Comparison of PPAR Ligands as Modulators of Resolution of Inflammation, via Their Influence on Cytokines and Oxylipins Release in Astrocytes
Abstract
:1. Introduction
2. Results
2.1. PPARs Ligands’ Effects on the Synthesis of Oxylipins by Naive and LPS-Stimulated Astrocytes
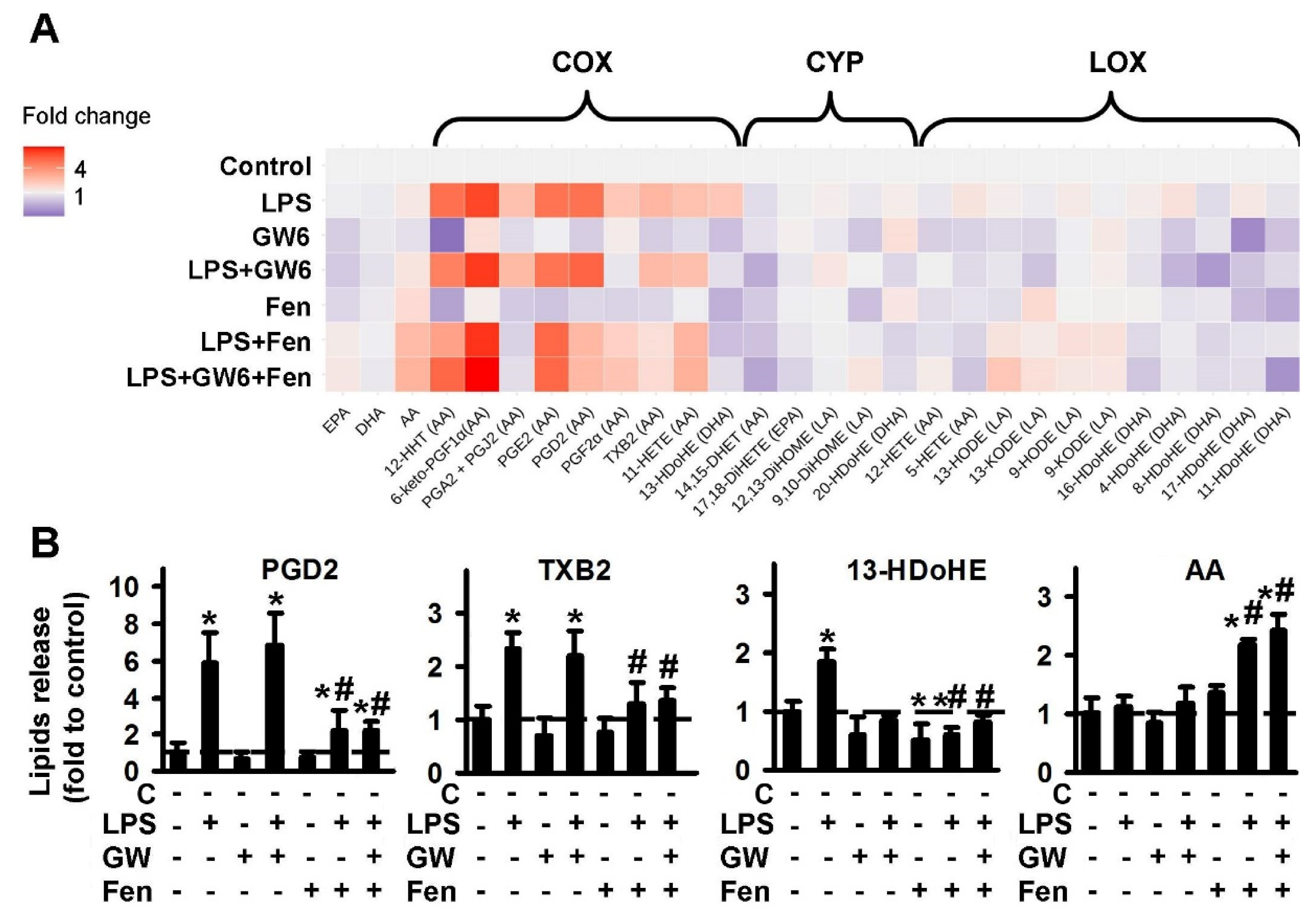
2.1.1. Comparison of PPARα Ligands: Agonist Fenofibrate (Fen), Antagonist GW6471 (GW6)
2.1.2. Comparison of PPARβ Ligands: Agonist GW501516 (GW5), Antagonist and Inverse agonist GSK0660 (GSK)
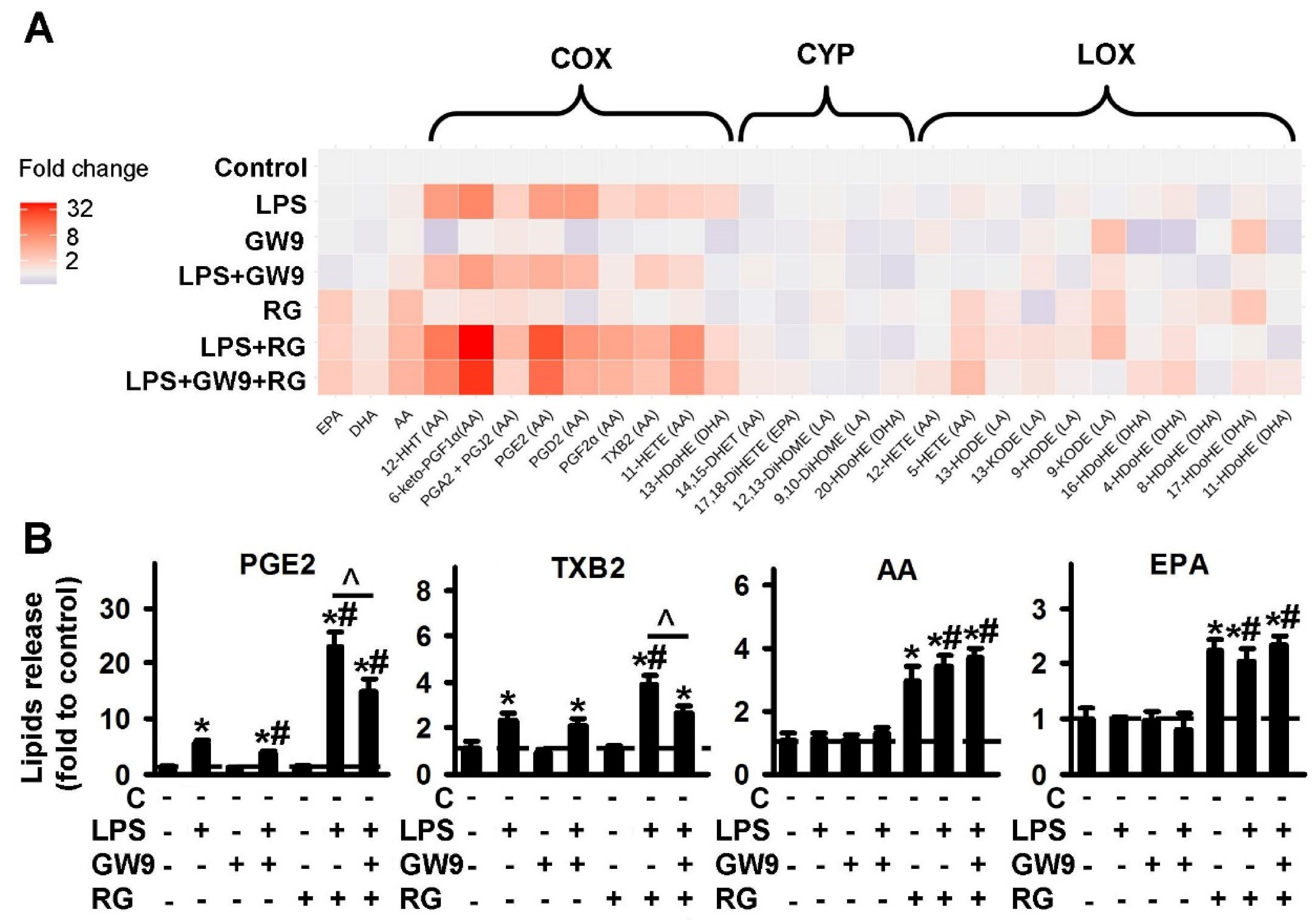
2.1.3. Comparison of PPARγ Ligands: Agonist Rosiglitazone (RG) and Antagonist GW9662 (GW9)
2.2. PPARs Ligands as Modulators of COX-2 Expression
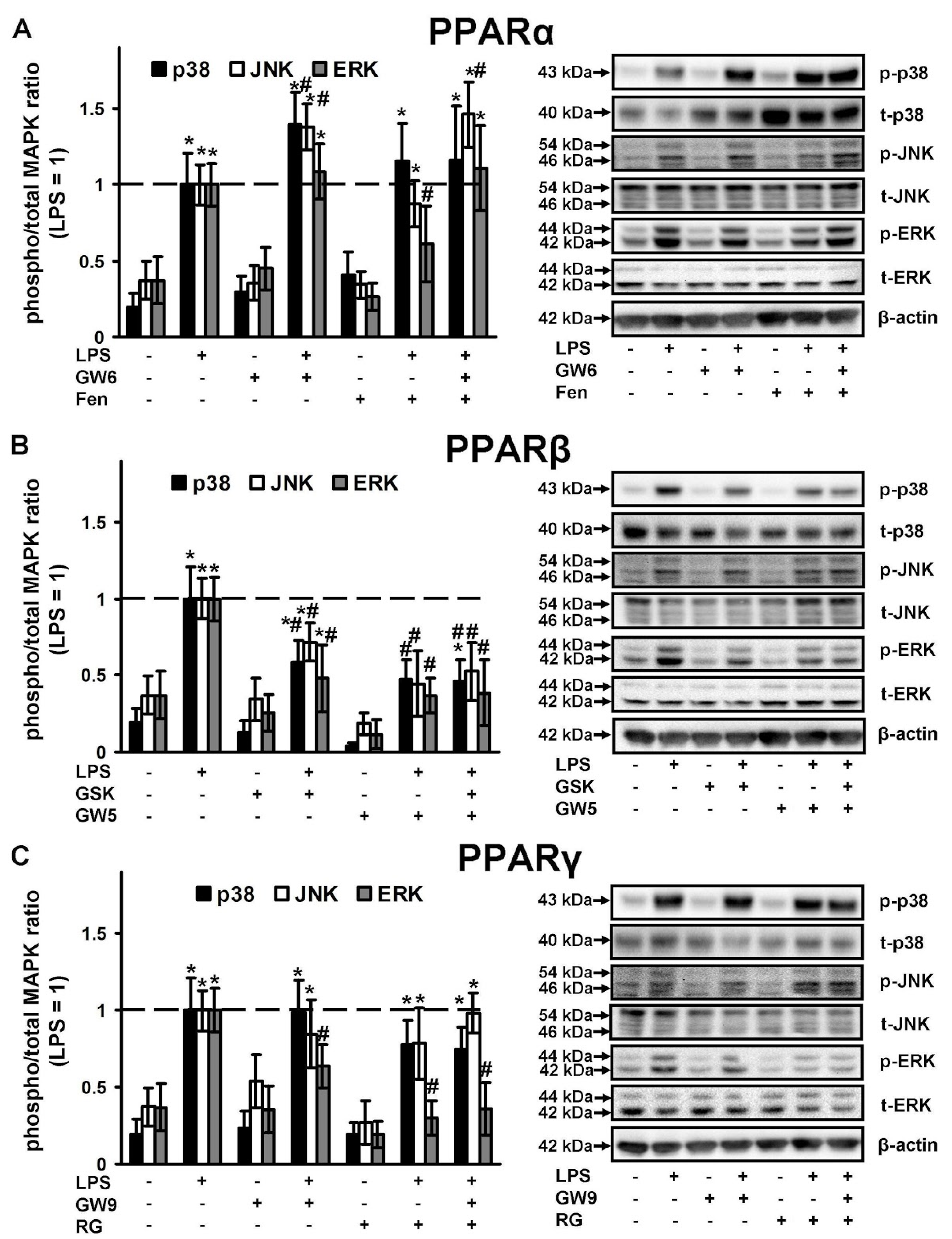
2.3. PPAR Ligands as Modulators of p38, JNK, ERK1\2 Mitogen Activated Protein Kinases (MAPKs)
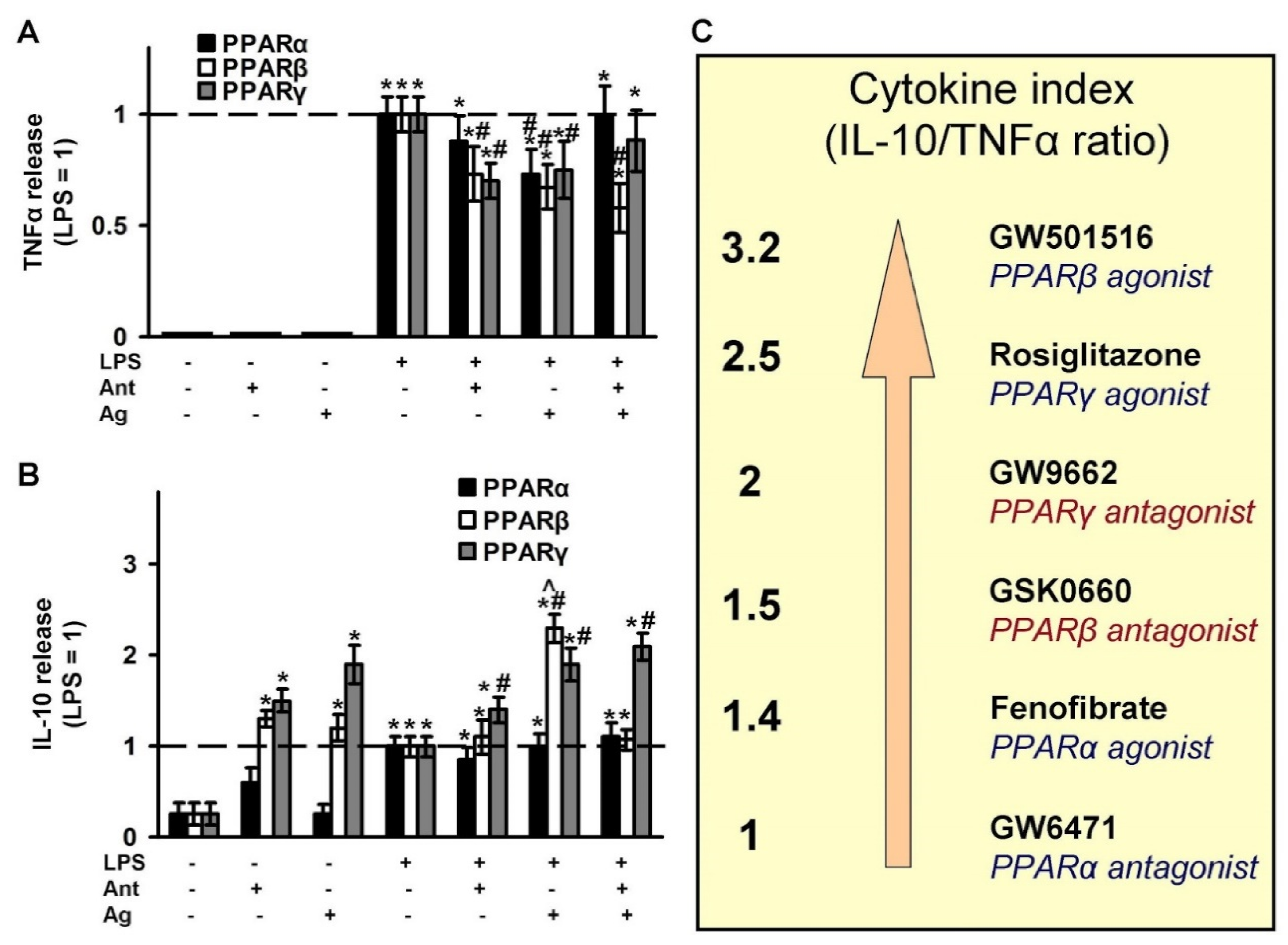
2.4. Effects of PPAR Ligands on Cytokine Markers of Inflammation (TNFα and IL-10)
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Primary Cell Culture
4.3. Western Blot Analysis
4.4. UPLC-MS/MS Conditions and Sample Preparation
4.5. Determination of TNFα, IL-10, IL-6 and Hyaluronic Acid by Enzyme-Linked Immunoassay
4.6. Experimental Data Analysis and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sofroniew, M.V.; Vinters, H.V. Astrocytes: Biology and pathology. Acta Neuropathol. 2010, 119, 7–35. [Google Scholar] [CrossRef] [Green Version]
- Farina, C.; Aloisi, F.; Meinl, E. Astrocytes are active players in cerebral innate immunity. Trends Immunol. 2007, 28, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Aleshin, S.; Strokin, M.; Sergeeva, M.; Reiser, G. Peroxisome proliferator-activated receptor (PPAR)β/δ, a possible nexus of PPARα- and PPARγ-dependent molecular pathways in neurodegenerative diseases: Review and novel hypotheses. Neurochem. Int. 2013, 63, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Sofroniew, M.V. Astrocyte barriers to neurotoxic inflammation. Nat. Rev. Neurosci. 2015, 16, 249–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ransohoff, R.M.; Schafer, D.; Vincent, A.; Blachère, N.E.; Bar-Or, A. Neuroinflammation: Ways in Which the Immune System Affects the Brain. Neurotherapeutics 2015, 12, 896–909. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, J.; Morales, L.; Barreto, G.E. Metabolic and Inflammatory Adaptation of Reactive Astrocytes: Role of PPARs. Mol. Neurobiol. 2017, 54, 2518–2538. [Google Scholar] [CrossRef]
