Phytopathogenic Cercosporoid Fungi—From Taxonomy to Modern Biochemistry and Molecular Biology
Abstract
:1. Introduction
2. Brief Taxonomical History of Cercosporoids
3. Molecular Investigations of Cercosporoids
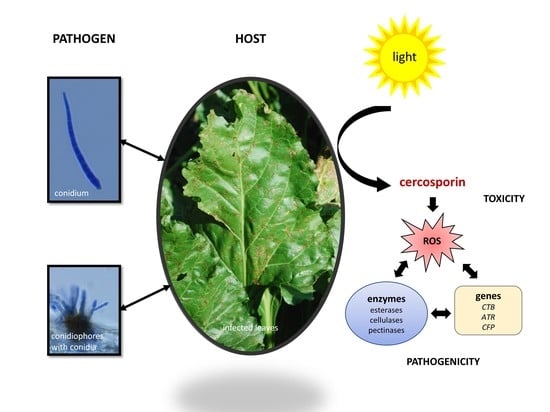
4. Cercosporin
4.1. Role in Disease
4.2. Cercosporin Production by Cercospora Species
4.3. Cercosporin Resistance
5. Importance of Redox Processes in Cercosporoid Physiology
6. Enzymes Involved in the Plant-Pathogen Interaction
7. Biotechnological Applications of Cercosporoid Fungi
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| (r)DNA | Ribosomal DNA |
| ACT | Actin |
| actA | Actin assembly-inducing protein |
| AFLP | Amplified Fragment Length Polymorphism |
| ATR2 | ABC transporter |
| CAL | Calmodulin |
| CAZymes | Carbohydrate-Active Enzymes |
| CBM1 | carbohydrate binding module protein |
| CMCase | Carboxymethyl cellulase |
| cmdA | Calmodulin-encoding gene |
| CRG1 | zinc cluster transcription factor |
| CTB | Polyketide synthase |
| CTB1 | polyketide synthase gene |
| CZK3 | MAP kinase kinase kinase |
| DNA | Deoxyribose Nucleic Acid |
| gapdh | glyceraldehyde-3-phosphate dehydrogenase |
| HIS | Histone H3 |
| his3 | histone H3 |
| ITS | Internal transcribed spacers |
| LME | Lignin-Modifying Enzymes |
| LSU | Large ribosomal subunit |
| MAP | Mitogen-activated protein kinases |
| MFS | Major Facilitator Superfamily transporter |
| NGS | next-generation sequencing |
| PCR | Polymerase Chain Reaction |
| PDT | Photodynamic therapy of cancer |
| PDX1 | Pancreatic and duodenal homeobox 1 |
| PDX2 | Pancreatic and duodenal homeobox 2 |
| PHI | pathogen-host interactions |
| PKS | polyketide synthase |
| REMI | Restriction Enzyme-Mediated Insertion |
| RFLP | Restriction Fragments Length Polymorphism |
| ROS | Reactive Oxygen Species |
| rpb2 | RNA polymerase II gene |
| s. lat. | Sensu lato |
| s. str. | Sensu stricto |
| tef1 | Translation elongation factor 1-alpha |
| TEF-1α | Translation elongation factor 1 alpha |
| TUB | Tubulin beta chain |
| tub2 | β-tubulin |
References
- Crous, P.W.; Braun, U. Mycosphaerella and Its Anamorphs: 1. Names Published in Cercospora and Passalora; Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre: Utrecht, The Netherlands, 2003. [Google Scholar]
- Braun, U.; Crous, P.W.; Schubert, K.; Shin, H.-D. Some reallocations of Stenella species to Zasmidium. Schlechtendalia 2010, 20, 99–104. [Google Scholar]
- Braun, U.; Crous, P.W.; Nakashima, C. Cercosporoid fungi (Mycosphaerellaceae) 5. Species on dicots (Anacardiaceae to Annonaceae). IMA Fungus 2016, 7, 161–216. [Google Scholar] [CrossRef] [PubMed]
- Braun, U.; Nakashima, C.; Crous, P.W. Cercosporoid fungi (Mycosphaerellaceae) 1. Species on other fungi, Pteridophyta and Gymnospermae. IMA Fungus 2013, 4, 265–345. [Google Scholar] [CrossRef] [PubMed]
- Groenewald, J.Z.; Nakashima, C.; Nishikawa, J.; Shin, H.D.; Park, J.H.; Jama, A.N.; Groenewald, M.; Braun, U.; Crous, P.W. Species concepts in Cercospora: Spotting the weeds among the roses. Stud. Mycol. 2013, 75, 115–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodwin, S.B.; Dunkle, L.D.; Zismann, V.L. Phylogenetic analysis of Cercospora and Mycosphaerella based on the internal transcribed spacer region of ribosomal DNA. Phytopathology 2001, 91, 648–658. [Google Scholar] [CrossRef] [Green Version]
- Świderska-Burek, U. Cercosporoid Fungi of Poland; Polish Botanical Society: Wrocław, Poland, 2015; Volume 105. [Google Scholar]
- Crous, P.W.; Braun, U.; Hunter, G.C.; Wingfield, M.J.; Verkley, G.J.; Shin, H.D.; Nakashima, C.; Groenewald, J.Z. Phylogenetic lineages in Pseudocercospora. Stud. Mycol. 2013, 75, 37–114. [Google Scholar] [CrossRef] [Green Version]
- Bakhshi, M.; Arzanlou, M.; Babai-Ahari, A.; Groenewald, J.Z.; Braun, U.; Crous, P.W. Application of the consolidated species concept to Cercospora spp. from Iran. Persoonia 2015, 34, 65–86. [Google Scholar] [CrossRef] [Green Version]
