Myoblast Migration and Directional Persistence Affected by Syndecan-4-Mediated Tiam-1 Expression and Distribution
Abstract
:1. Introduction
2. Results
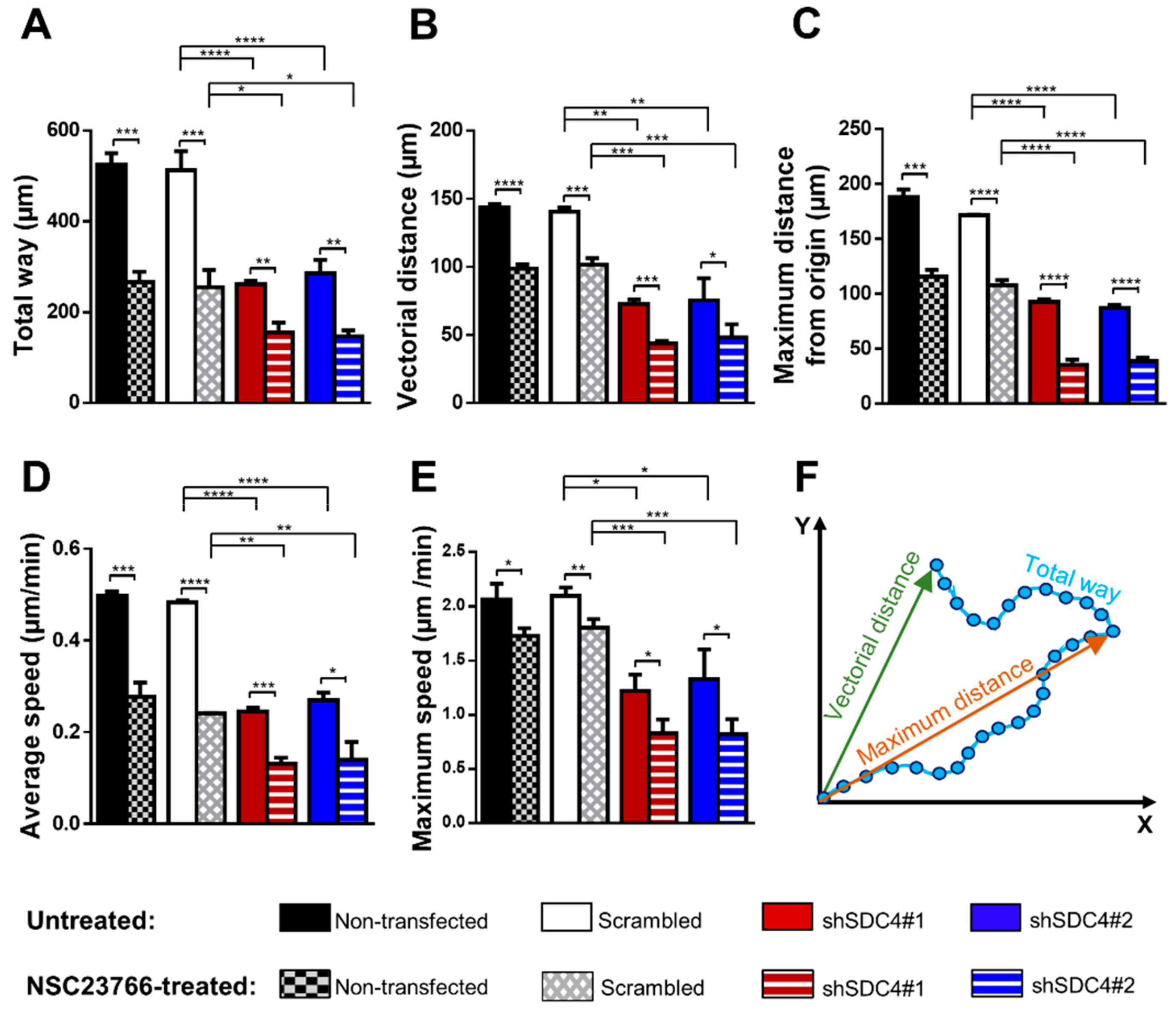
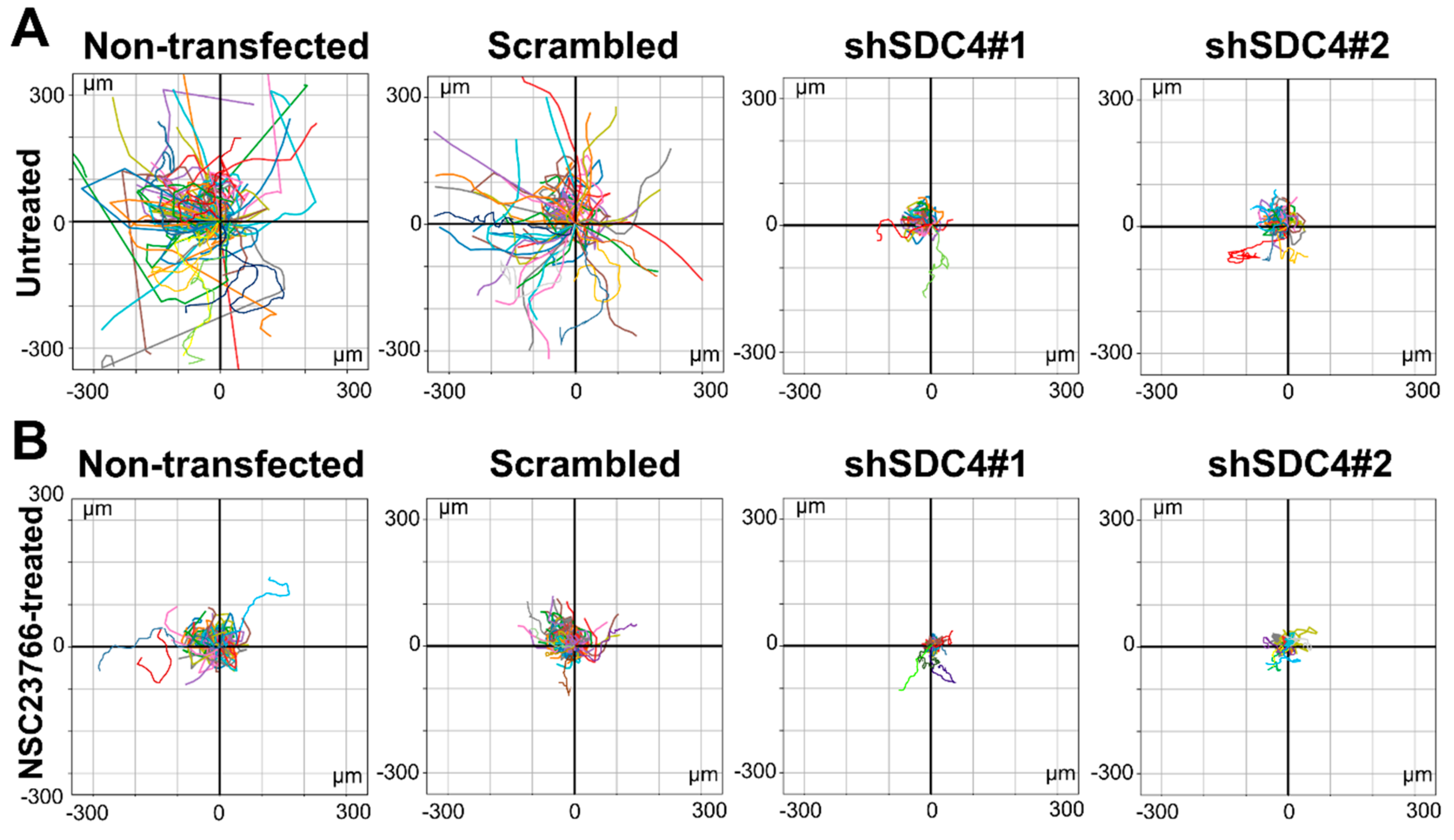
2.1. Syndecan-4 Silencing Reduces Myoblast Migration in a Random Migration Assay
2.2. Inhibition of Rac1 Does Not Restore the Defective Migratory Phenotype of Syndecan-4 Knockdown Cells
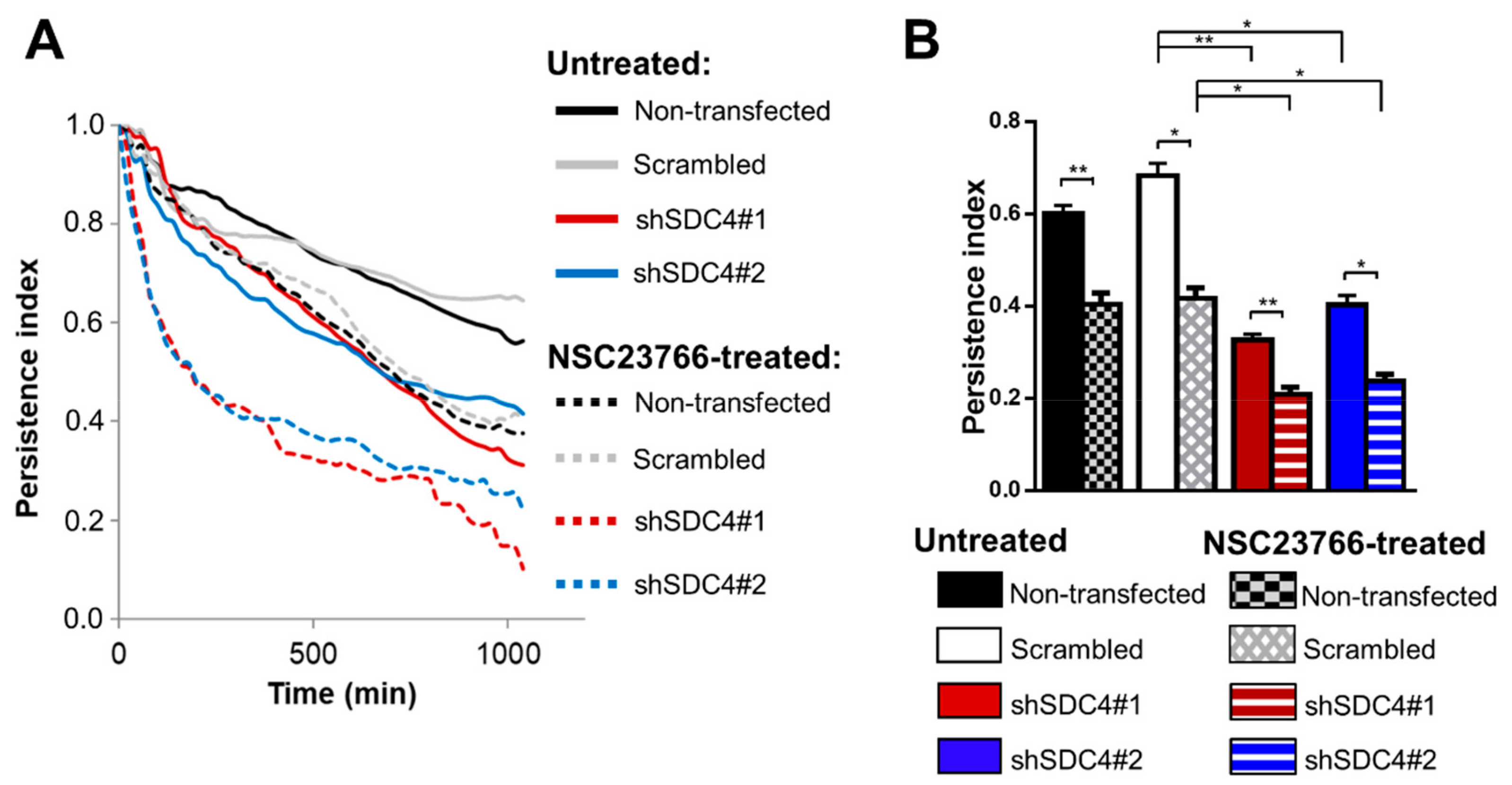
2.3. Syndecan-4 Knockdown Affects the Directional Persistence of Migration
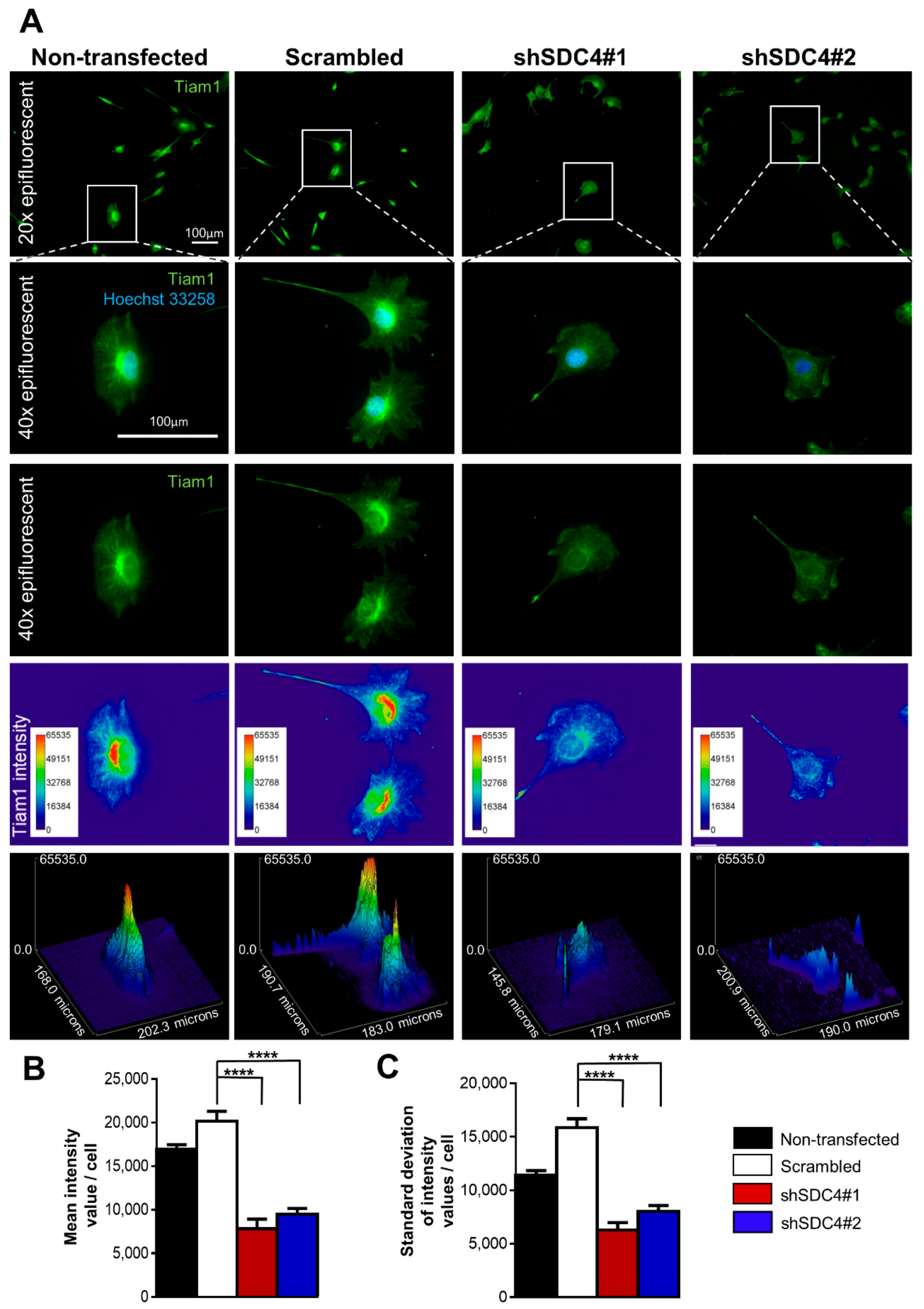
2.4. Syndecan-4 Affects Tiam1 Expression and Localization
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Plasmids
4.2. Time-Lapse Imaging of Live Cells
4.3. Single-Cell Tracking of Cultured Myoblasts
4.4. Trajectories and Wind-Rose Diagrams
4.5. Fluorescent Staining and Microscopy
4.6. Evaluation of Tiam1 Immunostaining
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GDI | Guanine dissociation inhibitors |
| GEF | Guanine nucleotide exchange factor |
| GAP | GTPase activating protein |
| Par | Partitioning defective |
| PKCα | Protein kinase C α |
| Rac1 | Ras-related C3 botulinum toxin substrate 1 |
| SDC4 | Syndecan-4 |
| Tiam1 | T-lymphoma invasion and metastasis-1 |
References
- Mauro, A.; Adams, W.R. The structure of the sarcolemma of the frog skeletal muscle fiber. J. Biophys. Biochem. Cytol. 1961, 10, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Chang, N.C.; Rudnicki, M.A. Satellite cells: The architects of skeletal muscle. Curr. Top. Dev. Biol. 2014, 107, 161–181. [Google Scholar] [CrossRef]
