Plant RNA Binding Proteins as Critical Modulators in Drought, High Salinity, Heat, and Cold Stress Responses: An Updated Overview
Abstract
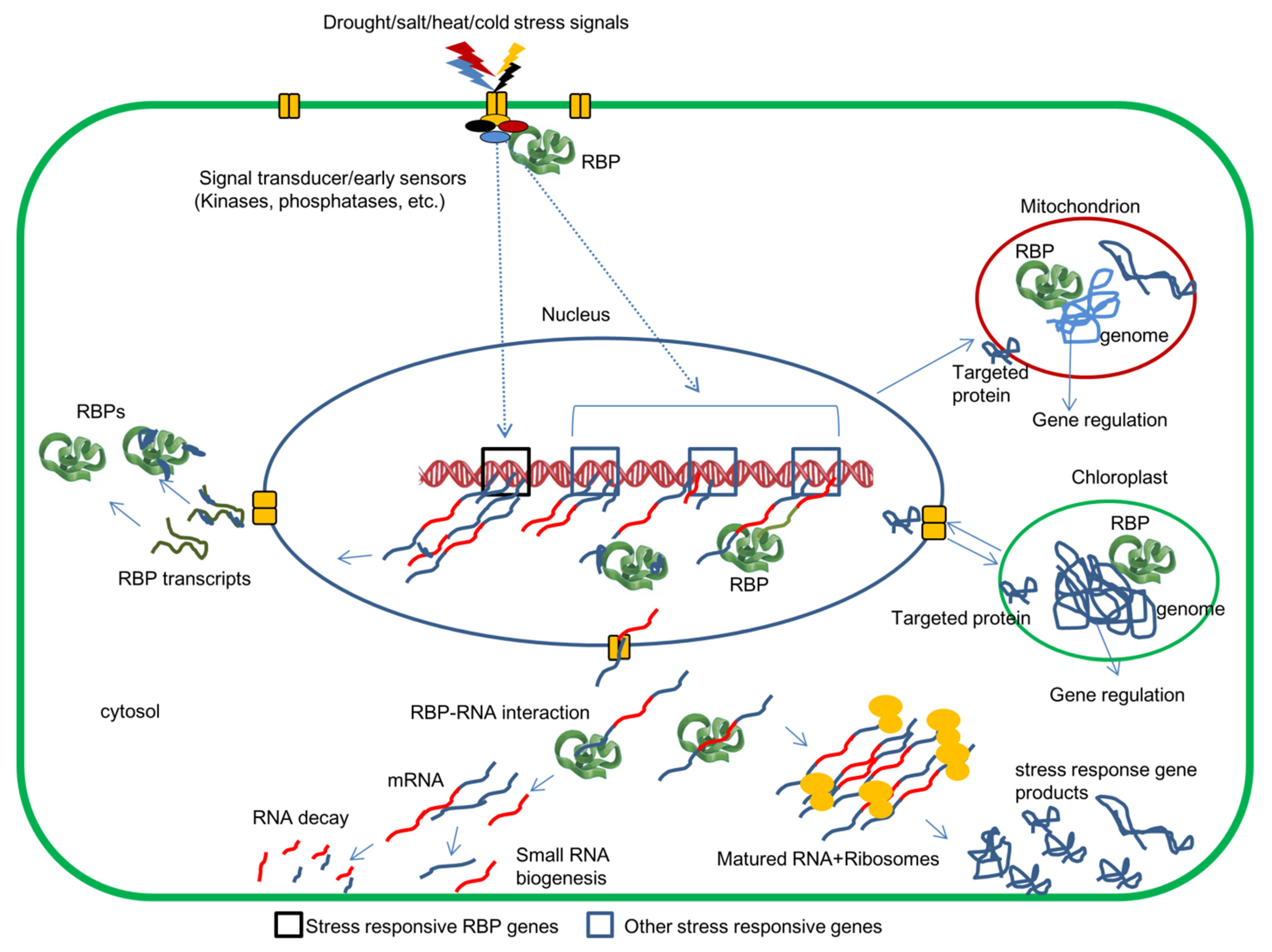
:1. Introduction
2. Regulation of RBPs
| RBPs | Study | Total Genes | Species | Drought | Modulation | Ref. | |
|---|---|---|---|---|---|---|---|
| Upregulated | Downregulated | ||||||
| YTH | - | 39 | T. aestivum L | - | responsive | [39] | |
| - | 10 | C. sinensis | - | CitYTH2, 3, 5, 7, 8 | CitYTH1, CitYTH4 | [40] | |
| MhYTP1 | 26 | M. domestica | T | - | - | [41] | |
| - | 26 | M. domestica | - | responsive | [42] | ||
| - | 13 | A. thaliana | - | - | AtYTH10 | [18] | |
| - | 12 | O. sativa | - | OsYTH01, 2, 3,5,12 | OsYTH10, 11 | [18] | |
| SR | - | 18 | M. esculenta Crantz | - | MeRS40 | MeSR20, Z21a, 34a, RSZ22a, SC35 | [43] |
| - | 18 | B. distachyon | - | responsive | [44] | ||
| SR45a | B. rapa | T | SR45a | - | [45] | ||
| GR-RBP | GRP7 | 8 | A. thaliana | S | - | - | [46] |
| - | 37-Ga 32-Gr | G. arboretum G. raimondii | - | GaRB-GRP4, 9,17 | - | [47] | |
| OsGRP3 | - | O. sativa | T | OsGRP3 | - | [48] | |
| BrRZ1, 2, 3 | - | B. rapa | S | BrRZ1, BrRZ2, BrRZ3 | - | [21] | |
| TaRZ2 or 3 | - | T. aestivum | S | - | - | [49] | |
| CSDP1 | - | A. thaliana | S | - | - | [50] | |
| CspA | - | E. coli/ A. thaliana | T | - | - | [51] | |
| PPR | SOAR1 | - | A. thaliana | T | - | - | [52] |
| - | 491 | O. sativa | - | LOC_Os02g46980, LOC_Os04g01990, LOC_Os03g53170 | - | [53] | |
| GmPPR4 | 179 DYW | G. max | T | GmPPR4 | - | [54] | |
| POCO1 | - | A. thaliana | S (M) | - | - | [55] | |
| DEAD-RH | SlDEAD31 | - | S. lycopersicum | T | SlDEAD31 | - | [56] |
| OsRH58 | - | O. sativa | T | OsRH58 | - | [32] | |
| OsABP | O. sativa | - | OsABP | [57] | |||
| ARP1 | - | A. thaliana | S | - | - | [28] | |
| PUF | APUM5 | A. thaliana | S | [58] | |||
