Identification of Recurrent Mutations in the microRNA-Binding Sites of B-Cell Lymphoma-Associated Genes in Follicular Lymphoma
Abstract
1. Introduction
2. Results
2.1. Identification of Somatic Mutations in miRNA-Binding Sites of FL
2.2. Genes Showing Recurrent Mutations in miRNA-Binding Sites Are Highly Enriched for Germinal-Center-Like B-Cell Lymphoma Genes
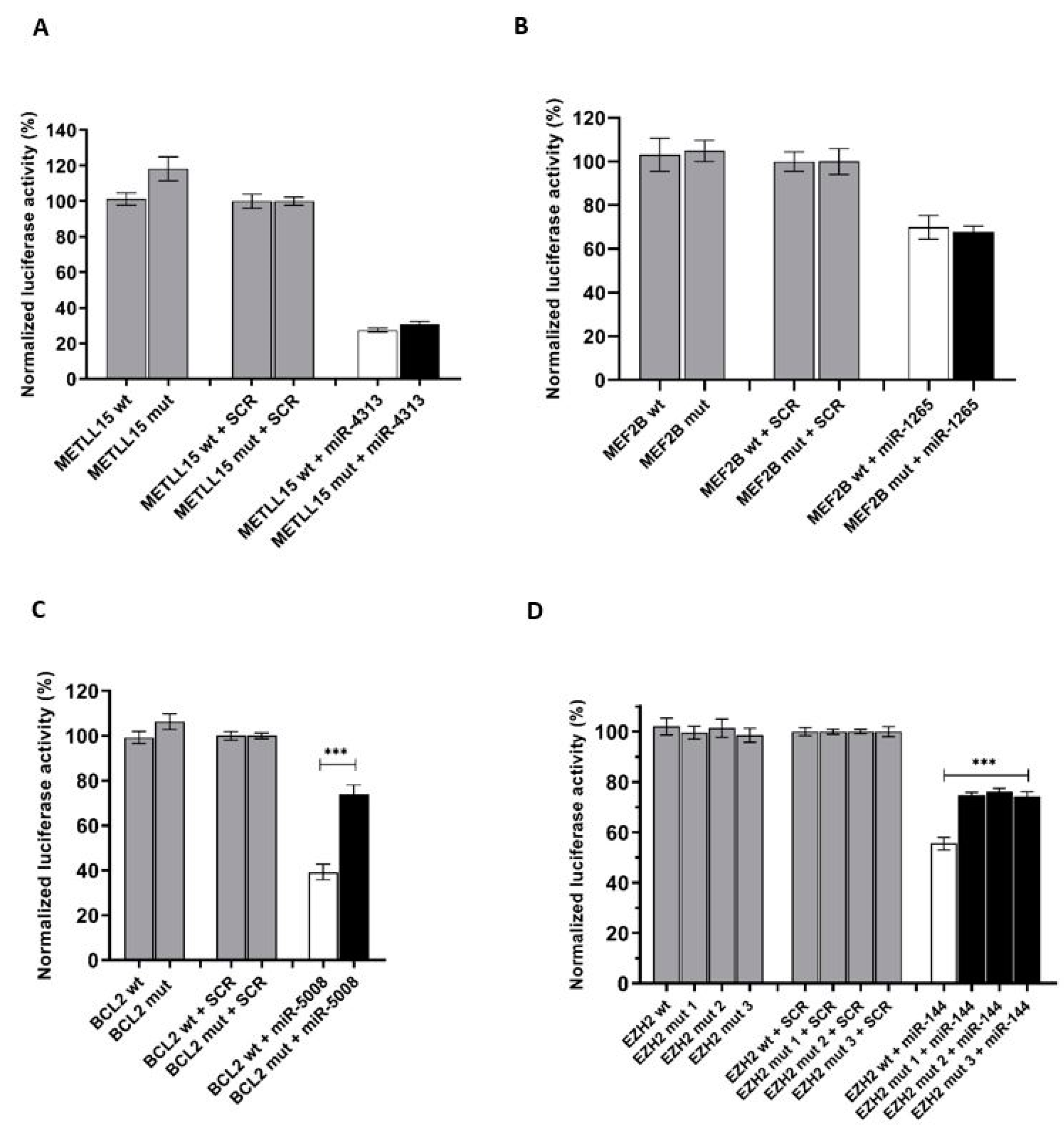
2.3. In Vitro Luciferase Assays Demonstrated That Mutations in miRNA Binding Sites Interfere with the miRNA Activity
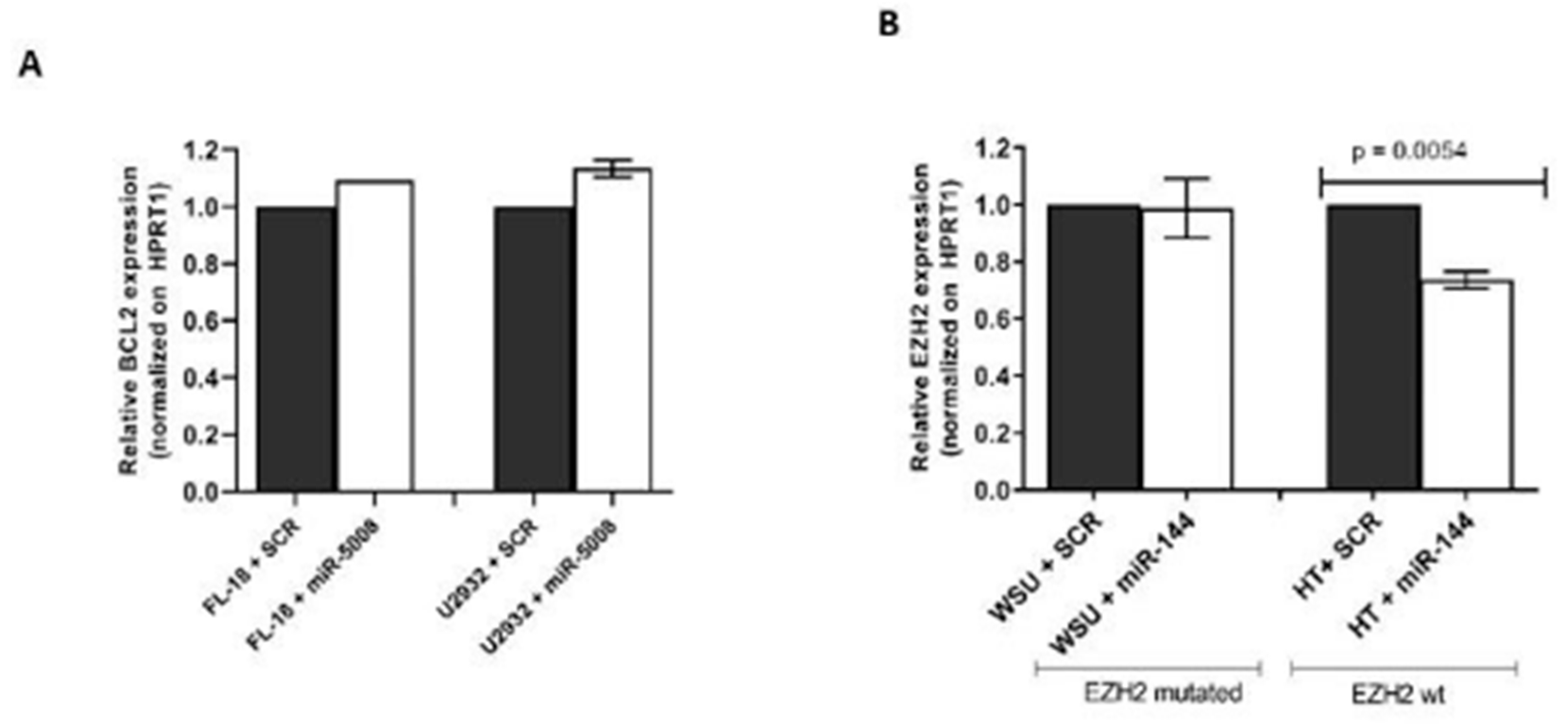
2.4. Testing the Effect of miR-5008 and miR-144 on Basal Target Gene Expression
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. Bioinformatic Identification of miRNA-Binding Site Mutations
4.3. Ampliseq (Ion Torrent) Panel NGS
4.4. Luciferase Assays
4.5. Cell Culture and Transfection
4.6. qRT-PCR
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Smith, A.; Crouch, S.; Lax, S.; Li, J.; Painter, D.; Howell, D.; Patmore, R.; Jack, A.; Roman, E. Lymphoma incidence, survival and prevalence 2004–2014: Sub-type analyses from the UK’s Haematological Malignancy Research Network. Br. J. Cancer 2015, 112, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Kahl, B.S.; Yang, D.T. Follicular lymphoma: Evolving therapeutic strategies. Blood 2016, 127, 2055–2063. [Google Scholar] [CrossRef] [PubMed]
- Provencio, M.; Sabin, P.; Gomez-Codina, J.; Torrente, M.; Calvo, V.; Llanos, M.; Guma, J.; Quero, C.; Blasco, A.; Cruz, M.A.; et al. Impact of treatment in long-term survival patients with follicular lymphoma: A Spanish Lymphoma Oncology Group registry. PLoS ONE 2017, 12, e0177204. [Google Scholar] [CrossRef]
- Lossos, I.S.; Gascoyne, R.D. Transformation of follicular lymphoma. Best Pract. Res. Clin. Haematol. 2011, 24, 147–163. [Google Scholar] [CrossRef]
- Wagner-Johnston, N.D.; Link, B.K.; Byrtek, M.; Dawson, K.L.; Hainsworth, J.; Flowers, C.R.; Friedberg, J.W.; Bartlett, N.L. Outcomes of transformed follicular lymphoma in the modern era: A report from the National LymphoCare Study (NLCS). Blood 2015, 126, 851–857. [Google Scholar] [CrossRef]
- Federico, M.; Caballero Barrigon, M.D.; Marcheselli, L.; Tarantino, V.; Manni, M.; Sarkozy, C.; Alonso-Alvarez, S.; Wondergem, M.; Cartron, G.; Lopez-Guillermo, A.; et al. Rituximab and the risk of transformation of follicular lymphoma: A retrospective pooled analysis. Lancet Haematol. 2018, 5, e359–e367. [Google Scholar] [CrossRef]