- Trepat, X.; Chen, Z.; Jacobson, K. Cell migration. Compr. Physiol. 2012, 2, 2369–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridley, A.J.; Schwartz, M.A.; Burridge, K.; Firtel, R.A.; Ginsberg, M.H.; Borisy, G.; Parsons, J.T.; Horwitz, A.R. Cell migration: integrating signals from front to back. Sci. (New York N.Y.) 2003, 302, 1704–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, M.; Gautreau, A. Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 2014, 15, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Manzanares, M.; Webb, D.J.; Horwitz, A.R. Cell migration at a glance. J. Cell Sci. 2005, 118, 4917–4919. [Google Scholar] [CrossRef] [Green Version]
- Mardakheh, F.K.; Self, A.; Marshall, C.J. RHO binding to FAM65A regulates Golgi reorientation during cell migration. J. Cell Sci. 2016, 129, 4466–4479. [Google Scholar] [CrossRef] [Green Version]
- Iden, S.; Collard, J.G. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat. Rev. Mol. Cell Biol. 2008, 9, 846–859. [Google Scholar] [CrossRef]
- Hall, A. Rho family GTPases. Biochem. Soc. Trans. 2012, 40, 1378–1382. [Google Scholar] [CrossRef] [Green Version]
- Hodge, R.G.; Ridley, A.J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 2016, 17, 496–510. [Google Scholar] [CrossRef]
