Zika M Oligopeptide ZAMP Confers Cell Death-Promoting Capability to a Soluble Tumor-Associated Antigen through Caspase-3/7 Activation
Abstract
1. Introduction
2. Results
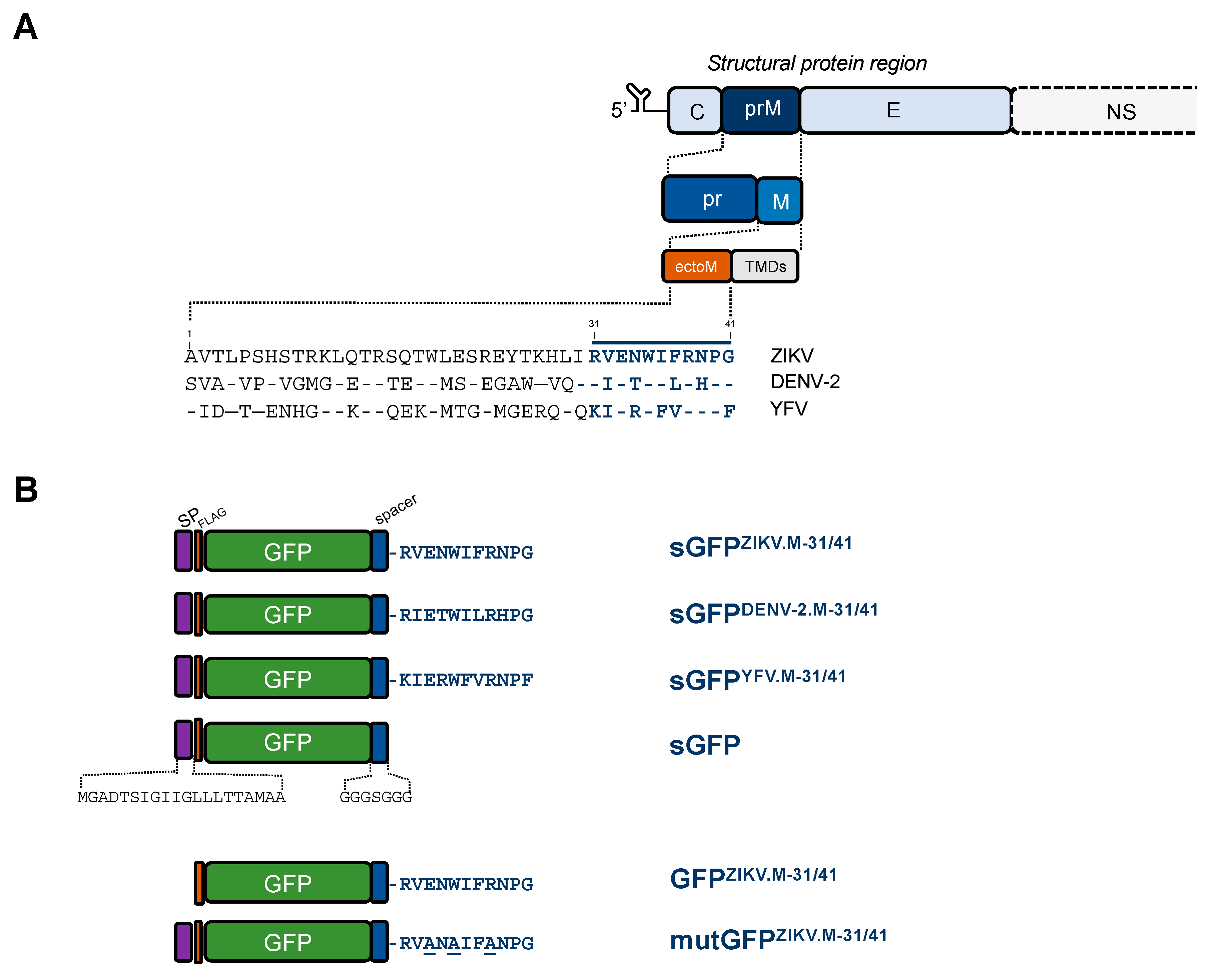
2.1. Apoptosis-Inducing Ability of a Recombinant GFP Protein Carrying the ZIKV Residues M-31/41
2.2. ZIKV M Oligopeptide Confers Death-Promoting Capability to MPF
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Antibodies
4.2. Vector Plasmids Expressing GFP and MPF Constructs
4.3. Flow Cytometry Analysis
4.4. Lactate Dehydrogenase Assay
4.5. MTT Assay
4.6. Caspase 3/7 Enzymatic Activity
4.7. Early Apoptosis Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ApoptoM | Dengue M apoptotic oligopeptide |
| C | Capsid protein |
| CNS | Central nervous system |
| DENV | Dengue virus |
| DENV-2 | Dengue virus type 2 |
| DMSO | Dimethyl sulfoxide |
| E | Envelope protein |
| ectoM | M ectodomain |
| ER | Endoplasmic reticulum |
| FBS | Fetal bovine serum |
| GFP | Green fluorescent protein |
| LDH | Lactate dehydrogenase |
| M | Membrane protein |
| MEM | Minimum Essential Media |
| MFI | Mean fluorescence intensity |
| MPF | Megakaryocyte-potentiating factor |
| MSLN | Mesothelin |
| NS | Nonstructural protein |
| PBS | Phosphate buffered saline |
| PFA | Paraformaldehyde |
| prM | Intracellular precursor of membrane protein |
| PS | Phosphatidylserine |
| sGFP | Soluble green fluorescent protein |
| SP | Signal peptide |
| SPHK2 | Sphingosine kinase 2 |
| TAA | Tumor-associated antigen |
| WNV | West Nile virus |
| YFV | Yellow Fever virus |
| ZAMP | Zika Apoptosis M Peptide |
| ZIKV | Zika virus |
References
- Solomon, I.H.; Milner, D.A.; Folkerth, R.D. Neuropathology of Zika Virus Infection. J. Neuroinfect. Dis. 2016, 7. [Google Scholar] [CrossRef]
- Cao-Lormeau, V.M.; Blake, A.; Mons, S.; Lastere, S.; Roche, C.; Vanhomwegen, J.; Dub, T.; Baudouin, L.; Teissier, A.; Larre, P.; et al. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet 2016, 387, 1531–1539. [Google Scholar] [CrossRef]
- Rubin, E.J.; Greene, M.F.; Baden, L.R. Zika Virus and Microcephaly. N. Engl. J. Med. 2016, 374, 984–985. [Google Scholar] [CrossRef]
- Haddow, A.D.; Schuh, A.J.; Yasuda, C.Y.; Kasper, M.R.; Heang, V.; Huy, R.; Guzman, H.; Tesh, R.B.; Weaver, S.C. Genetic characterization of Zika virus strains: Geographic expansion of the Asian lineage. PLoS Negl. Trop. Dis. 2012, 6, e1477. [Google Scholar] [CrossRef]