- Sole, C.; Larrea, E.; Di Pinto, G.; Tellaetxe, M.; Lawrie, C.H. miRNAs in B-cell lymphoma: Molecular mechanisms and biomarker potential. Cancer Lett. 2017, 405, 79–89. [Google Scholar] [CrossRef]
- Musilova, K.; Devan, J.; Cerna, K.; Seda, V.; Pavlasova, G.; Sharma, S.; Oppelt, J.; Pytlik, R.; Prochazka, V.; Prouzova, Z.; et al. miR-150 downregulation contributes to the high-grade transformation of follicular lymphoma by upregulating FOXP1 levels. Blood 2018, 132, 2389–2400. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Chi, J.; Taylor, S.; Tramonti, D.; Ballabio, E.; Palazzo, S.; Saunders, N.J.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Expression of microRNAs in diffuse large B cell lymphoma is associated with immunophenotype, survival and transformation from follicular lymphoma. J. Cell. Mol. Med. 2009, 13, 1248–1260. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Soneji, S.; Marafioti, T.; Cooper, C.D.; Palazzo, S.; Paterson, J.C.; Cattan, H.; Enver, T.; Mager, R.; Boultwood, J.; et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int. J. Cancer 2007, 121, 1156–1161. [Google Scholar] [CrossRef]
- Wang, W.; Corrigan-Cummins, M.; Hudson, J.; Maric, I.; Simakova, O.; Neelapu, S.S.; Kwak, L.W.; Janik, J.E.; Gause, B.; Jaffe, E.S.; et al. MicroRNA profiling of follicular lymphoma identifies microRNAs related to cell proliferation and tumor response. Haematologica 2012, 97, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, N.; Gollub, W.; Stassek, B.; Bernard, V.; Rades, D.; Feller, A.C.; Thorns, C. MicroRNA signatures in subtypes of follicular lymphoma. Anticancer Res. 2014, 34, 2105–2111. [Google Scholar] [PubMed]
- Malpeli, G.; Barbi, S.; Greco, C.; Zupo, S.; Bertolaso, A.; Scupoli, M.T.; Krampera, M.; Kamga, P.T.; Croce, C.M.; Scarpa, A.; et al. MicroRNA signatures and Foxp3+ cell count correlate with relapse occurrence in follicular lymphoma. Oncotarget 2018, 9, 19961–19979. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thompson, M.A.; Edmonds, M.D.; Liang, S.; McClintock-Treep, S.; Wang, X.; Li, S.; Eischen, C.M. miR-31 and miR-17-5p levels change during transformation of follicular lymphoma. Hum. Pathol. 2016, 50, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Hezaveh, K.; Kloetgen, A.; Bernhart, S.H.; Mahapatra, K.D.; Lenze, D.; Richter, J.; Haake, A.; Bergmann, A.K.; Brors, B.; Burkhardt, B.; et al. Alterations of microRNA and microRNA-regulated messenger RNA expression in germinal center B-cell lymphomas determined by integrative sequencing analysis. Haematologica 2016, 101, 1380–1389. [Google Scholar] [CrossRef]