- Nguanhom, J.; Cheewangkoon, R.; Groenewald, J.Z.; Braun, U.; To-Anun, C.; Crous, P.W. Taxonomy and phylogeny of Cercospora spp. from Northern Thailand. Phytotaxa 2015, 233, 27–48. [Google Scholar] [CrossRef] [Green Version]
- Guatimosim, E.; Schwartsburd, P.B.; Barreto, R.W.; Crous, P.W. Novel fungi from an ancient niche: Cercosporoid and related sexual morphs on ferns. Persoonia 2016, 37, 106–141. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Choi, I.Y.; Lee, W.H.; Lee, K.J.; Galea, V.; Shin, H.D. Identification and Characterization of Cercospora malayensis Causing Leaf Spot on Kenaf. Mycobiology 2017, 45, 114–118. [Google Scholar] [CrossRef]
- Bakhshi, M.; Arzanlou, M.; Babai-Ahari, A.; Groenewald, J.Z.; Crous, P.W. Novel primers improve species delimitation in Cercospora. IMA Fungus 2018, 9, 299–332. [Google Scholar] [CrossRef] [PubMed]
- Groenewald, M.; Groenewald, J.Z.; Crous, P.W. Distinct Species Exist within the Cercospora apii Morphotype. Phytopathology 2005, 95, 951–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chupp, C. A Monograph of the Fungus Genus Cercospora; Ithaca: New York, NY, USA, 1954; p. 667. [Google Scholar]
- Chand, R.; Singh, V.; Kumar, P.; Pal, C.; Chowdappa, P. The Genus Cercospora: Biology and Taxonomy. In Fungal Leaf Spot Diseases of Anual and Perennial Crops; Chowdappa, P., Singh, H.P., Eds.; Westville Publishing House: New Delhi, India, 2012; pp. 1–290. [Google Scholar]
- Braun, U.; Crous, P. (2415) Proposal to conserve the name Cercospora (Ascomycota: Mycosphaerellaceae) with a conserved type. Taxon 2016, 65, 185. [Google Scholar] [CrossRef]
- Fries, E.M. Summa Vegetabilium Scandinaviae; Typographia Academica: Uppsala, Sweden, 1849; Volume 2. [Google Scholar]
- Spegazzini, C. Mycetes Argentinenses; Anales del Museo Nacional de Historia Natural: Buenos Aires, Argentina, 1910. [Google Scholar]
- Deighton, F.C. Studies on Cercospora and allied genera. II. Passalora, Cercosporidium and some species of Fusicladium on Euphorbia. Mycol. Pap. 1967, 112, 1–80. [Google Scholar]
- Deighton, F.C. Studies on Cercospora and allied genera. III. Centrospora. Mycol. Pap. 1971, 124, 1–13. [Google Scholar]
- Deighton, F.C. Studies on Cercospora and allied genera. IV. Cercosporella Sacc., Pseudocercosporella gen. nov. and Pseudocercosporidium gen. nov. Mycol. Pap. 1973, 133, 1–63. [Google Scholar]
- Deighton, F.C. Studies on Cercospora and allied genera. V. Mycovellosiella Rangel, and a new species of Ramulariopsis. Mycol. Pap. 1974, 137, 1–75. [Google Scholar]
- Deighton, F.C. Studies on Cercospora and allied genera. VI. Pseudocercospora Speg., Pantospora Cif. and Cercoseptoria Petr. Mycol. Pap. 1976, 140, 1–168. [Google Scholar]
- Deighton, F.C. Studies on Cercospora and allied genera. VII. New species and redispositions. Mycol. Pap. 1979, 14, 1–56. [Google Scholar]
- Deighton, F.C. Studies on Cercospora and allied genera. VIII. Further notes on Cercoseptoria and some new species and redispositions. Mycol. Pap. 1983, 151, 1–13. [Google Scholar]
- Pollack, F.G. An Annotated Compilation of Cercospora Names; J. Cramer: Berlin, Germany, 1987; Volume 12, p. 212. [Google Scholar]
- Pons, N.; Sutton, B.C. Cercospora and similar fungi on yams (Dioscorea species). Mycol. Pap. 1988, 160, 1–78. [Google Scholar]
- Braun, U. Taxonomic Notes on Some Species of the Cercospora-Complex. Nova Hedwig. 1992, 55, 211–221. [Google Scholar]
- Braun, U. Taxonomic Notes on Some Species of the Cercospora Complex (III). Mycotaxon 1993, 48, 275–298. [Google Scholar] [CrossRef]
- Braun, U. A Monograph of Cercosporella, Ramularia and Allied Genera (Phytopathogenic Hyphomycetes); IHW-Verlag: Eching bei München, Germany, 1995; Volume 1. [Google Scholar]
- Braun, U.; Melnik, V.A. Cercosporoid fungi from Russia and Adjacent Countries. In Trudy Botanicheskogo Instituta im. V.L. Komarova; Rossijskaya Akademiya Nauk: St Petersburg, Russia, 1997; Volume 20, pp. 1–130. [Google Scholar]