- Medzhitov, R.; Schneider, D.S.; Soares, M.P. Disease tolerance as a defense strategy. Science 2012, 335, 936–941. [Google Scholar] [CrossRef] [Green Version]
- Nathan, C.; Ding, A. Nonresolving Inflammation. Cell 2010, 140, 871–882. [Google Scholar] [CrossRef] [Green Version]
- Serhan, C.N. Treating inflammation and infection in the 21st century: New hints from decoding resolution mediators and mechanisms. FASEB J. 2017, 31, 1273–1288. [Google Scholar] [CrossRef] [Green Version]
- Serhan, C.N.; Brain, S.D.; Buckley, C.D.; Gilroy, D.W.; Haslett, C.; O’Neill, L.A.J.; Perretti, M.; Rossi, A.G.; Wallace, J.L. Resolution of inflammation: State of the art, definitions and terms. FASEB J. 2007, 21, 325–332. [Google Scholar] [CrossRef] [Green Version]
- Hos, D.; Bucher, F.; Regenfuss, B.; Dreisow, M.L.; Bock, F.; Heindl, L.M.; Eming, S.A.; Cursiefen, C. IL-10 Indirectly Regulates Corneal Lymphangiogenesis and Resolution of Inflammation via Macrophages. Am. J. Pathol. 2016, 186, 159–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serhan, C.N. Resolution phase of inflammation: Novel endogenous anti-inflammatory and proresolving lipid mediators and pathways. Annu. Rev. Immunol. 2007, 25, 101–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilroy, D.W.; Bishop-Bailey, D. Lipid mediators in immune regulation and resolution. Br. J. Pharmacol. 2019, 176, 1009–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabbs, M.; Leng, S.; Devassy, J.G.; Monirujjaman, M.; Aukema, H.M. Advances in Our Understanding of Oxylipins Derived from Dietary PUFAs. Adv. Nutr. 2015, 6, 513–540. [Google Scholar] [CrossRef] [Green Version]
- Colas, R.A.; Shinohara, M.; Dalli, J.; Chiang, N.; Serhan, C.N. Identification and signature profiles for pro-resolving and inflammatory lipid mediators in human tissue. Am. J. Physiol. Cell Physiol. 2014, 307, C39–C54. [Google Scholar] [CrossRef] [Green Version]
- Dennis, E.A. Liberating Chiral Lipid Mediators, Inflammatory Enzymes, and LIPID MAPS from Biological Grease. J. Biol. Chem. 2016, 291, 24431–24448. [Google Scholar] [CrossRef] [Green Version]
- Dennis, E.A.; Norris, P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015, 15, 511–523. [Google Scholar] [CrossRef] [Green Version]
- Trépanier, M.-O.; Eiden, M.; Morin-Rivron, D.; Bazinet, R.P.; Masoodi, M. High-resolution lipidomics coupled with rapid fixation reveals novel ischemia-induced signaling in the rat neurolipidome. J. Neurochem. 2017, 140, 766–775. [Google Scholar] [CrossRef] [Green Version]