- Pan, P.; Zhang, Q.; Liu, W.; Wang, W.; Lao, Z.; Zhang, W.; Shen, M.; Wan, P.; Xiao, F.; Liu, F.; et al. Dengue Virus M Protein Promotes NLRP3 Inflammasome Activation To Induce Vascular Leakage in Mice. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Catteau, A.; Kalinina, O.; Wagner, M.C.; Deubel, V.; Courageot, M.P.; Despres, P. Dengue virus M protein contains a proapoptotic sequence referred to as ApoptoM. J. Gen. Virol. 2003, 84 Pt 10, 2781–2793. [Google Scholar] [CrossRef]
- Catteau, A.; Roue, G.; Yuste, V.J.; Susin, S.A.; Despres, P. Expression of dengue ApoptoM sequence results in disruption of mitochondrial potential and caspase activation. Biochimie 2003, 85, 789–793. [Google Scholar] [CrossRef]
- Chang, K.; Pastan, I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc. Natl. Acad. Sci. USA 1996, 93, 136–140. [Google Scholar] [CrossRef]
- Morello, A.; Sadelain, M.; Adusumilli, P.S. Mesothelin-Targeted CARs: Driving T Cells to Solid Tumors. Cancer Discov. 2016, 6, 133–146. [Google Scholar] [CrossRef]
- Ho, M.; Bera, T.K.; Willingham, M.C.; Onda, M.; Hassan, R.; FitzGerald, D.; Pastan, I. Mesothelin expression in human lung cancer. Clin. Cancer Res. 2007, 13, 1571–1575. [Google Scholar] [CrossRef]
- Chong, H.Y.; Leow, C.Y.; Abdul Majeed, A.B.; Leow, C.H. Flavivirus infection-A review of immunopathogenesis, immunological response, and immunodiagnosis. Virus Res. 2019, 274, 197770. [Google Scholar] [CrossRef]
- Pryor, M.J.; Azzola, L.; Wright, P.J.; Davidson, A.D. Histidine 39 in the dengue virus type 2 M protein has an important role in virus assembly. J. Gen. Virol. 2004, 85 Pt 12, 3627–3636. [Google Scholar] [CrossRef]
- Yoshii, K.; Igarashi, M.; Ichii, O.; Yokozawa, K.; Ito, K.; Kariwa, H.; Takashima, I. A conserved region in the prM protein is a critical determinant in the assembly of flavivirus particles. J. Gen. Virol. 2012, 93 Pt 1, 27–38. [Google Scholar] [CrossRef]
- Hsieh, S.C.; Wu, Y.C.; Zou, G.; Nerurkar, V.R.; Shi, P.Y.; Wang, W.K. Highly conserved residues in the helical domain of dengue virus type 1 precursor membrane protein are involved in assembly, precursor membrane (prM) protein cleavage, and entry. J. Biol. Chem. 2014, 289, 33149–33160. [Google Scholar] [CrossRef]
- Peng, J.G.; Wu, S.C. Glutamic acid at residue 125 of the prM helix domain interacts with positively charged amino acids in E protein domain II for Japanese encephalitis virus-like-particle production. J. Virol. 2014, 88, 8386–8396. [Google Scholar] [CrossRef]