- To-Anun, C.; Hidayat, I.; Meeboon, J. Genus Cercospora in Thailand: Taxonomy and phylogeny (with a dichotomous key to species). Plant Pathol. Quar. 2011, 1, 11–87. [Google Scholar] [CrossRef]
- Braun, U.; Crous, P.W.; Nakashima, C. Cercosporoid fungi (Mycosphaerellaceae) 2. Species on monocots (Acoraceae to Xyridaceae, excluding Poaceae). IMA Fungus 2014, 5, 203–390. [Google Scholar] [CrossRef]
- Braun, U.; Crous, P.W.; Nakashima, C. Cercosporoid fungi (Mycosphaerellaceae) 3. Species on monocots (Poaceae, true grasses). IMA Fungus 2015, 6, 25–97. [Google Scholar] [CrossRef]
- Braun, U.; Crous, P.W.; Nakashima, C. Cercosporoid fungi (Mycosphaerellaceae) 4. Species on dicots (Acanthaceae to Amaranthaceae). IMA Fungus 2015, 6, 373–469. [Google Scholar] [CrossRef]
- Crous, P.W.; Braun, U.; Groenewald, J.Z. Mycosphaerella is polyphyletic. Stud. Mycol. 2007, 58, 1–32. [Google Scholar] [CrossRef]
- Crous, P.W.; Summerell, B.A.; Carnegie, A.J.; Wingfield, M.J.; Hunter, G.C.; Burgess, T.I.; Andjic, V.; Barber, P.A.; Groenewald, J.Z. Unravelling Mycosphaerella: Do you believe in genera? Persoonia 2009, 23, 99–118. [Google Scholar] [CrossRef] [Green Version]
- Crous, P.W.; Schoch, C.L.; Hyde, K.D.; Wood, A.R.; Gueidan, C.; de Hoog, G.S.; Groenewald, J.Z. Phylogenetic lineages in the Capnodiales. Stud. Mycol. 2009, 64, 17–47. [Google Scholar] [CrossRef]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.-K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.-Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Arzanlou, M.; Groenewald, J.Z.; Gams, W.; Braun, U.; Shin, H.D.; Crous, P.W. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud. Mycol. 2007, 58, 57–93. [Google Scholar] [CrossRef] [PubMed]
- Montenegro-Calderon, J.G.; Martinez-Alvarez, J.A.; Vieyra-Hernandez, M.T.; Rangel-Macias, L.I.; Razzo-Soria, T.; Chavez-Herrera, R.; Ponce-Noyola, P.; Leal-Morales, C.A. Molecular identification of two strains of Cercospora rodmanii isolated from water hyacinth present in Yuriria lagoon, Guanajuato, Mexico and identification of new hosts for several other strains. Fungal Biol. 2011, 115, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.L.; Liu, Z.; Crous, P.W.; Szabo, L.J. Phylogenetic relationships among some cercosporoid anamorphs of Mycosphaerella based on rDNA sequence analysis. Mycol. Res. 1999, 103, 1491–1499. [Google Scholar] [CrossRef] [Green Version]
- Bakhshi, M.; Arzanlou, M.; Babai-Ahari, A.; Groenewald, J.Z.; Crous, P.W. Is morphology in Cercospora a reliable reflection of generic affinity? Phytotaxa 2015, 213, 22–34. [Google Scholar] [CrossRef] [Green Version]
- Zeng, F.; Lian, X.; Zhang, G.; Yu, X.; Bradley, C.A.; Ming, R. A comparative genome analysis of Cercospora sojina with other members of the pathogen genus Mycosphaerella on different plant hosts. Genom. Data 2017, 13, 54–63. [Google Scholar] [CrossRef] [PubMed]
- De Coninck, B.M.A.; Amand, O.; Delaure, S.L.; Lucas, S.; Hias, N.; Weyens, G.; Mathys, J.; De Bruyne, E.; Cammue, B.P.A. The use of digital image analysis and real-time PCR fine-tunes bioassays for quantification of Cercospora leaf spot disease in sugar beet breeding. Plant Pathol. 2012, 61, 76–84. [Google Scholar] [CrossRef]
- Dekkers, K.L.; You, B.J.; Gowda, V.S.; Liao, H.L.; Lee, M.H.; Bau, H.H.; Ueng, P.P.; Chung, K.R. The Cercospora nicotianae gene encoding dual O-methyltransferase and FAD-dependent monooxygenase domains mediates cercosporin toxin biosynthesis. Fungal Genet. Biol. 2007, 44, 444–454. [Google Scholar] [CrossRef] [Green Version]
- Beseli, A.; Noar, R.; Daub, M.E. Characterization of Cercospora nicotianae Hypothetical Proteins in Cercosporin Resistance. PLoS ONE 2015, 10, e0140676. [Google Scholar] [CrossRef]