- Okosun, J.; Bodor, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014, 46, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Marine, R.L.; Magana, L.C.; Castro, C.J.; Zhao, K.; Montmayeur, A.M.; Schmidt, A.; Diez-Valcarce, M.; Fan Ng, T.F.; Vinje, J.; Burns, C.C.; et al. Comparison of Illumina MiSeq and the Ion Torrent PGM and S5 platforms for whole-genome sequencing of picornaviruses and caliciviruses. J. Virol. Methods 2020. [Google Scholar] [CrossRef]
- Song, L.; Huang, W.; Kang, J.; Huang, Y.; Ren, H.; Ding, K. Comparison of error correction algorithms for Ion Torrent PGM data: Application to hepatitis B virus. Sci. Rep. 2017. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Bodor, C.; Grossmann, V.; Popov, N.; Okosun, J.; O’Riain, C.; Tan, K.; Marzec, J.; Araf, S.; Wang, J.; Lee, A.M.; et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood 2013, 122, 3165–3168. [Google Scholar] [CrossRef]
- Donaldson-Collier, M.C.; Sungalee, S.; Zufferey, M.; Tavernari, D.; Katanayeva, N.; Battistello, E.; Mina, M.; Douglass, K.M.; Rey, T.; Raynaud, F.; et al. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat. Genet. 2019, 51, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wu, L.; Zhu, M.; Wang, Z.; Wu, M.; Li, P.; Nie, Y.; Lin, X.; Hu, J.; Eskilsson, E.; et al. miRNA Mediated Noise Making of 3′UTR Mutations in Cancer. Genes 2018, 9, 545. [Google Scholar] [CrossRef] [PubMed]
- Ziebarth, J.D.; Bhattacharya, A.; Cui, Y. Integrative analysis of somatic mutations altering microRNA targeting in cancer genomes. PLoS ONE 2012, 7, e47137. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Ramos, C.M.; Barros, B.P.; Koyama, F.C.; Carpinetti, P.A.; Pezuk, J.; Doimo, N.T.S.; Habr-Gama, A.; Perez, R.O.; Parmigiani, R.B. E2F1 somatic mutation within miRNA target site impairs gene regulation in colorectal cancer. PLoS ONE 2017, 12, e0181153. [Google Scholar] [CrossRef]
- Ramsingh, G.; Koboldt, D.C.; Trissal, M.; Chiappinelli, K.B.; Wylie, T.; Koul, S.; Chang, L.W.; Nagarajan, R.; Fehniger, T.A.; Goodfellow, P.; et al. Complete characterization of the microRNAome in a patient with acute myeloid leukemia. Blood 2010, 116, 5316–5326. [Google Scholar] [CrossRef]
- Kridel, R.; Sehn, L.H.; Gascoyne, R.D. Pathogenesis of follicular lymphoma. J. Clin. Investig. 2012, 122, 3424–3431. [Google Scholar] [CrossRef]
