diseases. In Protein Modificomics; Elsevier BV: Amsterdam, The Netherlands, 2019; pp. 1–35. [Google Scholar]
- König, B.; Osheroff, N.; Wilms, J.; Muijsers, A.; Dekker, H.L.; Margoliash, E. Mapping of the interaction domain for purified cytochrome c 1 on cytochrome c. FEBS Lett. 1980, 111, 395–398. [Google Scholar] [CrossRef] [Green Version]
- Smith, H.T.; Ahmed, A.J.; Millett, F. Electrostatic interaction of cytochrome c with cytochrome c1 and cytochrome oxidase. J. Biol. Chem. 1981, 256, 4984–4990. [Google Scholar] [PubMed]
- Kokhan, O.; Wraight, C.A.; Tajkhorshid, E. The Binding Interface of Cytochrome c and Cytochrome c1 in the bc1 Complex: Rationalizing the Role of Key Residues. Biophys. J. 2010, 99, 2647–2656. [Google Scholar] [CrossRef] [Green Version]
- Godoy, L.C.; Muñoz-Pinedo, C.; Castro, L.; Cardaci, S.; Schonhoff, C.M.; King, M.; Tórtora, V.; Marín, M.; Miao, Q.; Jiang, J.F.; et al. Disruption of the M80-Fe ligation stimulates the translocation of cytochromecto the cytoplasm and nucleus in nonapoptotic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2653–2658. [Google Scholar] [CrossRef] [Green Version]
- Flatmark, T. On the heterogeneity of beef heart cytochrome C. I. Separation and isolation of subfractions by dise electrophoresis and column chromatography. Acta Chem. Scand. 1964, 18, 1656–1666. [Google Scholar] [CrossRef]
- Robinson, A.B.; McKerrow, J.H.; Legaz, M. Sequence dependent deamidation rates for model peptides of cytochrome C. Int. J. Pept. Protein Res. 1974, 6, 31–35. [Google Scholar] [CrossRef]
- Flatmark, T.; Sletten, K. Life span of rat kidney c. In Structure and Function of Cytochromes; Okunuti, K., Kamen, M.D., Sekuzu, I., Eds.; University Press: Tokyo, Japan, 1968; pp. 413–421. [Google Scholar]
- Flatmark, T.; Sletten, K. Multiple forms of cytochrome c in the rat. Precursor-product relationship between the main component Cy I and the minor components Cy II and Cy 3 in vivo. J. Biol. Chem. 1968, 243, 1623–1629. [Google Scholar] [PubMed]
- Parakra, R.D.; Kleffmann, T.; Jameson, G.N.L.; Ledgerwood, E.C. The proportion of Met80-sulfoxide dictates peroxidase activity of human cytochrome c. Dalton Trans. 2018, 47, 9128–9135. [Google Scholar] [CrossRef]
- Wang, L.; Kallenbach, N.R. Proteolysis as a measure of the free energy difference between cytochrome c and its derivatives. Protein Sci. 1998, 7, 2460–2464. [Google Scholar] [CrossRef] [Green Version]
- Birk, A.V.; Chao, W.M.; Liu, S.; Soong, Y.; Szeto, H.H. Disruption of cytochrome c heme coordination is responsible for mitochondrial injury during ischemia. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2015, 1847, 1075–1084. [Google Scholar] [CrossRef] [Green Version]
- Dalle-Donne, I.; Aldini, G.; Carini, M.; Colombo, R.; Rossi, R.; Milzani, A. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med. 2006, 10, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Jeong, M.S.; Choi, S.Y.; Kang, J.H. Oxidative modification of cytochrome c by hydrogen peroxide. Mol. Cells 2006, 22, 220–227. [Google Scholar] [PubMed]