- Luo, X.; Cao, J.; Huang, J.; Wang, Z.; Guo, Z.; Chen, Y.; Ma, S.; Liu, J. Genome sequencing and comparative genomics reveal the potential pathogenic mechanism of Cercospora sojina Hara on soybean. DNA Res. 2017, 25, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Bluhm, B.H.; Dhillon, B.; Lindquist, E.A.; Kema, G.H.; Goodwin, S.B.; Dunkle, L.D. Analyses of expressed sequence tags from the maize foliar pathogen Cercospora zeae-maydis identify novel genes expressed during vegetative, infectious, and reproductive growth. BMC Genom. 2008, 9, 523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuyama, S.; Tamura, T. Cercosporin. A pigment of Cercospora kikuchii Matsumoto et Tomoyasu. II. Physical and chemical properties of cercosporin and its derivatives. J. Am. Chem. Soc. 1957, 79, 5726–5729. [Google Scholar] [CrossRef]
- Yamazaki, S.; Ogawa, T. The chemistry and stereochemistry of cercosporin. Agric. Biol. Chem. 1972, 36, 1707–1718. [Google Scholar] [CrossRef]
- Yamazaki, S.; Okube, A.; Akiyama, Y.; Fuwa, K. Cercosporin, a novel photodynamic pigment isolated from Cercospora kikuchii. Agric. Biol. Chem. 1975, 39, 287–288. [Google Scholar] [CrossRef] [Green Version]
- Daub, M.E.; Ehrenshaft, M. The Photoactivated Cercospora Toxin Cercosporin: Contributions to Plant Disease and Fundamental Biology. Annu. Rev. Phytopathol. 2000, 38, 461–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daub, M.E.; Herrero, S.; Chung, K.R. Photoactivated perylenequinone toxins in fungal pathogenesis of plants. FEMS Microbiol. Lett. 2005, 252, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Daub, M.E.; Chung, K.R. Photoactivated perylenequinone toxins in plant pathogenesis. In The Mycota V. Plant Relationships, 2nd ed.; Deising, H., Ed.; Springer: Berlin, Germany, 2009; pp. 201–219. [Google Scholar]
- Spikes, J.D. Photosensitization. In The Science of Photobiology, 2nd ed.; Smith, K.C., Ed.; Plenum Press: New York, NY, USA, 1989; pp. 79–110. [Google Scholar]
- Moan, J.; Berg, K.; Kvam, E.; Western, A.; Malik, Z.; Ruck, A.; Schneckenburger, H. Intracellular localization of photosensitizers. In Photosensitizing Compounds: Their Chemistry, Biology and Clinical Use; Wiley: Chichester, UK, 1998; Volume 146, pp. 95–111. [Google Scholar]
- Daub, M.E.; Chung, K.-R. Cercosporin: A photoactivated toxin in plant disease. Online APSnet Features 2007, 10, 2007-0207. [Google Scholar] [CrossRef]
- Fuller, K.K.; Dunlap, J.C.; Loros, J.J. Fungal Light Sensing at the Bench and Beyond. Adv. Genet. 2016, 96, 1–51. [Google Scholar] [CrossRef]
- Daub, M.E. Peroxidation of tobacco membrane lipids by the photosensitizing toxin, cercosporin. Plant Physiol. 1982, 69, 1361–1364. [Google Scholar] [CrossRef] [Green Version]
- Daub, M.E.; Briggs, S.P. Changes in tobacco cell membrane composition and structure caused by cercosporin. Plant Physiol. 1983, 71, 763–766. [Google Scholar] [CrossRef] [Green Version]
- Calpouzos, L.; Stalknecht, G.F. Symptoms of Cercospora leaf spot of sugar beets influenced by light intensity. Phytopathology 1967, 57, 799–800. [Google Scholar]
- Souza, A.G.C.; Maffia, L.A.; SIlva, F.F.; Mizubuti, E.S.G.; Teixeira, H. A time series analysis of brown eye spot progress in conventional and organic coffee production systems. Plant Pathol. 2015, 64, 157–166. [Google Scholar] [CrossRef]
- Fajola, A.O. Cercosporin, a phytotoxin from Cercospora species. Physiol. Plant Pathol. 1978, 79, 157–164. [Google Scholar] [CrossRef]
- Steinkamp, M.P.; Martin, S.S.; Hoefert, L.L.; Ruppel, E.G. Ultrastructure of lesions produced in leaves of Beta vulgaris. Physiol. Plant Pathol. 1979, 15, 13–16. [Google Scholar] [CrossRef]
- Choquer, M.; Dekkers, K.L.; Chen, H.Q.; Cao, L.; Ueng, P.P.; Daub, M.E.; Chung, K.R. The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae. Mol. Plant Microbe 2005, 18, 468–476. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Lee, M.H.; Daub, M.E.; Chung, K.R. Molecular analysis of the cercosporin biosynthetic gene cluster in Cercospora nicotianae. Mol. Microbiol. 2007, 64, 755–770. [Google Scholar] [CrossRef]
- Weiland, J.J.; Chung, K.R.; Suttle, J.C. The role of cercosporin in virulence of Cercospora spp. to plant hosts. In Cercospora Leaf Spot of Sugar Beet and Related Species; Lartey, R.T., Weiland, J.J., Panella, L., Crous, P.W., Windels, C.E., Eds.; APS Press: Saint Paul Minessota, MN, USA; IS-MPMI: Jeju, Korea, 2010; pp. 109–117. [Google Scholar]