- Beguelin, W.; Popovic, R.; Teater, M.; Jiang, Y.; Bunting, K.L.; Rosen, M.; Shen, H.; Yang, S.N.; Wang, L.; Ezponda, T.; et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 2013, 23, 677–692. [Google Scholar] [CrossRef]
- Sungalee, S.; Mamessier, E.; Morgado, E.; Gregoire, E.; Brohawn, P.Z.; Morehouse, C.A.; Jouve, N.; Monvoisin, C.; Menard, C.; Debroas, G.; et al. Germinal center reentries of BCL2-overexpressing B cells drive follicular lymphoma progression. J. Clin. Investig. 2014, 124, 5337–5351. [Google Scholar] [CrossRef]
- Roulland, S.; Faroudi, M.; Mamessier, E.; Sungalee, S.; Salles, G.; Nadel, B. Early steps of follicular lymphoma pathogenesis. Adv. Immunol. 2011, 111, 1–46. [Google Scholar] [CrossRef]
- Castellino, A.; Santambrogio, E.; Nicolosi, M.; Botto, B.; Boccomini, C.; Vitolo, U. Follicular Lymphoma: The Management of Elderly Patient. Mediterr. J. Hematol. Infect. Dis. 2017, 9, e2017009. [Google Scholar] [CrossRef]
- Leich, E.; Hoster, E.; Wartenberg, M.; Unterhalt, M.; Siebert, R.; Koch, K.; Klapper, W.; Engelhard, M.; Puppe, B.; Horn, H.; et al. Similar clinical features in follicular lymphomas with and without breaks in the BCL2 locus. Leukemia 2016, 30, 854–860. [Google Scholar] [CrossRef] [PubMed]
- Correia, C.; Schneider, P.A.; Dai, H.; Dogan, A.; Maurer, M.J.; Church, A.K.; Novak, A.J.; Feldman, A.L.; Wu, X.; Ding, H.; et al. BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma. Blood 2015, 125, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Sneeringer, C.J.; Scott, M.P.; Kuntz, K.W.; Knutson, S.K.; Pollock, R.M.; Richon, V.M.; Copeland, R.A. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA 2010, 107, 20980–20985. [Google Scholar] [CrossRef] [PubMed]
- Yap, D.B.; Chu, J.; Berg, T.; Schapira, M.; Cheng, S.W.; Moradian, A.; Morin, R.D.; Mungall, A.J.; Meissner, B.; Boyle, M.; et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 2011, 117, 2451–2459. [Google Scholar] [CrossRef]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F.; et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef]
- Sahasrabuddhe, A.A.; Chen, X.; Chung, F.; Velusamy, T.; Lim, M.S.; Elenitoba-Johnson, K.S. Oncogenic Y641 mutations in EZH2 prevent Jak2/beta-TrCP-mediated degradation. Oncogene 2015, 34, 445–454. [Google Scholar] [CrossRef]
- Guo, Y.; Ying, L.; Tian, Y.; Yang, P.; Zhu, Y.; Wang, Z.; Qiu, F.; Lin, J. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013, 280, 4531–4538. [Google Scholar] [CrossRef]