- Moraes, L.A.; Piqueras, L.; Bishop-Bailey, D. Peroxisome proliferator-activated receptors and inflammation. Pharmacol. Ther. 2006, 110, 371–385. [Google Scholar] [CrossRef]
- Bensinger, S.J.; Tontonoz, P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 2008, 454, 470–477. [Google Scholar] [CrossRef]
- Wahli, W.; Michalik, L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 2012, 23, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Gardner, O.S.; Dewar, B.J.; Graves, L.M. Activation of mitogen-activated protein kinases by peroxisome proliferator-activated receptor ligands: An example of nongenomic signaling. Mol. Pharmacol. 2005, 68, 933–941. [Google Scholar] [CrossRef] [Green Version]
- Bright, J.J.; Kanakasabai, S.; Chearwae, W.; Chakraborty, S. PPAR Regulation of Inflammatory Signaling in CNS Diseases. PPAR Res. 2008, 2008, 658520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berezowski, V.; Fukuda, A.M.; Cecchelli, R.; Badaut, J. Endothelial cells and astrocytes: A concerto en duo in ischemic pathophysiology. Int. J. Cell Biol. 2012, 2012, 176287. [Google Scholar] [CrossRef]
- Schnegg, C.I.; Robbins, M.E. Neuroprotective Mechanisms of PPARdelta: Modulation of Oxidative Stress and Inflammatory Processes. PPAR Res. 2011, 2011, 373560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drew, P.D.; Xu, J.; Storer, P.D.; Chavis, J.A.; Racke, M.K. Peroxisome proliferator-activated receptor agonist regulation of glial activation: Relevance to CNS inflammatory disorders. Neurochem. Int. 2006, 49, 183–189. [Google Scholar] [CrossRef]
- Deplanque, D.; Gelé, P.; Pétrault, O.; Six, I.; Furman, C.; Bouly, M.; Nion, S.; Dupuis, B.; Leys, D.; Fruchart, J.C.; et al. Peroxisome proliferator-activated receptor-α activation as a mechanism of preventive neuroprotection induced by chronic fenofibrate treatment. J. Neurosci. 2003, 23, 6264–6271. [Google Scholar] [CrossRef] [Green Version]
- Sundararajan, S.; Jiang, Q.; Heneka, M.; Landreth, G. PPARgamma as a therapeutic target in central nervous system diseases. Neurochem. Int. 2006, 49, 136–144. [Google Scholar] [CrossRef]
- Dana, N.; Vaseghi, G.; Javanmard, S.H. Crosstalk between peroxisome proliferator-activated receptors and toll-like receptors: A systematic review. Adv. Pharm. Bull. 2019, 9, 12–21. [Google Scholar] [CrossRef]
- Song, G.J.; Nam, Y.; Jo, M.; Jung, M.; Koo, J.Y.; Cho, W.; Koh, M.; Park, S.B.; Suk, K. A novel small-molecule agonist of PPAR-γ potentiates an anti-inflammatory M2 glial phenotype. Neuropharmacology 2016, 109, 159–169. [Google Scholar] [CrossRef]