- Newman, A.G.; Townsend, C.A. Molecular Characterization of the Cercosporin Biosynthetic Pathway in the Fungal Plant Pathogen Cercospora nicotianae. J. Am. Chem. Soc. 2016, 138, 4219–4228. [Google Scholar] [CrossRef] [Green Version]
- de Jonge, R.; Ebert, M.K.; Huitt-Roehl, C.R.; Pal, P.; Suttle, J.C.; Spanner, R.E.; Neubauer, J.D.; Jurick, W.M.I.; Stott, K.A.; Secor, G.A.; et al. Gene cluster conservation provides insight into cercosporin biosynthesis and extends production to the genus Colletotrichum. Proc. Natl. Acad. Sci. USA 2018, 115, E5459–E5466. [Google Scholar] [CrossRef] [Green Version]
- Callahan, T.; Rose, M.; Meade, M.; Ehrenshaft, M.; Upchurch, R. CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Mol. Plant Microbe 1999, 12, 901–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amnuaykanjanasin, A.; Daub, M.E. The ABC transporter ATR1 is necessary for efflux of the toxin cercosporin in the fungus Cercospora nicotianae. Fungal Genet. Biol. 2009, 46, 146–158. [Google Scholar] [CrossRef]
- Choquer, M.; Lee, M.H.; Bau, H.J.; Chung, K.R. Deletion of a MFS transporter-like gene in Cercospora nicotianae reduces cercosporin toxin accumulation and fungal virulence. FEBS Lett. 2007, 581, 489–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staerkel, C.; Boenisch, M.J.; Kroger, C.; Bormann, J.; Schafer, W.; Stahl, D. CbCTB2, an O-methyltransferase is essential for biosynthesis of the phytotoxin cercosporin and infection of sugar beet by Cercospora beticola. BMC Plant Biol. 2013, 13, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shim, W.B.; Dunkle, L.D. CZK3, a MAP kinase kinase kinase homolog in Cercospora zeae-maydis, regulates cercosporin biosynthesis, fungal development, and pathogenesis. Mol. Plant Microbe 2003, 16, 760–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assante, G.; Locci, R.; Camarda, L.; Merlini, L.; Nasini, G. Screening of the genus Cercospora for secondary metabolites. Phytochemistry 1977, 16, 243–247. [Google Scholar] [CrossRef]
- Lynch, F.J.; Geoghegan, M.J. Production of cercosporin by Cercospora species. Trans. Br. Mycol. Soc. 1977, 69, 496–498. [Google Scholar] [CrossRef]
- Goodwin, S.B.; Dunkle, L.D. Cercosporin production in Cercospora and related anamorphs of Mycosphaerella. In Cercospora Leaf Spot of Sugar Beet and Related Species; Lartey, R.L., Weiland, J.J., Panella, L., Crous, P.W., Windels, C.E., Eds.; APS Press: St. Paul, MN, USA, 2010; pp. 97–108. [Google Scholar]
- Arnone, A.; Assante, G.; Di Modugno, V.; Merlini, L.; Nasini, G. Perylenequinones from cucumber seedlings infected with Cladosporium cucumerinum. Phytochemistry 1988, 6, 1675–1678. [Google Scholar] [CrossRef]
- Jenns, A.E.; Daub, M.E.; Upchurch, R.G. Regulation of cercosporin accumulation in culture by medium and temperature manipulation. Phytopathology 1989, 79, 213–219. [Google Scholar] [CrossRef]
- You, B.J.; Lee, M.H.; Chung, K.R. Production of cercosporin toxin by the phytopathogenic Cercospora fungi is affected by diverse environmental signals. Can. J. Microbiol. 2008, 54, 259–269. [Google Scholar] [CrossRef]
- Gunasinghe, N.; You, P.Y.; Cawthray, G.R.; Barbetti, M.J. Cercosporin from Pseudocercosporella capsellae and its critical role in white leaf spote development. Plant Dis. 2016, 100, 1521–1531. [Google Scholar] [CrossRef] [Green Version]
- Ahonsi, M.O.; Maurhofer, M.; Boss, D.; Défago, G. Relationship between aggressiveness of Stagonospora sp. isolates on field and hedge bindweeds, and in vitro production of fungal metabolites cercosporin, elsinochrome A and leptosphaerodione. Plant Pathol. 2005, 111, 203–215. [Google Scholar] [CrossRef]
- Daub, M.E.; Herrero, S.; Taylor, T.V. Strategies for the development of resistance to cercosporin, a toxin produced by Cercospora species. In Cercospora Leaf Spot of Sugar Beet and Related Species; Lartey, R.T., Weiland, J.J., Panella, L., Crous, P.W., Windels, C.E., Eds.; APS Press: St. Paul, MN, USA, 2010; pp. 157–172. [Google Scholar]
