Antimicrobial Resistance in Veterinary Medicine: An Overview
Abstract
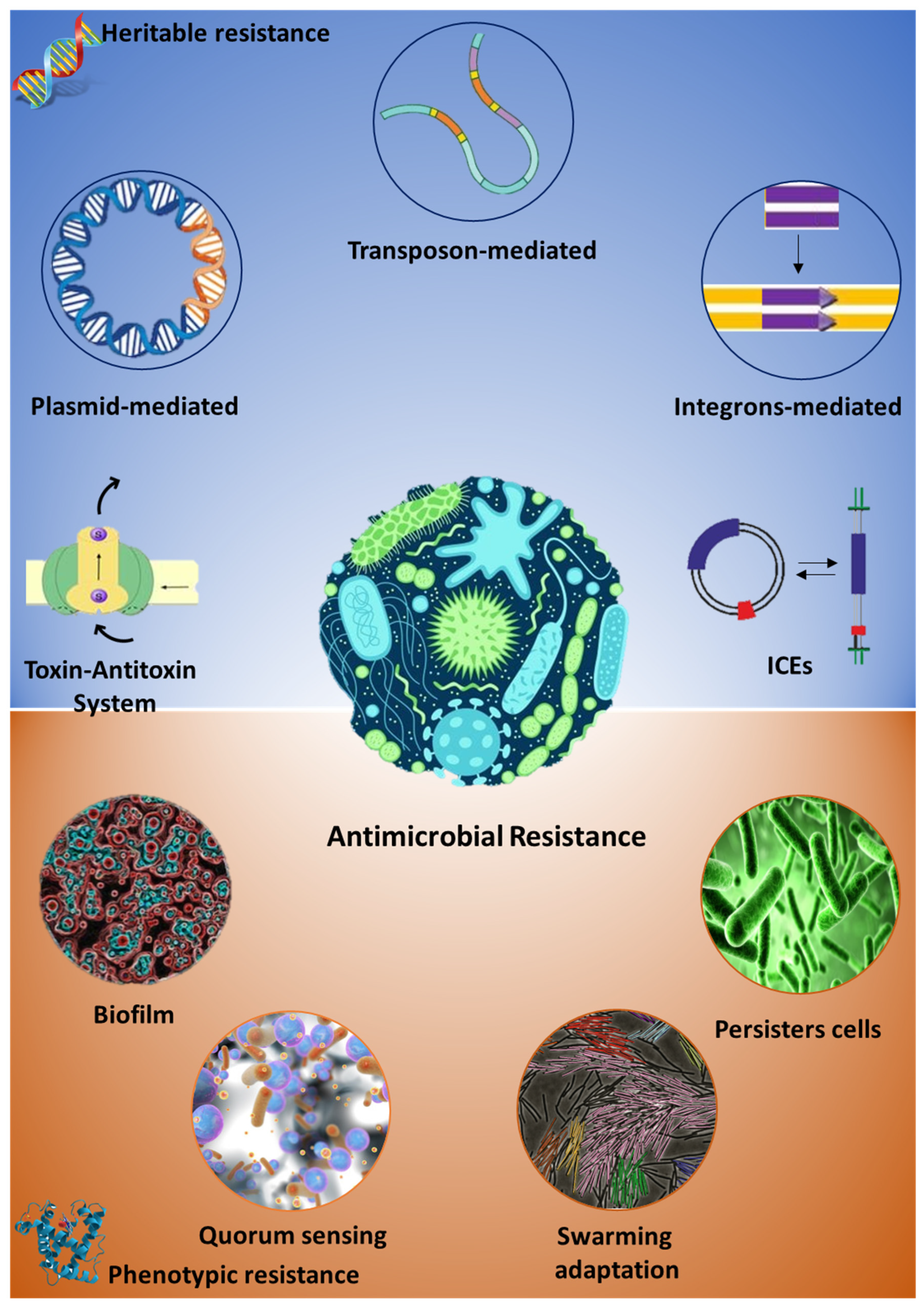
:1. Antibiotic Resistance: Origin and Diffusion
2. One-Health Approach to Face AMR
3. The Contribution of Companion Animals in AMR Spread
4. Aquaculture and AMR
5. Domestic Animal Husbandry Influence on AMR
6. Microorganisms Involved in Relevant Domestic Animal Infections
6.1. Campylobacter spp.
6.2. Salmonella spp.
6.3. Staphylococcus spp.
6.4. Enterococcus spp.
7. Latest Frontiers on Veterinary AMR Research
8. Future Perspective for Preventing Insurgence and/or Development of AMR
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Giedraitiene, A.; Vitkauskiene, A.; Naginiene, R.; Pavilonis, A. Antibiotic resistance mechanisms of clinically important bacteria. Medicina 2011, 47, 137–146. [Google Scholar] [CrossRef]
- Perry, J.; Waglechner, N.; Wright, G. The prehistory of antibiotic resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025197. [Google Scholar] [CrossRef] [PubMed]
- Sommer, M.O.; Church, G.M.; Dantas, G. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence 2010, 4, 299–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T.; Mondal, A.H.; Haq, Q.M.R. Antibiotics, resistome and resistance mechanisms: A bacterial perspective. Front. Microbiol. 2018, 9, 2066. [Google Scholar] [CrossRef] [Green Version]
- Olivares, J.; Bernardini, A.; Garcia-Leon, G.; Corona, F.; Sanchez, M.B.; Martinez, J.L. The intrinsic resistome of bacterial pathogens. Front. Microbiol. 2013, 4, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, L.; Hancock, R.E. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 4, 661–681. [Google Scholar] [CrossRef] [Green Version]
- Martínez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- Andersen, S.R.; Sandaa, R.A. Distribution of tetracycline resistance determinants among gram-negative bacteria isolated from polluted and unpolluted marine sediments. Appl. Environ. Microbiol. 1994, 60, 908–912. [Google Scholar] [CrossRef] [Green Version]
- Stokes, H.W.; Gillings, M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008, 9, 411–412. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. Mobile DNA and evolution in the 21st century. Mob. DNA 2010, 1, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, P.M. Plasmid encoded antibiotic resistance: Acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 2008, 153, S347–S357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cambray, G.; Guerout, A.M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef]
- Burrus, V.; Pavlovic, G.; Decaris, B.; Guédon, G. Conjugative transposons: The tip of the iceberg. Mol. Microbiol. 2002, 46, 601–610. [Google Scholar] [CrossRef]
- Drenkard, E.; Ausubel, F.M. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 2002, 416, 740–743. [Google Scholar] [CrossRef]
- He, J.; Baldini, R.L.; Déziel, E.; Saucier, M.; Zhang, Q.; Liberati, N.T.; Lee, D.; Urbach, J.; Goodman, H.M.; Rahme, L.G. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. USA 2004, 101, 2530–2535. [Google Scholar] [CrossRef] [Green Version]
- Davies, M.R.; Shera, J.; Van Domselaar, J.H.; Sriprakash, K.S.; McMillan, D.J. A Novel Integrative Conjugative Element Mediates Genetic Transfer from Group G Streptococcus to Other ß-Hemolytic Streptococci. J. Bacteriol. 2009, 191, 2257–2265. [Google Scholar] [CrossRef] [Green Version]
- Whittle, G.; Shoemaker, N.B.; Salyers, A.A. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 2002, 59, 2044–2054. [Google Scholar] [CrossRef]
- Bi, D.; Xu, Z.; Harrison, E.M.; Tai, C.; Wei, Y.; He, X.; Jia, S.; Deng, Z.; Rajakumar, K.; Ou, H.Y. ICEberg: A web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 2012, 40, D621–D626. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wei, Y.; Shen, Y.; Li, X.; Zhou, H.; Tai, C.; Deng, Z.; Ou, H.Y. TADB 2.0: An updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 2018, 46, D749–D753. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef] [PubMed]