- Chistyakov, D.V.; Aleshin, S.; Sergeeva, M.G.; Reiser, G. Regulation of peroxisome proliferator-activated receptor β/δ expression and activity levels by toll-like receptor agonists and MAP kinase inhibitors in rat astrocytes. J. Neurochem. 2014, 130, 563–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chistyakov, D.V.; Gavrish, G.E.; Goriainov, S.V.; Chistyakov, V.V.; Astakhova, A.A.; Azbukina, N.V.; Sergeeva, M.G. Oxylipin profiles as functional characteristics of acute inflammatory responses in astrocytes pre-treated with IL-4, IL-10, or LPS. Int. J. Mol. Sci. 2020, 21, 1780. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Wei, X.; Chen, L.; Liu, H.; Liu, X.; Wang, T.; Zhang, X. Anti-inflammatory effect of acetylpuerarin on eicosanoid signaling pathway in primary rat astrocytes. J. Mol. Neurosci. 2014, 52, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Livne-Bar, I.; Wei, J.; Liu, H.-H.; Alqawlaq, S.; Won, G.-J.; Tuccitto, A.; Gronert, K.; Flanagan, J.G.; Sivak, J.M. Astrocyte-derived lipoxins A4 and B4 promote neuroprotection from acute and chronic injury. J. Clin. Investig. 2017, 2, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chistyakov, D.V.; Grabeklis, S.; Goriainov, S.V.; Chistyakov, V.V.; Sergeeva, M.G.; Reiser, G. Astrocytes synthesize primary and cyclopentenone prostaglandins that are negative regulators of their proliferation. Biochem. Biophys. Res. Commun. 2018, 500, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Defaux, A.; Zurich, M.G.; Braissant, O.; Honegger, P.; Monnet-Tschudi, F. Effects of the PPAR-β agonist GW501516 in an in vitro model of brain inflammation and antibody-induced demyelination. J. Neuroinflamm. 2009, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Derogis, P.B.M.C.; Freitas, F.P.; Marques, A.S.F.; Cunha, D.; Appolinário, P.P.; de Paula, F.; Lourenço, T.C.; Murgu, M.; Di Mascio, P.; Medeiros, M.H.G.; et al. The Development of a Specific and Sensitive LC-MS-Based Method for the Detection and Quantification of Hydroperoxy- and Hydroxydocosahexaenoic Acids as a Tool for Lipidomic Analysis. PLoS ONE 2013, 8, e77561. [Google Scholar] [CrossRef] [Green Version]
- Matsunobu, T.; Okuno, T.; Yokoyama, C.; Yokomizo, T. Thromboxane A synthase-independent production of 12- hydroxyheptadecatrienoic acid, a BLT2 ligand. J. Lipid Res. 2013, 54, 2979–2987. [Google Scholar] [CrossRef] [Green Version]