- Ehrenshaft, M.; Jenns, A.E.; Daub, M.E. Targeted gene disruption of carotenoid biosynthesis in Cercospora nicotianae reveals no role for cartenoids in photosensitizer resistance. Mol. Plant Microbe 1995, 8, 569–575. [Google Scholar] [CrossRef]
- Daub, M.E.; Leisman, G.B.; Clark, R.A.; Bowden, E.F. Reductive detoxification as a mechanism of fungal resistance to singlet-oxygen-generating photosensitizers. Proc. Natl. Acad. Sci. USA 1992, 89, 9588–9592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leisman, G.B.; Daub, M.E. Singlet oxygen yields, optical properties, and phototoxicity of reduced derivatives of the photosensitizer cercosporin. Photochem. Photobiol. 1992, 55, 373–379. [Google Scholar] [CrossRef]
- Sollod, C.C.; Jenns, A.E.; Daub, M.E. Cell surface redox potential as a mechanism of defense against photosensitizers in fungi. Appl. Environ. Microb. 1992, 58, 444–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrenshaft, M.; Bilski, P.; Li, M.; Chignell, C.F.; Daub, M.E. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 9374–9378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilski, P.; Li, M.; Ehrenshaft, M.; Daub, M.; Chignell, C. Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem. Photobiol. 2000, 71, 129–134. [Google Scholar] [CrossRef]
- Chung, K.R.; Daub, M.E.; Kuchler, K.; Schuller, C. The CRG1 gene required for resistance to the singlet oxygen-generating cercosporin toxin in Cercospora nicotianae encodes a putative fungal transcription factor. Biochem. Biophys. Res. Commun. 2003, 302, 302–310. [Google Scholar] [CrossRef]
- Herrero, S.; Amnuaykanjanasin, A.; Daub, M.E. Identification of genes differentially expressed in the phytopathogenic fungus Cercospora nicotianae between cercosporin toxin-resistant and -susceptible strains. FEMS Microbiol. Lett. 2007, 275, 326–337. [Google Scholar] [CrossRef] [Green Version]
- Beseli, A.; Amnuaykanjanasin, A.; Herrero, S.; Thomas, E.; Daub, M.E. Membrane transporters in self resistance of Cercospora nicotianae to the photoactivated toxin cercosporin. Curr. Genet. 2015, 61, 601–620. [Google Scholar] [CrossRef]
- Beseli, A.; da Silva, M.G.; Daub, M.E. The role of Cercospora zeae-maydis homologs of Rhodobacter sphaeroides 1O2-resistance genes in resistance to the photoactivated toxin cercosporin. FEMS Microbiol. Lett. 2015, 362, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Vanacker, H.; Carver, T.L.; Foyer, C.H. Early H2O2 accumulation in mesophyll cells leads to induction of glutathione during the hyper-sensitive response in the barley-powdery mildew interaction. Plant Physiol. 2000, 123, 1289–1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolwell, G.P.; Wojtaszek, P. Mechanisms for the generation of reactive oxygen species in plant defence—A broad perspective. Physiol. Mol. Plant Pathol. 1997, 51, 347–366. [Google Scholar] [CrossRef]
- Schopfer, P. Histochemical Demonstration and Localization of H2O2 in Organs of Higher Plants by Tissue Printing on Nitrocellulose Paper. Plant Physiol. 1994, 104, 1269–1275. [Google Scholar] [CrossRef] [Green Version]
- Papadakis, A.K.; Roubelakis-Angelakis, K.A. The generation of active oxygen species differs in tobacco and grapevine mesophyll protoplasts. Plant Physiol. 1999, 121, 197–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamza, A.; El-Mogazy, S.; Derbalah, A. Fenton reagent and titanium dioxide nanoparticles as antifungal agents to control leaf spot of sugar beet under field conditions. J. Plant Prot. Res. 2016, 56, 270–278. [Google Scholar] [CrossRef]
- Gille, G.; Sigler, K. Oxidative stress and living cells. Folia Microbiol. 1995, 40, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Jaszek, M.; Grzywnowicz, K.; Malarczyk, E.; Leonowicz, A. Enhanced extracellular laccase activity as a part of the response system of white rot fungi: Trametes versicolor and Abortiporus biennis to paraquat-caused oxidative stress conditions. Pestic. Biochem. Phys. 2006, 85, 147–154. [Google Scholar] [CrossRef]