- Cheverton, A.M.; Gollan, B.; Przydacz, M.; Wong, C.T.; Mylona, A.; Hare, S.A.; Helaine, S. A Salmonella Toxin Promotes Persister Formation through Acetylation of tRNA. Mol. Cell 2016, 63, 86–96. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Tai, C.; Deng, Z.; Gan, J.; Oggioni, M.R.; Ou, H.Y. Identification and characterization of chromosomal relBE toxin-Antitoxin locus in Streptomyces cattleya DSM46488. Sci. Rep. 2016, 6, 32047. [Google Scholar] [CrossRef] [Green Version]
- Goormaghtigh, F.; Fraikin, N.; Putrinš, M.; Hallaert, T.; Hauryliuk, V.; Garcia-Pino, A.; Sjödin, A.; Kasvandik, S.; Udekwu, K.; Tenson, T.; et al. Reassessing the role of type II toxin-antitoxin systems in formation of escherichia coli type II persister cells. MBio 2018, 9, e00640-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maisonneuve, E.; Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 2014, 157, 539–548. [Google Scholar] [CrossRef] [Green Version]
- Ramisetty, B.C.M.; Ghosh, D.; Chowdhury, M.R.; Santhosh, R.S. What is the link between stringent response, endoribonuclease encoding type II toxin-antitoxin systems and persistence? Front. Microbiol. 2016, 8, 458. [Google Scholar] [CrossRef] [Green Version]
- Harms, A.; Fino, C.; Sørensen, M.A.; Semsey, S.; Gerdes, K. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. MBio 2017, 8, e01964-17. [Google Scholar] [CrossRef] [Green Version]
- Wright, G.D. The antibiotic resistome. Expert Opin. Drug Discov. 2010, 5, 779–788. [Google Scholar] [CrossRef]
- Gerdes, K.; Maisonneuve, E. Bacterial Persistence and Toxin-Antitoxin Loci. Annu. Rev. Microbiol. 2012, 66, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Vulić, M.; Keren, I.; Lewis, K. Role of oxidative stress in persister tolerance. Antimicrob. Agents Chemother. 2012, 56, 4922–4926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhang, Y. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 2007, 51, 2092–2099. [Google Scholar] [CrossRef] [Green Version]
- Amini, S.; Hottes, A.K.; Smith, L.E.; Tavazoie, S. Fitness landscape of antibiotic tolerance in pseudomonas aeruginosa biofilms. PLoS Pathog. 2011, 7, e1002298. [Google Scholar] [CrossRef]
- Rani, S.A.; Pitts, B.; Beyenal, H.; Veluchamy, R.A.; Lewandowski, Z.; Davison, W.M.; Buckingham-Meyer, K.; Stewart, P.S. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J. Bacteriol. 2007, 189, 4223–4233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overhage, J.; Bains, M.; Brazas, M.D.; Hancock, R.E.W. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 2008, 190, 2671–2679. [Google Scholar] [CrossRef] [Green Version]
- Kindrachuk, K.N.; Fernández, L.; Bains, M.; Hancock, R.E.W. Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 1874–1882. [Google Scholar] [CrossRef] [Green Version]
- Ramos, J.L.; Levesque, R.C. Pseudomonas, 1st ed.; Springer: Boston, MA, USA, 2006. [Google Scholar]
- Copeland, M.F.; Weibel, D.B. Bacterial swarming: A model system for studying dynamic self-assembly. Soft Matter 2009, 5, 1174–1187. [Google Scholar] [CrossRef]
- Hwang, I.Y.; Tan, M.H.; Koh, E.; Ho, C.L.; Poh, C.L.; Chang, M.W. Reprogramming microbes to be pathogen-Seeking killers. ACS Synth. Biol. 2014, 3, 228–237. [Google Scholar] [CrossRef]
- Jeffery, C.J. Why study moonlighting proteins? Front. Genet. 2015, 6, 211. [Google Scholar] [CrossRef] [Green Version]