- Van’T Erve, T.J.; Lih, F.B.; Kadiiska, M.B.; Deterding, L.J.; Eling, T.E.; Mason, R.P. Reinterpreting the best biomarker of oxidative stress: The 8-iso-PGF2α/PGF2α ratio distinguishes chemical from enzymatic lipid peroxidation. Free Radic. Biol. Med. 2015, 83, 245–251. [Google Scholar] [CrossRef] [Green Version]
- Picq, M.; Chen, P.; Perez, M.; Michaud, M.; Véricel, E.; Guichardant, M.; Lagarde, M. DHA metabolism: Targeting the brain and lipoxygenation. Mol. Neurobiol. 2010, 42, 48–51. [Google Scholar] [CrossRef] [Green Version]
- Guichardant, M.; Véricel, E.; Lagarde, M. Biological relevance of double lipoxygenase products of polyunsaturated fatty acids, especially within blood vessels and brain. Biochimie 2019, 159, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Pickens, C.A.; Sordillo, L.M.; Zhang, C.; Fenton, J.I. Obesity is positively associated with arachidonic acid-derived 5- and 11-hydroxyeicosatetraenoic acid (HETE). Metabolism 2017, 70, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.S.; Chan, H.F.; Leung, H.H.; Galano, J.M.; Oger, C.; Durand, T.; Lee, J.C.Y. Short-time UVA exposure to human keratinocytes instigated polyunsaturated fatty acid without inducing lipid peroxidation. Free Radic. Res. 2017, 51, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Omeragic, A.; Kara-Yacoubian, N.; Kelschenbach, J.; Sahin, C.; Cummins, C.L.; Volsky, D.J.; Bendayan, R. Peroxisome Proliferator-Activated Receptor-gamma agonists exhibit anti-inflammatory and antiviral effects in an EcoHIV mouse model. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Chistyakov, D.V.; Azbukina, N.V.; Astakhova, A.A.; Goriainov, S.V.; Chistyakov, V.V.; Sergeeva, M.G. Sex-mediated differences in lps induced alterations of TNFα, IL-10 expression, and prostaglandin synthesis in primary astrocytes. Int. J. Mol. Sci. 2018, 19, 2793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astakhova, A.A.; Chistyakov, D.V.; Pankevich, E.V.; Sergeeva, M.G. Regulation of cyclooxygenase 2 expression by agonists of PPAR nuclear receptors in the model of endotoxin tolerance in astrocytes. Biochemistry 2015, 80, 1262–1270. [Google Scholar] [CrossRef]
- Kaminska, B.; Gozdz, A.; Zawadzka, M.; Ellert-Miklaszewska, A.; Lipko, M. MAPK signal transduction underlying brain inflammation and gliosis as therapeutic target. Anat. Rec. 2009, 292, 1902–1913. [Google Scholar] [CrossRef]
- Chistyakov, D.V.; Aleshin, S.E.; Astakhova, A.A.; Sergeeva, M.G.; Reiser, G. Regulation of peroxisome proliferator-activated receptors (PPAR) α and -γ of rat brain astrocytes in the course of activation by toll-like receptor agonists. J. Neurochem. 2015, 134, 113–124. [Google Scholar] [CrossRef]