- Brown, E.; Brown, E.; Bentham, M.; Beaumont, H.; Bloy, A.; Thompson, R.; McKimmie, C.; Foster, R.; Ranson, N.; Macdonald, A.; et al. Flavivirus membrane (M) proteins as potential ion channel antiviral. Microbiology 2019, 1. [Google Scholar] [CrossRef]
- Brabant, M.; Baux, L.; Casimir, R.; Briand, J.P.; Chaloin, O.; Porceddu, M.; Buron, N.; Chauvier, D.; Lassalle, M.; Lecoeur, H.; et al. A flavivirus protein M-derived peptide directly permeabilizes mitochondrial membranes, triggers cell death and reduces human tumor growth in nude mice. Apoptosis 2009, 14, 1190–1203. [Google Scholar] [CrossRef]
- Souza, B.S.; Sampaio, G.L.; Pereira, C.S.; Campos, G.S.; Sardi, S.I.; Freitas, L.A.; Figueira, C.P.; Paredes, B.D.; Nonaka, C.K.; Azevedo, C.M.; et al. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci. Rep. 2016, 6, 39775. [Google Scholar] [CrossRef]
- Li, C.; Xu, D.; Ye, Q.; Hong, S.; Jiang, Y.; Liu, X.; Zhang, N.; Shi, L.; Qin, C.F.; Xu, Z. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell 2016, 19, 672. [Google Scholar] [CrossRef]
- Li, G.; Poulsen, M.; Yashiroda, Y.; Fenyvuesvolgyi, C.; Yashiroda, Y.; Yoshida, M.; Simard, J.M.; Gallo, R.C.; Zhao, R.Y. Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast. Proc. Natl. Acad. Sci. USA 2017, 114, E376–E385. [Google Scholar] [CrossRef]
- Rosa-Fernandes, L.; Cugola, F.R.; Russo, F.B.; Kawahara, R.; de Melo Freire, C.C.; Leite, P.E.C.; Bassi Stern, A.C.; Angeli, C.B.; de Oliveira, D.B.L.; Melo, S.R.; et al. Zika Virus Impairs Neurogenesis and Synaptogenesis Pathways in Human Neural Stem Cells and Neurons. Front. Cell. Neurosci. 2019, 13, 64. [Google Scholar] [CrossRef]
- Liu, J.; Li, Q.; Li, X.; Qiu, Z.; Li, A.; Liang, W.; Chen, H.; Cai, X.; Chen, X.; Duan, X.; et al. Zika Virus Envelope Protein induces G2/M Cell Cycle Arrest and Apoptosis via an Intrinsic Cell Death Signaling Pathway in Neuroendocrine PC12 Cells. Int. J. Biol. Sci. 2018, 14, 1099–1108. [Google Scholar] [CrossRef]
- de Wispelaere, M.; Khou, C.; Frenkiel, M.P.; Despres, P.; Pardigon, N. A Single Amino Acid Substitution in the M Protein Attenuates Japanese Encephalitis Virus in Mammalian Hosts. J. Virol. 2015, 90, 2676–2689. [Google Scholar] [CrossRef]
- Frumence, E.; Viranaicken, W.; Bos, S.; Alvarez-Martinez, M.T.; Roche, M.; Arnaud, J.D.; Gadea, G.; Despres, P. A Chimeric Zika Virus between Viral Strains MR766 and BeH819015 Highlights a Role for E-glycan Loop in Antibody-mediated Virus Neutralization. Vaccines 2019, 7, 55. [Google Scholar] [CrossRef]