- Jeffery, C.J. Intracellular/surface moonlighting proteins that aid in the attachment of gut microbiota to the host. AIMS Microbiol. 2019, 5, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Gancedo, C.; Flores, C.L.; Gancedo, J.M. The Expanding Landscape of Moonlighting Proteins in Yeasts. Microbiol. Mol. Biol. Rev. 2016, 80, 765–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huijbers, P.M.; Blaak, H.; De Jong, M.C.; Graat, E.A.; Vandenbroucke-Grauls, C.M.; De Roda Husman, A.M. Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef] [PubMed]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef] [Green Version]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Antimicrob. Resist. Bact. Livest. Companion Anim. 2018, 6, 521–547. [Google Scholar]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Wegener, H.C.; Collignon, P. Resistance in bacteria of the food chain: Epidemiology and control strategies. Expert Rev. Anti. Infect. Ther. 2008, 6, 733–750. [Google Scholar] [CrossRef]
- Rowan, A.; Kartal, T. Dog population & dog sheltering trends in the United States of America. Animals 2018, 8, 68. [Google Scholar]
- Messenger, A.M.; Barnes, A.N.; Gray, G.C. Reverse zoonotic disease transmission (Zooanthroponosis): A systematic review of seldom-documented human biological threats to animals. PLoS ONE 2014, 9, e89055. [Google Scholar] [CrossRef] [Green Version]
- Klous, G.; Huss, A.; Heederik, D.J.J.; Coutinho, R.A. Human-livestock contacts and their relationship to transmission of zoonotic pathogens, a systematic review of literature. One Health 2016, 2, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Pomba, C.; Rantala, M.; Greko, C.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Mateus, A.; Moreno, M.A.; Pyörälä, S.; Ružauskas, M.; et al. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 2017, 72, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.O.; Rookt, K.A.; Shofer, F.S.; Rankin, S.C. Screening of Staphylococcus aureus, Staphylococcus intermedius, and Staphylococcus schleiferi isolates obtained from small companion animals for antimicrobial resistance: A retrospective review of 749 isolates (2003–2004). Vet. Dermatol. 2006, 17, 332–337. [Google Scholar] [CrossRef]
- Rendle, D.I.; Page, S.W. Antimicrobial resistance in companion animals. Equine Vet. J. 2018, 50, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, A.; Lloyd, D.H. Companion animals: A reservoir for methicillin-resistant Staphylococcus aureus in the community? Epidemiol. Infect. 2010, 138, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Manian, F.A. Asymptomatic Nasal Carriage of Mupirocin-Resistant, Methicillin-Resistant Staphylococcus aureus (MRSA) in a Pet Dog Associated with MRSA Infection in Household Contacts. Clin. Infect. Dis. 2003, 36, e26–e28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weese, J.S.; Lefebvre, S.L. Risk factors methicillin-resistant staphylococcus aureus colonization in horses admitted to a veterinary teaching hospital. Can. Vet. J. 2007, 48, 921–926. [Google Scholar]
- Davis, M.F.; Iverson, S.A.; Baron, P.; Vasse, A.; Silbergeld, E.K.; Lautenbach, E.; Morris, D.O. Household transmission of methicillin-resistant Staphylococcus aureus and other staphylococci. Lancet Infect. Dis. 2012, 12, 703–716. [Google Scholar] [CrossRef]
- Van Duijkeren, E.; Ikawaty, R.; Broekhuizen-Stins, M.J.; Jansen, M.D.; Spalburg, E.C.; de Neeling, A.J.; Allaart, J.G.; van Nes, A.; Wagenaar, J.A.; Fluit, A.C. Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Vet. Microbiol. 2008, 126, 383–389. [Google Scholar] [CrossRef]
- Santos, L.; Ramos, F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents 2018, 52, 135–143. [Google Scholar] [CrossRef]
- Aich, N.; Ahmed, N.; Paul, A. Issues of Antibiotic Resistance in Aquaculture Industry and Its Way Forward. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 26–41. [Google Scholar] [CrossRef]
- ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals. EFSA J. 2017, 15.