- Chistyakov, D.V.; Azbukina, N.V.; Astakhova, A.A.; Polozhintsev, A.I.; Sergeeva, M.G.; Reiser, G. Toll-like receptors control p38 and JNK MAPK signaling pathways in rat astrocytes differently, when cultured in normal or high glucose concentrations. Neurochem. Int. 2019, 131, 104513. [Google Scholar] [CrossRef]
- Chistyakov, D.V.; Nikolskaya, A.I.; Goriainov, S.V.; Astakhova, A.A.; Sergeeva, M.G. Inhibitor of hyaluronic acid synthesis 4-methylumbelliferone as an anti-inflammatory modulator of lps-mediated astrocyte responses. Int. J. Mol. Sci. 2020, 21, 8203. [Google Scholar] [CrossRef]
- Burmeister, A.R.; Marriott, I. The interleukin-10 family of cytokines and their role in the CNS. Front. Cell. Neurosci. 2018, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpentier, P.A.; Duncan, D.S.; Miller, S.D. Glial toll-like receptor signaling in central nervous system infection and autoimmunity. Brain. Behav. Immun. 2008, 22, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Chistyakov, D.V.; Azbukina, N.V.; Lopachev, A.V.; Kulichenkova, K.N.; Astakhova, A.A.; Sergeeva, M.G. Rosiglitazone as a Modulator of TLR4 and TLR3 Signaling Pathways in Rat Primary Neurons and Astrocytes. Int. J. Mol. Sci. 2018, 19, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schumann, R.R.; Pfeil, D.; Freyer, D.; Buerger, W.; Lamping, N.; Kirschning, C.J.; Goebel, U.B.; Weber, J.R. Lipopolysaccharide and pneumococcal cell wall components activate the mitogen activated protein kinases (MAPK) erk-1, erk-2, and p38 in astrocytes. Glia 1998, 22, 295–305. [Google Scholar] [CrossRef]
- Cristiano, L.; Bernardo, A.; Cerù, M.P. Peroxisome proliferator-activated receptors (PPARs) and peroxisomes in rat cortical and cerebellar astrocytes. J. Neurocytol. 2001, 30, 671–683. [Google Scholar] [CrossRef]
- Aleshin, S.; Grabeklis, S.; Hanck, T.; Sergeeva, M.; Reiser, G. Peroxisome proliferator-activated receptor (PPAR)-gamma positively controls and PPARalpha negatively controls cyclooxygenase-2 expression in rat brain astrocytes through a convergence on PPARbeta/delta via mutual control of PPAR expression levels. Mol. Pharmacol. 2009, 76, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chavis, J.A.; Racke, M.K.; Drew, P.D. Peroxisome proliferator-activated receptor-α and retinoid X receptor agonists inhibit inflammatory responses of astrocytes. J. Neuroimmunol. 