- Mertens, A.E.; Pegtel, D.M.; Collard, J.G. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 2006, 16, 308–316. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Ellenbroek, S.I.; Mertens, A.E.; van der Kammen, R.A.; de Rooij, J.; Collard, J.G. The Par-Tiam1 complex controls persistent migration by stabilizing microtubule-dependent front-rear polarity. Curr. Biol. Cb, 2007; 17, 1632–1634. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Watanabe, T.; Matsuzawa, K.; Katsumi, A.; Kakeno, M.; Matsui, T.; Ye, F.; Sato, K.; Murase, K.; Sugiyama, I.; et al. Tiam1 interaction with the PAR complex promotes talin-mediated Rac1 activation during polarized cell migration. J. Cell Biol. 2012, 199, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Pasten, C.; Cerda, J.; Jausoro, I.; Court, F.A.; Caceres, A.; Marzolo, M.P. ApoER2 and Reelin are expressed in regenerating peripheral nerve and regulate Schwann cell migration by activating the Rac1 GEF protein, Tiam1. Mol. Cell. Neurosci. 2015, 69, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cornelison, D.D.; Filla, M.S.; Stanley, H.M.; Rapraeger, A.C.; Olwin, B.B. Syndecan-3 and syndecan-4 specifically mark skeletal muscle satellite cells and are implicated in satellite cell maintenance and muscle regeneration. Dev. Biol. 2001, 239, 79–94. [Google Scholar] [CrossRef] [Green Version]