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.H.; Park, S.G.; Choi, S.M.; Hwang, Y.O.; Ham, H.J.; Kim, S.U.; Lee, Y.K.; Kim, M.S.; Park, G.Y.; Kim, K.S.; et al. Antimicrobial resistance and resistance genes in Escherichia coli strains isolated from commercial fish and seafood. Int. J. Food Microbiol. 2012, 152, 14–18. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Buschmann, A.H.; Dölz, H.J. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 2016, 16, e127–e133. [Google Scholar] [CrossRef]
- O’Neill, J. The O’Neill Review on Antimicrobial Resistance: Antimicrobials in Agriculture and the Environment. Rev. Antimicrob. Resist. 2015, 1–44. [Google Scholar]
- Burridge, L.; Weis, J.S.; Cabello, F.; Pizarro, J.; Bostick, K. Chemical use in salmon aquaculture: A review of current practices and possible environmental effects. Aquaculture 2010, 306, 7–23. [Google Scholar] [CrossRef]
- Marston, H.D.; Dixon, D.M.; Knisely, J.M.; Palmore, T.N.; Fauci, A.S. Antimicrobial resistance. JAMA 2016, 316, 1193–1204. [Google Scholar] [CrossRef] [Green Version]
- Varaldo, P.E.; Facinelli, B.; Bagnarelli, P.; Menzo, S.; Mingoia, M.; Brenciani, A.; Giacometti, A.; Barchiesi, F.; Brescini, L.; Cirioni, O.; et al. Antimicrobial Resistance: A Challenge for the Future. In The First Outstanding 50 Years of “Università Politecnica Delle Marche”; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial Food Animal Production, Antimicrobial Resistance, and Human Health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef]
- Economou, V.; Gousia, P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Stokstad, E.L.R.; Jukes, T.H. Further Observations on the “Animal Protein Factor” (17751). Proc. Soc. Exp. Biol. Med. 1950, 73, 1950. [Google Scholar] [CrossRef]
- Giguère, S.; Prescott, J.F.; Dowling, P.M. Antimicrobial Therapy in Veterinary Medicine, 5th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Alexander, T.W.; Yanke, L.J.; Topp, E.; Olson, M.E.; Read, R.R.; Morck, D.W.; McAllister, T.A. Effect of subtherapeutic administration of antibiotics on the prevalence of antibiotic-resistant Escherichia coli bacteria in feedlot cattle. Appl. Environ. Microbiol. 2008, 74, 4405–4416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woolums, A.R.; Karisch, B.B.; Frye, J.G.; Epperson, W.; Smith, D.R.; Blanton, J.; Austin, F.; Kaplan, R.; Hiott, L.; Woodley, T.; et al. Multidrug resistant Mannheimia haemolytica isolated from high-risk beef stocker cattle after antimicrobial metaphylaxis and treatment for bovine respiratory disease. Vet. Microbiol. 2018, 221, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Doster, E.; Rovira, P.; Noyes, N.R.; Burgess, B.A.; Yang, X.; Weinroth, M.D.; Lakin, S.M.; Dean, C.J.; Linke, L.; Magnuson, R.; et al. Investigating Effects of Tulathromycin Metaphylaxis on the Fecal Resistome and Microbiome of Commercial Feedlot Cattle Early in the Feeding Period. Front. Microbiol. 2018, 9, 1715. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Van Donkersgoed, J.; Bohaychuk, V.; Besser, T.; Song, X.M.; Wagner, B.; Hancock, D.; Renter, D.; Dargatz, D.; Morley, P.S. Antimicrobial drug use and antimicrobial resistance in enteric bacteria among cattle from alberta feedlots. Foodborne Pathog. Dis. 2010, 7, 449–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clifford, K.; Desai, D.; da Costa, C.P.; Meyer, H.; Klohe, K.; Winkler, A.; Rahman, T.; Islam, T.; Zaman, M.H. Antimicrobial resistance in livestock and poor quality veterinary medicines. Bull. World Health Organ 2018, 8, 662–664. [Google Scholar] [CrossRef]
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A review of antibiotic use in food animals: Perspective, policy, and potential. Public Health Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef] [Green Version]
- Barton, M.D. Impact of antibiotic use in the swine industry. Curr. Opin. Microbiol. 2014, 19, 9–15. [Google Scholar] [CrossRef]
- Varela, N.P.; Gadbois, P.; Thibault, C.; Gottschalk, M.; Dick, P.; Wilson, J. Antimicrobial resistance and prudent drug use for Streptococcus suis. Anim. Health Res. Rev. 2013, 14, 68–77. [Google Scholar] [CrossRef]
- Aarestrup, F.M. Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic Clin. Pharmacol. Toxicol. 2005, 96, 271–281. [Google Scholar] [CrossRef]
- Aarestrup, F.M. Other Pathogens. In Antimicrobial Resistance in Bacteria of Animal Origin; ASM Press: Washington, DC , USA, 2019. [Google Scholar]
- McEwen, S.A.; Fedorka-Cray, P.J. Antimicrobial Use and Resistance in Animals. Clin. Infect. Dis. 2002, 34, S93–S106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, A.; McAllister, T.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cromwell, G.L. Why and how antibiotics are used in swine production. Anim. Biotechnol. 2002, 13, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [Green Version]
- Gyles, C.L. Antimicrobial resistance in selected bacteria from poultry. Anim. Health Res. Rev. 2008, 9, 149–158. [Google Scholar] [CrossRef]