2006, 176, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Bazinet, R.P.; Layé, S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- Famitafreshi, H.; Karimian, M. Prostaglandins as the Agents That Modulate the Course of Brain Disorders. Degener. Neurol. Neuromuscul. Dis. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Tzeng, S.F.; Hsiao, H.Y.; Mak, O.T. Prostaglandins and cyclooxygenases in glial cells during brain inflammation. Curr. Drug Targets Inflamm. Allergy 2005, 4, 335–340. [Google Scholar] [CrossRef]
- Kim, E.J.; Kwon, K.J.; Park, J.Y.; Lee, S.H.; Moon, C.H.; Baik, E.J. Neuroprotective effects of prostaglandin E2 or cAMP against microglial and neuronal free radical mediated toxicity associated with inflammation. J. Neurosci. Res. 2002, 70, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Rothhammer, V.; Quintana, F.J. Control of autoimmune CNS inflammation by astrocytes. Semin. Immunopathol. 2015, 37, 625–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, W.; Zong, Y.; Mohammad, A.; Ajit, D.; Cui, J.; Han, D.; Hamilton, J.L.; Simonyi, A.; Sun, A.Y.; Gu, Z.; et al. Pro-inflammatory cytokines and lipopolysaccharide induce changes in cell morphology, and upregulation of ERK1/2, iNOS and sPLA2-IIA expression in astrocytes and microglia. J. Neuroinflamm. 2011, 8, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storer, P.D.; Xu, J.; Chavis, J.; Drew, P.D. Peroxisome proliferator-activated receptor-gamma agonists inhibit the activation of microglia and astrocytes: Implications for multiple sclerosis. J. Neuroimmunol. 2005, 161, 113–122. [Google Scholar] [CrossRef]
- Pankevich, E.V.V.; Astakhova, A.A.A.; Chistyakov, D.V.V.; Sergeeva, M.G.G. Antiinflammatory effect of rosiglitazone via modulation of mRNA stability of interleukin 10 and cyclooxygenase 2 in astrocytes. Biochemistry 2017, 82, 1276–1284. [Google Scholar] [CrossRef]
- Chawla, A.; Barak, Y.; Nagy, L.; Liao, D.; Tontonoz, P.; Evans, R.M. PPAR-γ dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat. Med. 2001, 7, 48–52. [Google Scholar] [CrossRef]
- Zhang, C.; Deng, J.; Liu, D.; Tuo, X.; Yu, Y.; Yang, H.; Wang, N. Nuciferine inhibits proinflammatory cytokines via the PPARs in LPS-induced RAW264.7 cells. Molecules 2018, 23, 2723. [Google Scholar] [CrossRef] [Green Version]
- Sergeeva, M.G.; Aleshin, S.E.; Grabeklis, S.; Reiser, G. PPAR activation has dichotomous control on the expression levels of cytosolic and secretory phospholipase A2 in astrocytes; inhibition in naïve, untreated cells and enhancement in LPS-stimulated cells. J. Neurochem. 2010, 115, 399–410. [Google Scholar] [CrossRef]