- Parquet, M.C.; Kumatori, A.; Hasebe, F.; Morita, K.; Igarashi, A. West Nile virus-induced bax-dependent apoptosis. FEBS Lett. 2001, 500, 17–24. [Google Scholar] [CrossRef]
- Parquet, M.C.; Kumatori, A.; Hasebe, F.; Mathenge, E.G.; Morita, K. St. Louis encephalitis virus induced pathology in cultured cells. Arch. Virol. 2002, 147, 1105–1119. [Google Scholar] [CrossRef]
- Suzuki, T.; Okamoto, T.; Katoh, H.; Sugiyama, Y.; Kusakabe, S.; Tokunaga, M.; Hirano, J.; Miyata, Y.; Fukuhara, T.; Ikawa, M.; et al. Infection with flaviviruses requires BCLXL for cell survival. PLoS Pathog. 2018, 14, e1007299. [Google Scholar] [CrossRef]
- Morchang, A.; Lee, R.C.H.; Yenchitsomanus, P.T.; Sreekanth, G.P.; Noisakran, S.; Chu, J.J.H.; Limjindaporn, T. RNAi screen reveals a role of SPHK2 in dengue virus-mediated apoptosis in hepatic cell lines. PLoS ONE 2017, 12, e0188121. [Google Scholar] [CrossRef]
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Levine, A.J.; Oren, M. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer 2009, 9, 749–758. [Google Scholar] [CrossRef]
- Lu, W.; Cheng, F.; Yan, W.; Li, X.; Yao, X.; Song, W.; Liu, M.; Shen, X.; Jiang, H.; Chen, J.; et al. Selective targeting p53(WT) lung cancer cells harboring homozygous p53 Arg72 by an inhibitor of CypA. Oncogene 2017, 36, 4719–4731. [Google Scholar] [CrossRef] [PubMed]
- Bressac, B.; Galvin, K.M.; Liang, T.J.; Isselbacher, K.J.; Wands, J.R.; Ozturk, M. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 1990, 87, 1973–1977. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Old, L.J. Cancer-testis antigens: Targets for cancer immunotherapy. Cancer J. Sci. Am. 1999, 5, 16–17. [Google Scholar] [PubMed]
- Ribas, A.; Weber, J.S.; Chmielowski, B.; Comin-Anduix, B.; Lu, D.; Douek, M.; Ragavendra, N.; Raman, S.; Seja, E.; Rosario, D.; et al. Intra-lymph node prime-boost vaccination against Melan A and tyrosinase for the treatment of metastatic melanoma: Results of a phase 1 clinical trial. Clin. Cancer Res. 2011, 17, 2987–2996. [Google Scholar] [CrossRef] [PubMed]
- van Meerten, T.; Hagenbeek, A. CD20-targeted therapy: The next generation of antibodies. Semin. Hematol. 2010, 47, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Dermime, S.; Gilham, D.E.; Shaw, D.M.; Davidson, E.J.; Meziane, E.-K.; Armstrong, A.; Hawkins, R.E.; Stern, P.L. Vaccine and antibody-directed T cell tumour immunotherapy. Biochim. Biophys. Acta 2004, 1704, 11–35. [Google Scholar] [CrossRef]