- Scott, L.C.; Menzies, P.I. Antimicrobial Resistance and Small Ruminant Veterinary Practice. Vet. Clin. N. Am. Food Anim. Pract. 2011, 27, 23–32. [Google Scholar] [CrossRef]
- Toledo, Z.; Simaluiza, R.J.; Astudillo, X.; Fernández, H. Occurrence and antimicrobial susceptibility of thermophilic campylobacter species isolated from healthy children attending municipal care centers in Southern Ecuador. Rev. Inst. Med. Trop. Sao Paulo 2017, 59, e77. [Google Scholar] [CrossRef] [Green Version]
- Bojanić, K.; Midwinter, A.C.; Marshall, J.C.; Rogers, L.E.; Biggs, P.J.; Acke, E. Isolation of Campylobacter spp. from Client-Owned Dogs and Cats, and Retail Raw Meat Pet Food in the Manawatu, New Zealand. Zoonoses Public Health 2017, 64, 438–449. [Google Scholar] [CrossRef]
- Koluman, A.; Dikici, A. Antimicrobial resistance of emerging foodborne pathogens: Status quo and global trends. Crit. Rev. Microbiol. 2013, 39, 57–69. [Google Scholar] [CrossRef]
- Padungton, P.; Kaneene, J.B. Campylobacter spp. in human, chickens, pigs and their antimicrobial resistance. J. Vet. Med. Sci. 2003, 65, 161–170. [Google Scholar] [CrossRef] [Green Version]
- Alfredson, D.A.; Korolik, V. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 2007, 277, 123–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Future Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, P. Use of fluoroquinolones and development of resistance. J. Vet. Pharmacol. Ther. 2009, 32, 52–53. [Google Scholar]
- Zhang, Q.; Plummer, P. Mechanisms of Antibiotic Resistance. In Campylobacter, 3rd ed.; ASM Press: Washington, DC, USA, 2008; pp. 263–276. [Google Scholar]
- Payot, S.; Bolla, J.M.; Corcoran, D.; Fanning, S.; Mégraud, F.; Zhang, Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 2006, 8, 1967–1971. [Google Scholar] [CrossRef] [PubMed]
- Ge, B.; McDermott, P.F.; White, D.G.; Meng, J. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 2005, 49, 3347–3354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, N.; Sahin, O.; Lin, J.; Michel, L.O.; Zhang, Q. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 2003, 47, 390–394. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, D.; Quinn, T.; Cotter, L.; Fanning, S. An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int. J. Antimicrob. Agents 2006, 27, 40–45. [Google Scholar] [CrossRef]
- Kurinčič, M.; Botteldoorn, N.; Herman, L.; Smole Možina, S. Mechanisms of erythromycin resistance of Campylobacter spp. isolated from food; animals and humans. Int. J. Food Microbiol. 2007, 120, 186–190. [Google Scholar] [CrossRef]
- Lin, J.; Yan, M.; Sahin, O.; Pereira, S.; Chang, Y.J.; Zhanq, Q. Effect of macrolide usage on emergence of erythromycin-resistant Campylobacter isolates in chickens. Antimicrob. Agents Chemother. 2007, 51, 1678–1686. [Google Scholar] [CrossRef] [Green Version]
- Cagliero, C.; Mouline, C.; Cloeckaert, A.; Payot, S. Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 2006, 50, 3893–3896. [Google Scholar] [CrossRef] [Green Version]
- Gibreel, A.; Wetsch, N.M.; Taylor, D.E. Contribution of the CmeABC efflux pump to macrolide and tetracycline resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 2007, 51, 3212–3216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantero, G.; Correa-Fiz, F.; Ronco, T.; Strube, M.; Cerdà-Cuéllar, M.; Pedersen, K. Characterization of Campylobacter jejuni and Campylobacter coli Broiler Isolates by Whole-Genome Sequencing. Foodborne Pathog. Dis. 2018, 15, 145–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deusch, S.; Tilocca, B.; Camarinha-Silva, A.; Seifert, J. News in livestock research—Use of Omics-technologies to study the microbiota in the gastrointestinal tract of farm animals. Comput. Struct. Biotechnol. J. 2015, 13, 55–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, L.M.; Sayed, A.S.M.; ElKader, H.A.A.; Faddan, N.H.A.; Al Hosary, A.A.T. Phylogenetic analysis of Salmonella species isolated from cows; buffaloes; and humans based on gyrB gene sequences. Trop. Anim. Health Prod. 2020, 1–6. [Google Scholar] [CrossRef]
- Heuzenroeder, M. Salmonella in Domestic Animals. Aust. Vet. J. 2000, 78, 570. [Google Scholar] [CrossRef]
- Cosby, D.E.; Cox, N.A.; Harrison, M.A.; Wilson, J.L.; Jeff Buhr, R.; Fedorka-Cray, P.J. Salmonella and antimicrobial resistance in broilers: A review. J. Appl. Poult. Res. 2015, 24, 408–426. [Google Scholar] [CrossRef]
- Holmberg, S.D.; Wells, J.G.; Cohen, M.L. Animal-to-man transmission of antimicrobial-resistant Salmonella: Investigations of U.S. outbreaks, 1971–1983. Science 1984, 225, 833–835. [Google Scholar] [CrossRef]
- Spika, J.S.; Waterman, S.H.; Hoo, G.W.; St Louis, M.E.; Pacer, R.E.; James, S.M.; Bissett, M.L.; Mayer, L.W.; Chiu, J.Y.; Hall, B. Chloramphenicol-resistant Salmonella newport traced through hamburger to dairy farms. A major persisting source of human salmonellosis in California. N. Engl. J. Med. 1987, 316, 565–570. [Google Scholar] [CrossRef]
- Gebreyes, W.A.; Altier, C. Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine. J. Clin. Microbiol. 2002, 40, 2813–2822. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic resistance in salmonella typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef] [Green Version]