- Elfenbein, A.; Simons, M. Syndecan-4 signaling at a glance. J. Cell Sci. 2013, 126, 3799–3804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, B.K.; Jung, Y.S.; Shin, J.; Han, I.; Mortier, E.; Zimmermann, P.; Whiteford, J.R.; Couchman, J.R.; Oh, E.S.; Lee, W. Structural basis of syndecan-4 phosphorylation as a molecular switch to regulate signaling. J. Mol. Biol. 2006, 355, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Greene, D.K.; Tumova, S.; Couchman, J.R.; Woods, A. Syndecan-4 associates with alpha-actinin. J. Biol. Chem. 2003, 278, 7617–7623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Multhaupt, H.A.; Yoneda, A.; Whiteford, J.R.; Oh, E.S.; Lee, W.; Couchman, J.R. Syndecan signaling: when, where and why? J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2009, 60 Suppl 4, 31–38. [Google Scholar]
- Gopal, S.; Sogaard, P.; Multhaupt, H.A.; Pataki, C.; Okina, E.; Xian, X.; Pedersen, M.E.; Stevens, T.; Griesbeck, O.; Park, P.W.; et al. Transmembrane proteoglycans control stretch-activated channels to set cytosolic calcium levels. J. Cell Biol. 2015, 210, 1199–1211. [Google Scholar] [CrossRef] [Green Version]
- Bass, M.D.; Roach, K.A.; Morgan, M.R.; Mostafavi-Pour, Z.; Schoen, T.; Muramatsu, T.; Mayer, U.; Ballestrem, C.; Spatz, J.P.; Humphries, M.J. Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J. Cell Biol. 2007, 177, 527–538. [Google Scholar] [CrossRef]
- Keller-Pinter, A.; Ughy, B.; Domoki, M.; Pettko-Szandtner, A.; Letoha, T.; Tovari, J.; Timar, J.; Szilak, L. The phosphomimetic mutation of syndecan-4 binds and inhibits Tiam1 modulating Rac1 activity in PDZ interaction-dependent manner. PLoS ONE 2017, 12, e0187094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelison, D.D.; Wilcox-Adelman, S.A.; Goetinck, P.F.; Rauvala, H.; Rapraeger, A.C.; Olwin, B.B. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 2004, 18, 2231–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller-Pinter, A.; Szabo, K.; Kocsis, T.; Deak, F.; Ocsovszki, I.; Zvara, A.; Puskas, L.; Szilak, L.; Dux, L. Syndecan-4 influences mammalian myoblast proliferation by modulating myostatin signalling and G1/S transition. Febs Lett. 2018, 592, 3139–3151. [Google Scholar] [CrossRef] [PubMed]
- Brooks, R.; Williamson, R.; Bass, M. Syndecan-4 independently regulates multiple small GTPases to promote fibroblast migration during wound healing. Small Gtpases 2012, 3, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Saoncella, S.; Calautti, E.; Neveu, W.; Goetinck, P.F. Syndecan-4 regulates ATF-2 transcriptional activity in a Rac1-dependent manner. J. Biol. Chem. 2004, 279, 47172–47176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkachenko, E.; Elfenbein, A.; Tirziu, D.; Simons, M. Syndecan-4 clustering induces cell migration in a PDZ-dependent manner. Circ. Res. 2006, 98, 1398–1404. [Google Scholar] [CrossRef] [Green Version]
- Matthews, H.K.; Marchant, L.; Carmona-Fontaine, C.; Kuriyama, S.; Larrain, J.; Holt, M.R.; Parsons, M.; Mayor, R. Directional migration of neural crest cells in vivo is regulated by Syndecan-4/Rac1 and non-canonical Wnt signaling/RhoA. Dev. (Camb. Engl.) 