- Cao, J.; Han, X.; Qi, X.; Jin, X.; Li, X. TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via miR-144-3p. Int. J. Oncol. 2017, 51, 1115–1123. [Google Scholar] [CrossRef]
- Lin, L.; Zheng, Y.; Tu, Y.; Wang, Z.; Liu, H.; Lu, X.; Xu, L.; Yuan, J. MicroRNA-144 suppresses tumorigenesis and tumor progression of astrocytoma by targeting EZH2. Hum. Pathol. 2015, 46, 971–980. [Google Scholar] [CrossRef]
- Wang, H.; Wang, A.; Hu, Z.; Xu, X.; Liu, Z.; Wang, Z. A Critical Role of miR-144 in Diffuse Large B-cell Lymphoma Proliferation and Invasion. Cancer Immunol. Res. 2016, 4, 337–344. [Google Scholar] [CrossRef]
- Huet, S.; Xerri, L.; Tesson, B.; Mareschal, S.; Taix, S.; Mescam-Mancini, L.; Sohier, E.; Carrere, M.; Lazarovici, J.; Casasnovas, O.; et al. EZH2 alterations in follicular lymphoma: Biological and clinical correlations. Blood Cancer J. 2017, 7, e555. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [PubMed]
- Kinsella, R.J.; Kahari, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database J. Biol. Databases Curation 2011, 2011, bar030. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Ohno, H.; Doi, S.; Fukuhara, S.; Nishikori, M.; Uchino, H.; Fujii, H. A newly established human lymphoma cell line, FL-18, carrying a 14;18 translocation. Jpn. J. Cancer Res. 1985, 76, 563–566. [Google Scholar]
- Quentmeier, H.; Amini, R.M.; Berglund, M.; Dirks, W.G.; Ehrentraut, S.; Geffers, R.; Macleod, R.A.; Nagel, S.; Romani, J.; Scherr, M.; et al. U-2932: Two clones in one cell line, a tool for the study of clonal evolution. Leukemia 2013, 27, 1155–1164. [Google Scholar] [CrossRef]


| Gene | Mutated Position | Mutation | Location | miRNA | Cases (n = 55) |
|---|---|---|---|---|---|
| EZH2 | chr7:148508727 | T/A | Exon | hsa-mir-144 | 8 |
| ARMC10 | chr7:102739179 | A/G | 3′ UTR | hsa-mir-222 | 18 |
| TUBB | chr6:30692754 | C/CTT | 3′ UTR | hsa-mir-1302 | 7 |
| MEF2B | chr19:19260045 | T/A | Exon | hsa-mir-1265 | 3 |
| METTL15 | chr11:28353434 | G/A | 3′ UTR | hsa-mir-4313 | 12 |
| ZNF195 | chr11:3380000 | t/TC | 3′ UTR | hsa-mir-1915 | 16 |
| BCL2 | chr18:60793447 | G/A | 3′ UTR | hsa-mir-5008 | 2 |
| THOC3 | chr5:175386586 | A/G | 3′ UTR | hsa-mir-371a | 15 |
| TXNDC2 | chr18:9887493 | T/C | Exon | hsa-mir-2110 | 3 |
| PCDH7 | chr4:30732983 | GTA/G | Intron | hsa-mir-329 | 4 |
| RC3H1 | chr1:173901940 | A/AAAT | 3′ UTR | hsa-mir-548an | 2 |
| AQP3 | chr9:33441702 | C/A | 3′ UTR | hsa-mir-146b | 8 |