- Zizzo, G.; Cohen, P.L. The PPAR-γ antagonist GW9662 elicits differentiation of M2c-like cells and upregulation of the MerTK/Gas6 axis: A key role for PPAR-γ in human macrophage polarization. J. Inflamm. 2015, 12. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Calvo, R.; Serrano, L.; Coll, T.; Moullan, N.; Sánchez, R.M.; Merlos, M.; Palomer, X.; Laguna, J.C.; Michalik, L.; Wahli, W.; et al. Activation of peroxisome proliferator-activated receptor β/δ inhibits lipopolysaccharide-induced cytokine production in adipocytes by lowering nuclear factor-κB activity via extracellular signal-related kinase 1/2. Diabetes 2008, 57, 2149–2157. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.; Wang, H.; Hajishengallis, G.N.; Martin, M. TLR-signaling networks: An integration of adaptor molecules, kinases, and cross-talk. J. Dent. Res. 2011, 90, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Chistyakov, D.V.; Astakhova, A.A.; Sergeeva, M.G. Resolution of inflammation and mood disorders. Exp. Mol. Pathol. 2018, 105, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Dougherty, E.J.; Danner, R.L. PPARγ signaling and emerging opportunities for improved therapeutics. Pharmacol. Res. 2016, 111, 76–85. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chistyakov, D.V.; Astakhova, A.A.; Goriainov, S.V.; Sergeeva, M.G. Comparison of PPAR Ligands as Modulators of Resolution of Inflammation, via Their Influence on Cytokines and Oxylipins Release in Astrocytes. Int. J. Mol. Sci. 2020, 21, 9577. https://doi.org/10.3390/ijms21249577
Chistyakov DV, Astakhova AA, Goriainov SV, Sergeeva MG. Comparison of PPAR Ligands as Modulators of Resolution of Inflammation, via Their Influence on Cytokines and Oxylipins Release in Astrocytes. International Journal of Molecular Sciences. 2020; 21(24):9577. https://doi.org/10.3390/ijms21249577
Chicago/Turabian StyleChistyakov, Dmitry V., Alina A. Astakhova, Sergei V. Goriainov, and Marina G. Sergeeva. 2020. "Comparison of PPAR Ligands as Modulators of Resolution of Inflammation, via Their Influence on Cytokines and Oxylipins Release in Astrocytes" International Journal of Molecular Sciences 21, no. 24: 9577. https://doi.org/10.3390/ijms21249577
APA StyleChistyakov, D. V., Astakhova, A. A., Goriainov, S. V., & Sergeeva, M. G. (2020). Comparison of PPAR Ligands as Modulators of Resolution of Inflammation, via Their Influence on Cytokines and Oxylipins Release in Astrocytes. International Journal of Molecular Sciences, 21(24), 9577. https://doi.org/10.3390/ijms21249577