- Daub, M.E. Resistance of fungi to the photosensitizing toxin, cercosporin. Phytopathology 1987, 77, 1515–1520. [Google Scholar] [CrossRef] [Green Version]
- Ehrenshaft, M.; Chung, K.R.; Jenns, A.E.; Daub, M.E. Functional characterization of SOR1, a gene required for resistance to photosensitizing toxins in the fungus Cercospora nicotianae. Curr. Genet. 1999, 34, 478–485. [Google Scholar] [CrossRef]
- Daub, M.E.; Hangarter, R.P. Production of singlet oxygen and superoxide by the fungal toxin, cercosporin. Plant Physiol. 1983, 73, 855–857. [Google Scholar] [CrossRef] [Green Version]
- Dobrowolski, D.C.; Foote, C.S. Chemistry of singlet oxygen 46. Quantum yield of cercosporin-sensitized singlet oxygen formation. Angew. Chem.-Ger. Ed. 1983, 95, 729–730. [Google Scholar] [CrossRef]
- Daub, M.E. Cercosporin, a photosensitizing toxin from Cercospora species. Phytopathology 1982, 72, 370–374. [Google Scholar] [CrossRef]
- Daub, M.E.; Li, M.; Bilski, P.; Chignell, C.F. Dihydrocercosporin singlet oxygen production and subcellular localization: A possible defense against cercosporin phototoxicity in Cercospora. Photochem. Photobiol. 2000, 71, 135–140. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkolazka, A.; Paszczynski, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef] [Green Version]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant Cell Wall–Degrading Enzymes and Their Secretion in Plant-Pathogenic Fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef]
- Berglund, L.; Brunstedt, J.; Nielsen, K.K.; Chen, Z.; Mikkelsen, J.D.; Marcker, K.A. A proline-rich chitinase from Beta vulgaris. Plant Mol. Biol. 1995, 27, 211–216. [Google Scholar] [CrossRef]
- Gnanamangai, B.M.; Ponmurugan, P.; Yazhini, R.; Pragadeesh, S.K. PR Enzyme Activities of Cercospora theae Causing Bird’s Eye Spot Disease in Tea Plants (Camellia sinensis (L.) O. Kuntze). Plant Pathol. J. 2011, 10, 13–21. [Google Scholar] [CrossRef]
- Nielsen, K.K.; Jorgensen, P.; Mikkelsen, J.D. Antifungal activity of sugar beet chitinase against Cercospora beticola: An autoradiographic study on cell wall degradation. Plant Pathol. 1994, 43, 979–986. [Google Scholar] [CrossRef]
- Weiland, J.; Koch, G. Sugarbeet leaf spot disease (Cercospora beticola Sacc.)†. Mol. Plant Pathol. 2004, 5, 157–166. [Google Scholar] [CrossRef]
- Wilson, D.B. Cellulases and biofuels. Curr. Opin. Biotechnol. 2009, 20, 295–299. [Google Scholar] [CrossRef]
- Srivastava, N.; Srivastava, M.; Mishra, P.K.; Gupta, V.K.; Molina, G.; Rodriguez-Couto, S.; Manikanta, A.; Ramteke, P.W. Applications of fungal cellulases in biofuel production: Advances and limitations. Renew. Sust. Energy Rev. 2018, 82, 2379–2386. [Google Scholar] [CrossRef]
- Alabi, R.O.; Naqvi, S.H.Z. Production of cellulolytic and pectolytic enzymes by Cercospora arachidicola. Trans. Br. Mycol. Soc. 1977, 68, 296–298. [Google Scholar] [CrossRef]
- Weiland, J. Light-induced secretion of esterase by Cercospora beticola Sacc. In Proceedings of the International Congress on Molecular Plant-Microbe Interaction, IS-MPMI, Saint Paul, MN, USA, 10–14 July 2001; pp. 109–117. [Google Scholar]
- Herrero, S.; Daub, M.E. Genetic manipulation of vitamin B-6 biosynthesis in tobacco and fungi uncovers limitations to up-regulation of the pathway. Plant Sci. 2007, 172, 609–620. [Google Scholar] [CrossRef]
- Lynch, F.J.; Geoghegan, M.J. Antibiotic activity of a fungal perylene-quinone and some of its derivatives. Trans. Br. Mycol. Soc. 1979, 72, 31–37. [Google Scholar] [CrossRef]
- Kumarihamy, M.; Khan, S.I.; Jacob, M.; Tekwani, B.L.; Duke, S.O.; Ferreira, D.; Nanayakkara, N.P. Antiprotozoal and antimicrobial compounds from the plant pathogen Septoria pistaciarum. J. Nat. Prod. 2012, 75, 883–889. [Google Scholar] [CrossRef] [Green Version]