- Tomášková, N.; Varinská, L.; Sedlák, E. Rate of oxidative modification of cytochrome c by hydrogen peroxide is modulated by Hofmeister anions. Gen. Physiol. Biophys. 2010, 29, 255–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaidi, S.; Hassan, I.; Islam, A.; Ahmad, F. The role of key residues in structure, function, and stability of cytochrome-c. Cell. Mol. Life Sci. 2013, 71, 229–255. [Google Scholar] [CrossRef] [PubMed]
- Maity, H.; Maity, M.; Englander, S.W. How Cytochrome c Folds, and Why: Submolecular Foldon Units and their Stepwise Sequential Stabilization. J. Mol. Biol. 2004, 343, 223–233. [Google Scholar] [CrossRef]
- Hu, W.; Kan, Z.-Y.; Mayne, L.; Englander, S.W. Cytochrome c folds through foldon-dependent native-like intermediates in an ordered pathway. Proc. Natl. Acad. Sci. USA 2016, 113, 3809–3814. [Google Scholar] [CrossRef] [Green Version]
- Navarro, J.A.; Hervás, M.; Genzor, C.; Cheddar, G.; Fillat, M.; De La Rosa, M.Á.; Gomezmoreno, C.; Cheng, H.; Xia, B.; Chae, Y.; et al. Site-Specific Mutagenesis Demonstrates That the Structural Requirements for Efficient Electron Transfer in Anabaena Ferredoxin and Flavodoxin Are Highly Dependent on the Reaction Partner: Kinetic Studies with Photosystem I, Ferredoxin:NADP+ Reductase, and Cytochrome c. Arch. Biochem. Biophys. 1995, 321, 229–238. [Google Scholar] [CrossRef]
- Frazão, C.; Enguita, F.J.; Coelho, R.; Sheldrick, G.M.; Navarro, J.A.; Hervás, M.; De La Rosa, M.Á.; Carrondo, M.A. Crystal structure of low-potential cytochrome c549 from Synechocystis sp. PCC 6803 at 1.21 A resolution. JBIC J. Biol. Inorg. Chem. 2001, 6, 324–332. [Google Scholar] [CrossRef]
- Navarro, J.A.; Hervás, M.; De La Cerda, B.; De La Rosa, M.Á. Purification and Physicochemical Properties of the Low Potential Cytochrome C549 from the Cyanobacterium Synechocystis Sp PCC 6803. Arch. Biochem. Biophys. 1995, 318, 46–52. [Google Scholar] [CrossRef]
- Medina, M.; Hervás, M.; Navarro, J.A.; De La Rosa, M.Á.; Gómez-Moreno, C.; Tollin, G.; Medina, M. A laser flash absorption spectroscopy study of Anabaena sp. PCC 7119 flavodoxin photoreduction by photosystem I particles from spinach. FEBS Lett. 1992, 313, 239–242. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Xu, W.; Hervás, M.; Navarro, J.A.; Rosa, M.A.; Chitnis, P.R. Oxidizing side of the cyanobacterial photosystem I. Evidence for interaction between the electron donor proteins and a luminal surface helix of the PsaB subunit. J. Biol. Chem. 1999, 274, 19048–19054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goñi, G.; Herguedas, B.; Hervás, M.; Peregrina, J.R.; De La Rosa, M.A.; Gómez-Moreno, C.; Navarro, J.A.; Hermoso, J.A.; Martínez-Júlvez, M.; Medina, M. Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP+ reductase. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2009, 1787, 144–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De La Cerda, B.; Navarro, J.A.; Herv́s, >M.; De La Rosa, M.Á. Changes in the Reaction Mechanism of Electron Transfer from Plastocyanin to Photosystem I in the CyanobacteriumSynechocystissp. PCC 6803 As Induced by Site-Directed Mutagenesis of the Copper Protein. Biochemistry 1997, 36, 10125–10130. [Google Scholar] [CrossRef] [PubMed]