2008, 135, 1771–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Dickerson, J.B.; Guo, F.; Zheng, J.; Zheng, Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA 2004, 101, 7618–7623. [Google Scholar] [CrossRef] [Green Version]
- Boissier, P.; Huynh-Do, U. The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling. Cell. Signal. 2014, 26, 483–491. [Google Scholar] [CrossRef]
- Carmona-Fontaine, C.; Matthews, H.; Mayor, R. Directional cell migration in vivo: Wnt at the crest. Cell Adhes. Migr. 2008, 2, 240–242. [Google Scholar] [CrossRef] [Green Version]
- Pankov, R.; Endo, Y.; Even-Ram, S.; Araki, M.; Clark, K.; Cukierman, E.; Matsumoto, K.; Yamada, K.M. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 2005, 170, 793–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller-Pinter, A.; Bottka, S.; Timar, J.; Kulka, J.; Katona, R.; Dux, L.; Deak, F.; Szilak, L. Syndecan-4 promotes cytokinesis in a phosphorylation-dependent manner. Cell Mol. Life Sci. 2010, 67, 1881–1894. [Google Scholar] [CrossRef] [PubMed]
- Adams, H.C.; Chen, R.; Liu, Z.; Whitehead, I.P. Regulation of breast cancer cell motility by T-cell lymphoma invasion and metastasis-inducing protein. Breast Cancer Res. Bcr 2010, 12, R69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, J.; Yang, F.; Wu, W. Tiam1 high expression is associated with poor prognosis in solid cancers: A meta-analysis. Medicine 2019, 98, e17529. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, R.; Gautreau, A. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 2014, 9, 1931–1943. [Google Scholar] [CrossRef]