| hnRNP-like | - | E. guineensis | - | EgRBP42 | - | [25] | |
| RBP (Domain) | Study | Total Genes | Species | Salinity | Modulation | Ref. | |
|---|---|---|---|---|---|---|---|
| UP | Down | ||||||
| YTH | - | 39 | T. aestivum L | - | responsive | [39] | |
| - | 10 | C. sinensis | - | CitYTH4 | CitYTH2 | [40] | |
| - | 5 | C. sativus | - | - | CsYTH1, 2, 3, 4, 5 | [38] | |
| - | 26 | M. domestica | - | responsive | [42] | ||
| - | 13 | A. thaliana | - | - | AtYTH10 | [18] | |
| SR | SR45a | A. thaliana | S | SR45a-1a, SR45a-1b | - | [34] | |
| SR45.1 | A. thaliana | T | [59] | ||||
| MeSR34 | 18 | M. esculenta Crantz | T | MeRS40, MeRS31, MeRS2Z33 | MeRSZ21a, MeSCL28, MeRS2Z33, MeSR34a, MeRSZ22a, MeSC35 | [43] | |
| - | 18 | B. distachyon | - | responsive | [44] | ||
| - | 28 | B. rapa | - | BrSR3, BrSCL2, BrSR-like 3 | - | [20] | |
| GR-RBP | NtRGP-1a, 1b, 2, 3 | N. tabacum | - | - | NtRGP-1a, 1b, 2, 3 | [60] | |
| AtGRDP2 | A. thaliana | T | - | - | [61] | ||
| GRP7 | 8 | A. thaliana | S | - | - | [46] | |
| - | 37-Ga 32-Gr | G. arboreum G. raimondii | - | GaRB-GRP4, 17, 9, GrRB-GRP31,9 | GrRB-GRP27,14 | [47] | |
| AtZFP1 | - | A. thaliana | T | AtZFP1 (At2g25900) | [62] | ||
| BrRZ1,2,3 | - | B. rapa | S | BrRZ1, BrRZ2, BrRZ3 | [21] | ||
| TaRZ1, 2,3 | - | T. aestivum | S | - | [49] | ||
| SRP1 | - | A. thaliana | S | - | [24] | ||
| CSDP1 | - | A. thaliana | S | - | [50] | ||
| CSDP2 | - | A. thaliana | T | - | [50] | ||
| CspA | - | E. coli/A. thaliana | T | - | [51] | ||
| PPR | wsl | 477 | O. sativa | S (M) | responsive | [63] | |
| PGN | - | A. thaliana | S (M) | - | - | [64] | |
| SOAR1 | - | A. thaliana | T | - | - | [52] | |
| PPR40 | A. thaliana | T | [65] | ||||
| - | 491 | O. sativa | LOC_Os05g47510, LOC_Os11g37330, LOC_Os03g53170 | - | [53] | ||
| PPR96 | 105-E-type | A. thaliana | S (M) | - | - | [66] | |
| - | 626 | poplar | PtrPPR5, 41,121, 185, 277, 481, 574 | PtrPPR8, 30, 119 | [67] | ||