- Adachi, J.; Hasegawa, M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J. Mol. Evol. 1996, 42, 459–468. [Google Scholar] [CrossRef]
- Gao, L.; Laude, K.; Cai, H. Mitochondrial Pathophysiology, Reactive Oxygen Species, and Cardiovascular Diseases. Vet. Clin. N. Am. Small Anim. Pract. 2008, 38, 137–155. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Berezin, C.; Glaser, F.; Rosenberg, J.; Paz, I.; Pupko, T.; Fariselli, P.; Casadio, R.; Ben-Tal, N. ConSeq: The identification of functionally and structurally important residues in protein sequences. Bioinformatics 2004, 20, 1322–1324. [Google Scholar] [CrossRef]
- Imai, M.; Saio, T.; Kumeta, H.; Uchida, T.; Inagaki, F.; Ishimori, K. Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c. Biochem. Biophys. Res. Commun. 2016, 469, 978–984. [Google Scholar] [CrossRef]
- Storz, P. Mitochondrial ROS – radical detoxification, mediated by protein kinase D. Trends Cell Biol. 2007, 17, 13–18. [Google Scholar] [CrossRef]
- Liesa, M.; Shirihai, O. Mitochondrial Dynamics in the Regulation of Nutrient Utilization and Energy Expenditure. Cell Metab. 2013, 17, 491–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hüttemann, M.; Lee, I.; Grossman, L.I.; Doan, J.W.; Sanderson, T.H. Phosphorylation of mammalian cytochrome c and cytochrome c oxidase in the regulation of cell destiny: Respiration, apoptosis, and human disease. Single Mol. Single Cell Seq. 2012, 748, 237–264. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, C.M.; Singh, A.T.K. Apoptosis: A Target for Anticancer Therapy. Int. J. Mol. Sci. 2018, 19, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacMillan-Crow, L.A. Mitochondrial tyrosine nitration precedes chronic allograft nephropathy. Free. Radic. Biol. Med. 2001, 31, 1603–1608. [Google Scholar] [CrossRef]
- Morison, I.M.; Bordé, E.M.C.; Cheesman, E.J.; Cheong, P.L.; Holyoake, A.J.; Fichelson, S.; Weeks, R.J.; Lo, A.; Davies, S.M.K.; Wilbanks, S.M.; et al. A mutation of human cytochrome c enhances the intrinsic apoptotic pathway but causes only thrombocytopenia. Nat. Genet. 2008, 40, 387–389. [Google Scholar] [CrossRef]
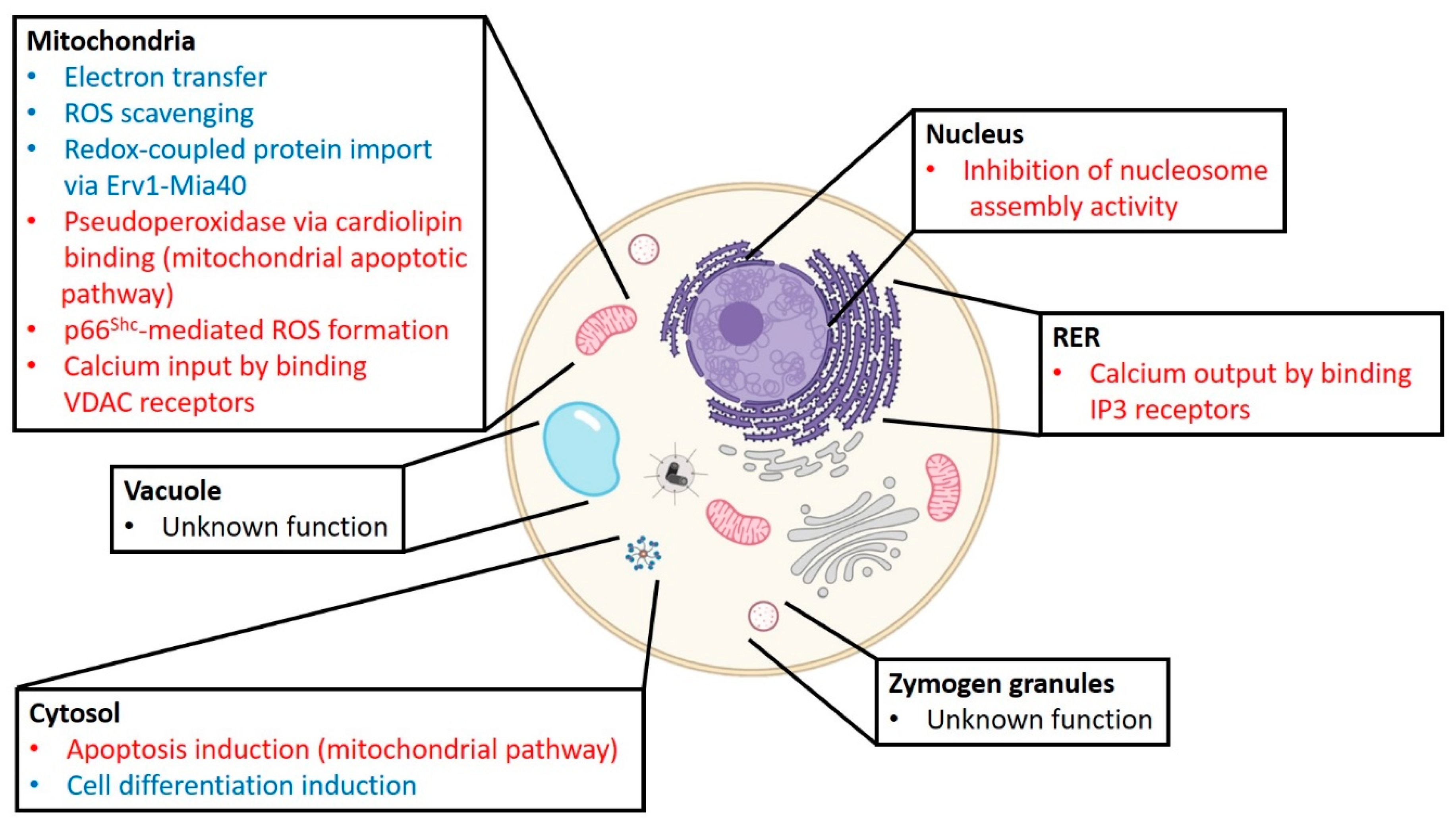

| Modification | Sites | Effects | References |
|---|---|---|---|
| Phosphorylation | Thr28, Tyr46, Ser47, Tyr48, Thr58 *, Tyr74, Tyr97 | Increase of Cc peroxidase activity. Decrease of electron transfer efficiency (Thr28, Ser47 and Tyr48). Increase of electron transfer efficiency under supercomplex formation (Tyr97). Modification of redox potential (Tyr48). Inhibition of caspase activation. | [53,55,77,78,79,80,81,82] |