 : increase;
: increase;  : decrease.
: decrease.
 : increase;
: increase;  : decrease.
: decrease.
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becsky, D.; Gyulai-Nagy, S.; Balind, A.; Horvath, P.; Dux, L.; Keller-Pinter, A. Myoblast Migration and Directional Persistence Affected by Syndecan-4-Mediated Tiam-1 Expression and Distribution. Int. J. Mol. Sci. 2020, 21, 823. https://doi.org/10.3390/ijms21030823
Becsky D, Gyulai-Nagy S, Balind A, Horvath P, Dux L, Keller-Pinter A. Myoblast Migration and Directional Persistence Affected by Syndecan-4-Mediated Tiam-1 Expression and Distribution. International Journal of Molecular Sciences. 2020; 21(3):823. https://doi.org/10.3390/ijms21030823
Chicago/Turabian StyleBecsky, Daniel, Szuzina Gyulai-Nagy, Arpad Balind, Peter Horvath, Laszlo Dux, and Aniko Keller-Pinter. 2020. "Myoblast Migration and Directional Persistence Affected by Syndecan-4-Mediated Tiam-1 Expression and Distribution" International Journal of Molecular Sciences 21, no. 3: 823. https://doi.org/10.3390/ijms21030823
APA StyleBecsky, D., Gyulai-Nagy, S., Balind, A., Horvath, P., Dux, L., & Keller-Pinter, A. (2020). Myoblast Migration and Directional Persistence Affected by Syndecan-4-Mediated Tiam-1 Expression and Distribution. International Journal of Molecular Sciences, 21(3), 823. https://doi.org/10.3390/ijms21030823