- Jaja, I.F.; Bhembe, N.L.; Green, E.; Oguttu, J.; Muchenje, V. Molecular characterisation of antibiotic-resistant Salmonella enterica isolates recovered from meat in South Africa. Acta Trop. 2019, 190, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Ren, S.Q.; Chang, M.X.; Chen, P.X.; Ding, H.Z.; Jiang, H.X. Resistance mechanisms and fitness of Salmonella Typhimurium and Salmonella Enteritidis mutants evolved under selection with ciprofloxacin in vitro. Sci. Rep. 2017, 7, 9113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, J.M.; Mondragon, A.C.; Martinez, B.; Guarddon, M.; Rodriguez, J.A. Prevalence and antimicrobial resistance patterns of Salmonella from different raw foods in Mexico. J. Food Prot. 2009, 72, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Drlica, K.; Hiasa, H.; Kerns, R.; Malik, M.; Mustaev, A.; Zhao, X. Quinolones: Action and Resistance Updated. Curr. Top. Med. Chem. 2009, 9, 981–998. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-mediated quinolone resistance: A multifaceted threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef] [Green Version]
- Gunell, M.; Webber, M.A.; Kotilainen, P.; Lilly, A.J.; Caddick, J.M.; Jalava, J.; Huovinen, P.; Siitonen, A.; Hakanen, A.J.; Piddock, L.J.V. Mechanisms of resistance in nontyphoidal Salmonella enterica strains exhibiting a nonclassical quinolone resistance phenotype. Antimicrob. Agents Chemother. 2009, 53, 3832–3836. [Google Scholar] [CrossRef] [Green Version]
- Bakkeren, E.; Huisman, J.S.; Fattinger, S.A.; Hausmann, A.; Furter, M.; Egli, A.; Slack, E.; Sellin, M.E.; Bonhoeffer, S.; Regoes, R.R.; et al. Salmonella persisters promote the spread of antibiotic resistance plasmids in the gut. Nature 2019, 573, 276–280. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis—The “accidental” pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Normanno, G.; Corrente, M.; La Salandra, G.; Dambrosio, A.; Quaglia, N.C.; Parisi, A.; Greco, G.; Bellacicco, A.L.; Virgilio, S.; Celano, G.V. Methicillin-resistant Staphylococcus aureus MRSA in foods of animal origin product in Italy. Int. J. Food Microbiol. 2007, 117, 219–222. [Google Scholar] [CrossRef]
- Ward, M.J.; Gibbons, C.L.; McAdam, P.R.; van Bunnik, B.A.D.; Girvan, E.K.; Edwards, G.F.; Fitzgerald, J.R.; Woolhouse, M.E.J. Time-scaled evolutionary analysis of the transmission and antibiotic resistance dynamics of Staphylococcus aureus clonal complex 398. Appl. Environ. Microbiol. 2014, 80, 7275–7282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuny, C.; Wieler, L.H.; Witte, W. Livestock-Associated MRSA: The impact on humans. Antibiotics 2015, 4, 521–543. [Google Scholar] [CrossRef] [PubMed]
- García-Álvarez, L.; Holden, M.T.G.; Lindsay, H.; Webb, C.R.; Brown, D.F.J.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C.; et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Li, Z.; Chlebowicz, M.A.; Tao, X.; Ni, M.; Hu, Y.; Li, Z.; Grundmann, H.; Murray, S.; Pascoe, B.; et al. Genetic features of livestock-associated Staphylococcus aureus ST9 isolates from Chinese pigs that carry the lsaE gene for quinupristin/dalfopristin resistance. Int. J. Med. Microbiol. 2016, 306, 722–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Li, X.; Osmundson, T.; Shi, L.; Ren, J.; Yan, H. WGS analysis of ST9-MRSA-XII isolates from live pigs in China provides insights into transmission among porcine; human and bovine hosts. J. Antimicrob. Chemother. 2018, 73, 2652–2661. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Lu, H.; Wang, X.; Gao, Q.; Dai, Y.; Shang, J.; Li, M. Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front. Cell. Infect. Microbiol. 2017, 7, 127. [Google Scholar] [CrossRef]
- Tröscher-Mußotter, J.; Tilocca, B.; Stefanski, V.; Seifert, J. Analysis of the bacterial and host proteins along and across the porcine gastrointestinal tract. Proteomes 2019, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Tilocca, B.; Witzig, M.; Rodehutscord, M.; Seifert, J. Variations of phosphorous accessibility causing changes in microbiome functions in the gastrointestinal tract of chickens. PLoS ONE 2016, 11, e0164735. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.; Pang, S.; Abraham, S.; Coombs, G.W. Antimicrobial-resistant CC17 Enterococcus faecium: The past; the present and the future. J. Glob. Antimicrob. Resist. 2019, 16, 36–47. [Google Scholar] [CrossRef] [Green Version]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal Infection—Treatment and Antibiotic Resistance. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Werner, G.; Coque, T.M.; Franz, C.M.A.P.; Grohmann, E.; Hegstad, K.; Jensen, L.; van Schaik, W.; Weaver, K. Antibiotic resistant enterococci-Tales of a drug resistance gene trafficker. Int. J. Med. Microbiol. 2013, 303, 360–379. [Google Scholar] [CrossRef]