| Nitration | Tyr46, Tyr48, Tyr67, Tyr74, Tyr97 (only nitration of Tyr74 and Tyr67 are detected in vivo) | Proteolytic degradation (Y46 and Y48). Increase of peroxidase activity (Y46, Y48 and Y74). Inhibition of caspase activation. | [83,84,85,86,87] |
| Nitrosylation | Heme and Met80 | Inhibition of Cc/CL complex peroxidase activity. Changes in protein conformation and heme coordination | [88,89] |
| Acetylation | Lysines (only acetylation of Lys8 and Lys53 are detected in vivo) | Decrease of electron transfer efficiency in the respiratory chain. Changes in the protein configuration. Inhibition of caspase activation | [90,91,92,93,94,95,96] |
| Glycosylation | Lysines 1 | Down-regulation of proteolytic degradation. Enhancement of thermodynamic stability. Inhibition of caspase activation. | [97,98] |
| Glycation | Arg91, Lys72, Lys87, Arg92 | Monomer aggregation. Reduction of conformational stability. Decrease of electron transfer efficiency in the respiratory chain. Decrease of ability to bind membrane. Enhance peroxidase activity. | [99,100,101,102] |
| Deamidation | Gln42, Asn31, Asn52 and Asn70 | Conformational changes. Modification of redox potential. | [103,104] |
| Sulfoxidation | Met80 | Loss of autoxidizable function. Decrease of electron transfer efficiency in the respiratory chain. Enhance peroxidase activity. Increase of apoptosis induction. | [105,106,107,108,109,110] |
| Homocysteinylation | Lys8 or Lys13, Lys86 or Lys87, Lys99, and Lys100 | Protein denaturation. Increase of resistance to proteolysis. Protein aggregation. Enhancement of peroxidase activity. | [111,112,113,114,115] |
| Carbonylation | Lys53, Lys55, Lys60 †, Lys72/Lys73 | Enhancement of peroxidase activity. Impairment of CL binding. Protein aggregation. | [116,117,118] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerra-Castellano, A.; Márquez, I.; Pérez-Mejías, G.; Díaz-Quintana, A.; De la Rosa, M.A.; Díaz-Moreno, I. Post-Translational Modifications of Cytochrome c in Cell Life and Disease. Int. J. Mol. Sci. 2020, 21, 8483. https://doi.org/10.3390/ijms21228483
Guerra-Castellano A, Márquez I, Pérez-Mejías G, Díaz-Quintana A, De la Rosa MA, Díaz-Moreno I. Post-Translational Modifications of Cytochrome c in Cell Life and Disease. International Journal of Molecular Sciences. 2020; 21(22):8483. https://doi.org/10.3390/ijms21228483
Chicago/Turabian StyleGuerra-Castellano, Alejandra, Inmaculada Márquez, Gonzalo Pérez-Mejías, Antonio Díaz-Quintana, Miguel A. De la Rosa, and Irene Díaz-Moreno. 2020. "Post-Translational Modifications of Cytochrome c in Cell Life and Disease" International Journal of Molecular Sciences 21, no. 22: 8483. https://doi.org/10.3390/ijms21228483
APA StyleGuerra-Castellano, A., Márquez, I., Pérez-Mejías, G., Díaz-Quintana, A., De la Rosa, M. A., & Díaz-Moreno, I. (2020). Post-Translational Modifications of Cytochrome c in Cell Life and Disease. International Journal of Molecular Sciences, 21(22), 8483. https://doi.org/10.3390/ijms21228483