- Elkord, E.; Burt, D.J.; Drijfhout, J.W.; Hawkins, R.E.; Stern, P.L. CD4+ T-cell recognition of human 5T4 oncofoetal antigen: Implications for initial depletion of CD25+ T cells. Cancer Immunol. Immunother. 2008, 57, 833–847. [Google Scholar] [CrossRef]
- Zhu, Z.; Gorman, M.J.; McKenzie, L.D.; Chai, J.N.; Hubert, C.G.; Prager, B.C.; Fernandez, E.; Richner, J.M.; Zhang, R.; Shan, C.; et al. Correction: Zika virus has oncolytic activity against glioblastoma stem cells. J. Exp. Med. 2017, 214, 3145. [Google Scholar] [CrossRef]
- Kaid, C.; Goulart, E.; Caires-Junior, L.C.; Araujo, B.H.S.; Soares-Schanoski, A.; Bueno, H.M.S.; Telles-Silva, K.A.; Astray, R.M.; Assoni, A.F.; Junior, A.F.R.; et al. Zika Virus Selectively Kills Aggressive Human Embryonal CNS Tumor Cells In Vitro and In Vivo. Cancer Res. 2018, 78, 3363–3374. [Google Scholar] [CrossRef]
- Chen, Q.; Wu, J.; Ye, Q.; Ma, F.; Zhu, Q.; Wu, Y.; Shan, C.; Xie, X.; Li, D.; Zhan, X.; et al. Erratum for Chen et al., “Treatment of Human Glioblastoma with a Live Attenuated Zika Virus Vaccine Candidate”. mBio 2019, 10. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanwalscappel, B.; Haddad, J.G.; Almokdad, R.; Decotter, J.; Gadea, G.; Desprès, P. Zika M Oligopeptide ZAMP Confers Cell Death-Promoting Capability to a Soluble Tumor-Associated Antigen through Caspase-3/7 Activation. Int. J. Mol. Sci. 2020, 21, 9578. https://doi.org/10.3390/ijms21249578
Vanwalscappel B, Haddad JG, Almokdad R, Decotter J, Gadea G, Desprès P. Zika M Oligopeptide ZAMP Confers Cell Death-Promoting Capability to a Soluble Tumor-Associated Antigen through Caspase-3/7 Activation. International Journal of Molecular Sciences. 2020; 21(24):9578. https://doi.org/10.3390/ijms21249578
Chicago/Turabian StyleVanwalscappel, Bénédicte, Juliano G. Haddad, Roba Almokdad, Jason Decotter, Gilles Gadea, and Philippe Desprès. 2020. "Zika M Oligopeptide ZAMP Confers Cell Death-Promoting Capability to a Soluble Tumor-Associated Antigen through Caspase-3/7 Activation" International Journal of Molecular Sciences 21, no. 24: 9578. https://doi.org/10.3390/ijms21249578
APA StyleVanwalscappel, B., Haddad, J. G., Almokdad, R., Decotter, J., Gadea, G., & Desprès, P. (2020). Zika M Oligopeptide ZAMP Confers Cell Death-Promoting Capability to a Soluble Tumor-Associated Antigen through Caspase-3/7 Activation. International Journal of Molecular Sciences, 21(24), 9578. https://doi.org/10.3390/ijms21249578