| DPY19L2 | chr12:63953768 | T/C | 3′ UTR | hsa-mir-1303 | 11 |
| MYO5B | chr18:47352742 | T/G | 3′ UTR | hsa-mir-216b | 55 |
| MYO5B | chr18:47352754 | A/G | 3′ UTR | hsa-mir-2681 | 55 |
| YY2 | chrX:21876221 | A/G | 3′ UTR | hsa-mir-448 | 36 |
| Gene | Predicted miRNA Binding Site | miRNA | Mutation | Total Patients (n = 55) (%) | tFL/ntFL |
|---|---|---|---|---|---|
| ARMC10 | chr7:102739177–102739198 | hsa-mir-222 | c.1320A > G | 18 (33) | 13/7 |
| BCL2 | chr18:60793436–60793458 | hsa-mir-5008 | c.3623C > T | 2 (4) | 2/0 |
| METTL15 | chr11:28353429–28353448 | hsa-mir-4313 | c.2588G > A | 12 (22) | 1/11 |
| EZH2 | chr7:148508722–148508742 | hsa-mir-144 | c.2115A > T | 8 (15) | 11/2 |
| EZH2 | chr7:148508722–148508742 | hsa-mir-144 | c.2114T > A | 5 (9) | 4/2 |
| MEF2B | chr19:19260038–19260055 | hsa-mir-1265 | c.336A > T | 3 (6) | 3/1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larrea, E.; Fernandez-Mercado, M.; Guerra-Assunção, J.A.; Wang, J.; Goicoechea, I.; Gaafar, A.; Ceberio, I.; Lobo, C.; Okosun, J.; Enright, A.J.; et al. Identification of Recurrent Mutations in the microRNA-Binding Sites of B-Cell Lymphoma-Associated Genes in Follicular Lymphoma. Int. J. Mol. Sci. 2020, 21, 8795. https://doi.org/10.3390/ijms21228795
Larrea E, Fernandez-Mercado M, Guerra-Assunção JA, Wang J, Goicoechea I, Gaafar A, Ceberio I, Lobo C, Okosun J, Enright AJ, et al. Identification of Recurrent Mutations in the microRNA-Binding Sites of B-Cell Lymphoma-Associated Genes in Follicular Lymphoma. International Journal of Molecular Sciences. 2020; 21(22):8795. https://doi.org/10.3390/ijms21228795
Chicago/Turabian StyleLarrea, Erika, Marta Fernandez-Mercado, José Afonso Guerra-Assunção, Jun Wang, Ibai Goicoechea, Ayman Gaafar, Izaskun Ceberio, Carmen Lobo, Jessica Okosun, Anton J. Enright, and et al. 2020. "Identification of Recurrent Mutations in the microRNA-Binding Sites of B-Cell Lymphoma-Associated Genes in Follicular Lymphoma" International Journal of Molecular Sciences 21, no. 22: 8795. https://doi.org/10.3390/ijms21228795
APA StyleLarrea, E., Fernandez-Mercado, M., Guerra-Assunção, J. A., Wang, J., Goicoechea, I., Gaafar, A., Ceberio, I., Lobo, C., Okosun, J., Enright, A. J., Fitzgibbon, J., & Lawrie, C. H. (2020). Identification of Recurrent Mutations in the microRNA-Binding Sites of B-Cell Lymphoma-Associated Genes in Follicular Lymphoma. International Journal of Molecular Sciences, 21(22), 8795. https://doi.org/10.3390/ijms21228795