- Hayes, J.R.; English, L.L.; Carter, P.J.; Proescholdt, T.; Lee, K.Y.; Wagner, D.D.; White, D.G. Prevalence and Antimicrobial Resistance of Enterococcus Species Isolated from Retail Meats. Appl. Environ. Microbiol. 2003, 69, 7153–7160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, F.; Atanassova, V.; Klein, G. Extended-spectrum ß-lactamase- and ampc-producing enterobacteria in healthy broiler chickens, Germany. Emerg. Infect. Dis. 2013, 19, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.A.; Westman, E.L.; Wright, G.D. The antibiotic resistome: What’s new? Curr. Opin. Microbiol. 2014, 21, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007, 5, 175–186. [Google Scholar] [CrossRef]
- Stenuit, B.; Agathos, S.N. Deciphering microbial community robustness through synthetic ecology and molecular systems synecology. Curr. Opin. Biotechnol. 2015, 33, 305–317. [Google Scholar] [CrossRef]
- Pitta, D.W.; Dou, Z.; Kumar, S.; Indugu, N.; Toth, J.D.; Vecchiarelli, B.; Bhukya, B. Metagenomic Evidence of the Prevalence and Distribution Patterns of Antimicrobial Resistance Genes in Dairy Agroecosystems. Foodborne Pathog. Dis. 2016, 13, 296–302. [Google Scholar] [CrossRef]
- Tilocca, B.; Costanzo, N.; Morittu, V.M.; Spina, A.A.; Soggiu, A.; Britti, D.; Roncada, P.; Piras, C. Milk microbiota: Characterization methods and role in cheese production. J. Proteom. 2020, 210, 103534. [Google Scholar] [CrossRef]
- Cohen, A.; Bont, L.; Engelhard, D.; Moore, E.; Fernández, D.; Kreisberg-Greenblatt, R.; Oved, K.; Eden, E.; Hays, J.P. A multifaceted “omics” approach for addressing the challenge of antimicrobial resistance. Future Microbiol. 2015, 10, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Boja, E.S.; Kinsinger, C.R.; Rodriguez, H.; Srinivas, P. Integration of omics sciences to advance biology and medicine. Clin. Proteom. 2014, 11, 45. [Google Scholar] [CrossRef] [Green Version]
- Thomas, T.; Gilbert, J.; Meyer, F. Metagenomics: A guide from sampling to data analysis. Microb. Inform. 2012, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Zoetendal, E.G.; Rajilić-Stojanovic, M.; De Vos, W.M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 2008, 57, 1605–1615. [Google Scholar] [CrossRef] [PubMed]
- Del Chierico, F.; Vernocchi, P.; Bonizzi, L.; Carsetti, R.; Castellazzi, A.M.; Dallapiccola, B.; De Vos, W.; Guerzoni, M.E.; Manco, M.; Marseglia, G.L.; et al. Early-life gut microbiota under physiological and pathological conditions: The central role of combined meta-omics-based approaches. J. Proteom. 2012, 75, 4580–4587. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R. An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinforma 2013, 42, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.; Davidovich, M.; Rokney, A.; Valinsky, L.; Rahav, G.; Gal-Mor, O. Emergence of new variants of antibiotic resistance genomic islands among multidrug-resistant Salmonella enterica in poultry. Environ. Microbiol. 2020, 22, 413–432. [Google Scholar] [CrossRef] [PubMed]
- Rafique, M.; Potter, R.F.; Ferreiro, A.; Wallace, M.A.; Rahim, A.; Ali Malik, A.; Siddique, N.; Abbas, M.A.; D’Souza, A.W.; Burnham, C.D.; et al. Genomic Characterization of Antibiotic Resistant Escherichia coli Isolated From Domestic Chickens in Pakistan. Front. Microbiol. 2020, 10, 3052. [Google Scholar] [CrossRef]
- Sabino, Y.N.V.; Santana, M.F.; Oyama, L.B.; Santos, F.G.; Moreira, A.J.S.; Huws, S.A.; Mantovani, H.C. Characterization of antibiotic resistance genes in the species of the rumen microbiota. Nat. Commun. 2019, 10, 5252. [Google Scholar] [CrossRef]
- Parisi, A.; Capozzi, L.; Bianco, A.; Caruso, M.; Latorre, L.; Costa, A.; Giannico, A.; Ridolfi, D.; Bulzacchelli, C.; Santagada, G. Identification of virulence and antibiotic resistance factors in arcobacter butzleri isolated from bovine milk by whole genome sequencing. Ital. J. Food Saf. 2019, 8, 7840. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Field, D.; Huang, Y.; Edwards, R.A.; Li, W.; Gilna, P.; Joint, I. Detection of Large Numbers of Novel Sequences in the Metatranscriptomes of Complex Marine Microbial Communities. PLoS ONE 2011, 3, 277–286. [Google Scholar]
- Vieites, J.M.; Guazzaroni, M.E.; Beloqui, A.; Golyshin, P.N.; Ferrer, M. Metagenomics approaches in systems microbiology. FEMS Microbiol. Rev. 2009, 33, 236–255. [Google Scholar] [CrossRef]
- Marcelino, V.R.; Wille, M.; Hurt, A.C.; González-Acuña, D.; Klaassen, M.; Schlub, T.E.; Eden, J.S.; Shi, M.; Iredell, J.R.; Sorrell, T.C.; et al. Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. 2019, 17, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Feng, M.; Yang, K.; Cao, Y.; Zhang, J.; Xu, J.; Hernández, S.H.; Wei, X.; Fan, M. Transcriptomic and metabolomic analyses reveal antibacterial mechanism of astringent persimmon tannin against Methicillin-resistant Staphylococcus aureus isolated from pork. Food Chem. 2020, 309, 125692. [Google Scholar] [CrossRef] [PubMed]