- Mastrangelopoulou, M.; Grigalavicius, M.; Berg, K.; Menard, M.; Theodossiou, T.A. Cytotoxic and Photocytotoxic Effects of Cercosporin on Human Tumor Cell Lines. Photochem. Photobiol. 2019, 95, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Costa-Silva, T.A.; Nogueira, M.A.; Souza, C.R.F.; Oliveira, W.P.; Said, S. Lipase Production by Endophytic Fungus Cercospora kikuchii: Stability of Enzymatic Activity after Spray Drying in the Presence of Carbohydrates. Dry. Technol. 2011, 29, 1112–1119. [Google Scholar] [CrossRef]
- Costa-Silva, T.A.; Souza, C.R.F.; Oliveira, W.P.; Said, S. Characterization and Spray Drying of Lipase Produced by the Endophytic Fungus Cercospora kikuchii. Braz. J. Chem. Eng. 2014, 31, 849–858. [Google Scholar] [CrossRef] [Green Version]
- Carneiro, L.A.B.C.; Costa-Silva, T.A.; Souza, C.R.F.; Bachmann, L.; Oliveira, W.P.; Said, S. Immobilization of Lipases Produced by the Endophytic Fungus Cercospora kikuchii on Chitosan Microparticles. Braz. Arch. Biol. Technol. 2014, 57, 578–586. [Google Scholar] [CrossRef] [Green Version]
- Thakur, V.; Kumar, P.; Verma, A.; Chand, D. Decolorization of dye by alginate immobilized laccase from Cercospora SPF-6: Using compact 5 stage plug flow reactor. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 183–200. [Google Scholar] [CrossRef]
- Pal, V.; Mukhopadhyay, A. Study of the cellulolytic and pectolytic enzymes in nine biological forms of Cercospora beticola Sacc. Acta Phytopathol. Hung. 1984, 19, 263–269. [Google Scholar]
- Orner, V.A.; Cantonwine, E.G.; Wang, X.M.; Abouelleil, A.; Bochicchio, J.; Nusbaum, C.; Culbreath, A.K.; Abdo, Z.; Arias, R.S. Draft Genome Sequence of Cercospora arachidicola, Causal Agent of Early Leaf Spot in Peanuts. Genome Announc. 2015, 3, e01281-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Enzyme | Source (Species Name) a | Citations | Potential Biotechnological Application |
|---|---|---|---|
| lipase | Cercospora kikuchii | [123,124,125] | food and chemical industry, biodiesel transesterification |
| esterase | C. beticola | [118] | wood industry and biofuel production |
| cellulases | C. beticola | [115,127] | wood industry and biofuel production |
| pectinases | C. beticola | [115,127] | food and textile industry |
| laccase | Cercospora SPF-6 | [126] | dye decolorization |
| endo-glucanase (CMCase) | C. theae | [112] | wood industry and biofuel production |
| invertase | C. theae | [112] | food industry, cosmetics and pharmaceutical industry |
| protease | C. theae | [112] | chemical and textile industry, food industry |
| pectinase | C. theae | [112] | food and textile industry |
| amylase | C. theae | [112] | food and textile industry, pharmaceutical and fine-chemical industry |
| CMCase | C. arachidicola | [117] | food industry and biofuel production |
| polygalacturonase (pectinase) | C. arachidicola | [117] | food and textile industry |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Świderska-Burek, U.; Daub, M.E.; Thomas, E.; Jaszek, M.; Pawlik, A.; Janusz, G. Phytopathogenic Cercosporoid Fungi—From Taxonomy to Modern Biochemistry and Molecular Biology. Int. J. Mol. Sci. 2020, 21, 8555. https://doi.org/10.3390/ijms21228555
Świderska-Burek U, Daub ME, Thomas E, Jaszek M, Pawlik A, Janusz G. Phytopathogenic Cercosporoid Fungi—From Taxonomy to Modern Biochemistry and Molecular Biology. International Journal of Molecular Sciences. 2020; 21(22):8555. https://doi.org/10.3390/ijms21228555
Chicago/Turabian StyleŚwiderska-Burek, Urszula, Margaret E. Daub, Elizabeth Thomas, Magdalena Jaszek, Anna Pawlik, and Grzegorz Janusz. 2020. "Phytopathogenic Cercosporoid Fungi—From Taxonomy to Modern Biochemistry and Molecular Biology" International Journal of Molecular Sciences 21, no. 22: 8555. https://doi.org/10.3390/ijms21228555
APA StyleŚwiderska-Burek, U., Daub, M. E., Thomas, E., Jaszek, M., Pawlik, A., & Janusz, G. (2020). Phytopathogenic Cercosporoid Fungi—From Taxonomy to Modern Biochemistry and Molecular Biology. International Journal of Molecular Sciences, 21(22), 8555. https://doi.org/10.3390/ijms21228555