- Hettich, R.L.; Pan, C.; Chourey, K.; Giannone, R.J. Metaproteomics: Harnessing the power of high performance mass spectrometry to identify the suite of proteins that control metabolic activities in microbial communities. Anal. Chem. 2013, 85, 4203–4214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siuti, N.; Kelleher, N.L. Decoding protein modifications using top-down mass spectrometry. Nat. Methods 2007, 4, 817–821. [Google Scholar] [CrossRef] [PubMed]
- VerBerkmoes, N.C.; Denef, V.J.; Hettich, R.L.; Banfield, J.F. Systems Biology: Functional analysis of natural microbial consortia using community proteomics. Nat. Rev. Microbiol. 2009, 7, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Soggiu, A.; Bonizzi, L.; Gaviraghi, A.; Deriu, F.; De Martino, L.; Iovane, G.; Amoresano, A.; Roncada, P. Comparative proteomics to evaluate multi drug resistance in Escherichia coli. Mol. Biosyst. 2012, 8, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Feng Zhang, D.; Li, H.; Min Lin, X.; Xian Peng, X. Outer membrane proteomics of kanamycin-resistant Escherichia coli identified MipA as a novel antibiotic resistance-related protein. FEMS Microbiol. Lett. 2015, 362, fnv074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piras, C.; Soggiu, A.; Greco, V.; Martino, P.A.; Del Chierico, F.; Putignani, L.; Urbani, A.; Nally, J.E.; Bonizzi, L.; Roncada, P. Mechanisms of antibiotic resistance to enrofloxacin in uropathogenic Escherichia coli in dog. J. Proteom. 2015, 127, 365–376. [Google Scholar] [CrossRef]
- Pérez-Llarena, F.J.; Bou, G. Proteomics as a tool for studying bacterial virulence and antimicrobial resistance. Front. Microbiol. 2016, 7, 410. [Google Scholar] [CrossRef] [Green Version]
- Park, A.J.; Krieger, J.R.; Khursigara, C.M. Survival proteomes: The emerging proteotype of antimicrobial resistance. FEMS Microbiol. Rev. 2016, 40, 323–342. [Google Scholar] [CrossRef] [Green Version]
- Tang, J. Microbial Metabolomics. Curr. Genom. 2011, 12, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Jordan, K.W.; Nordenstam, J.; Lauwers, G.Y.; Rothenberger, D.A.; Alavi, K.; Garwood, M.; Cheng, L.L. Metabolomic characterization of human rectal adenocarcinoma with intact tissue magnetic resonance spectroscopy. Dis. Colon Rectum 2009, 52, 520–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rochfort, S. Metabolomics reviewed: A new “omics” platform technology for systems biology and implications for natural products research. J. Nat. Prod. 2005, 68, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Li, W.; Sun, L.; Lin, Z.; Jiang, Y.; Ling, Y.; Lin, X. Comparative metabolomics shows the metabolic profiles fluctuate in multi-drug resistant Escherichia coli strains. J. Proteom. 2019, 207, 103468. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, G.; Zhang, S.; Fu, Y.; Jiang, Y.; Yang, X.; Lin, X. An integrated quantitative proteomic and metabolomics approach to reveal the negative regulation mechanism of LamB in antibiotics resistance. J. Proteom. 2019, 194, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Tilocca, B.; Burbach, K.; Heyer, C.M.E.; Hoelzle, L.E.; Mosenthin, R.; Stefanski, V.; Camarinha-Silva, A.; Seifert, J. Dietary changes in nutritional studies shape the structural and functional composition of the pigs’ fecal microbiome-from days to weeks. Microbiome 2017, 5, 144. [Google Scholar] [CrossRef] [Green Version]
| Resistant Bacteria | Isolation Source | Antimicrobial Compound(s) | Major Resistance Mechanism |
|---|---|---|---|
| Campylobacter spp. | Cattle Chickens Turkey Pig Sheep Dog Cat Horse | Quinolones Macrolides Lincosamides Chloramphenicol Aminoglycosides Tetracycline β-lactams Cotrimoxazole Tylosin | Point mutation on GyrA gene CmeABC multidrug efflux pump Methylation of ribosomal target Point mutation in the ribosomal target |
| Salmonella spp. | Human Chicken Turkey Pig Cat Dog Horse | Tetracyclines Sulfonamides Streptomycin Kanamycin Chloramphenicol β-lactams Amoxicillin/clavulanic acid Nalidixic acid Ceftriaxone | Multiple point mutations on GyrA and GyrB, parC, and parE genes AcrAB-TolC multidrug efflux pumps Changing outer membrane proteins qnr genes-containing plasmid Salmonella Pathogenicity Island-1 and -2 |
| Staphylococcus spp. | Human Farm animals Dog Cat Horse | Penicillin Methicillin Vancomycin | Staphylococcal Cassette Chromosome-mec genes (SCC-mec) |
| Enterococcus spp | Human Farm animals | Vancomycin, Aminoglycosides Penicillin Chloramphenicol Erythromycin Tetracyclines | lsa gene |
| Other Gram-negative | Human Farm animals Pet | β-lactams | Extended-spectrum β-lactamases |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma, E.; Tilocca, B.; Roncada, P. Antimicrobial Resistance in Veterinary Medicine: An Overview. Int. J. Mol. Sci. 2020, 21, 1914. https://doi.org/10.3390/ijms21061914
Palma E, Tilocca B, Roncada P. Antimicrobial Resistance in Veterinary Medicine: An Overview. International Journal of Molecular Sciences. 2020; 21(6):1914. https://doi.org/10.3390/ijms21061914
Chicago/Turabian StylePalma, Ernesto, Bruno Tilocca, and Paola Roncada. 2020. "Antimicrobial Resistance in Veterinary Medicine: An Overview" International Journal of Molecular Sciences 21, no. 6: 1914. https://doi.org/10.3390/ijms21061914