| - | 179 DYW-PPR | Glycine max | - | GmPPR4 | - | [54] | |
| DEAD-RH | AtRH17 | - | A. thaliana | T | - | AtRH9, AtRH25 | [68] |
| AtRH9, 25 | - | A. thaliana | S | - | - | [69] | |
| OsRH58 | - | A. thaliana | T | OsRH58 | - | [19] | |
| SlDEAD31 | - | S. lycopersicum | T | SlDEAD30, 31 | - | [56] | |
| STRS1, STRS2 | - | A. thaliana | S | - | - | [70] | |
| OsSUV3 | O. sativa | T | [71] | ||||
| OsABP | O. sativa | - | OsABP | [57] | |||
| KH | hos5-1 | A. thaliana | S (M) | - | - | ||
| PUF | APUM5 | A. thaliana | S | [58] | |||
| SAHY9/APUM23 | A. thaliana | S | [72] | ||||
| SDP | At1g12800 | - | A. thaliana | S (M) | - | - | [73] |
| OsRBD1 | - | O. sativa | T | - | - | [74] | |
| Ds-RBPs | FRY2/CPL1 | - | A. thaliana | S (M) | - | - | [75] |
| DRB2,3 | - | A. thaliana | T | - | - | [76] | |
| CRM | CFM4) | - | A. thaliana | S (M) | - | - | [77] |
| - | 14 | O. sativa | - | Os04g0464800, Os11g0592400, Os08g036010, Os05g0551900, Os09g0363100, Os06g0304500, Os01g0958400, Os04g0492900, Os05g0145500, Os08g0188000, Os08g0174900, Os01g0323300, Os01g0495900, Os10g0512500 | [78] | ||
| hnRNP | - | - | E. guineensis | - | EgRBP42 | - | [25] |
| At3g54770 | - | thaliana | S | - | - | [28] | |
| Tudor-SN | - | S (M) | - | - | [79] | ||
| RBP Domain | Study | Total Genes | Species | Heat | Modulation | Ref. | |
|---|---|---|---|---|---|---|---|
| Upregulated | Downregulated | ||||||
| YTH | - | 39 | T. aestivum L | - | responsive | [39] | |
| - | 10 | C. sinensis | - | CitYTH2, 4, 5, 9 | CitYTH3, 6, 7, 10 | [40] | |
| - | 26 | M. domestica | - | responsive | [42] | ||
| - | 13 | A. thaliana | - | AtYTH07, 10 | AtYTH08 | [18] | |
| - | 12 | O. sativa | - | OsYTH08 | OsYTH11, 12 | [18] | |
| SR | - | 19 | S. lycopersicum | - | RSZ and RS2Z subfamilies, Sl-RS28, 29, 42, 46a | Sl-RS41 | [36] |
| - | 18 | B. distachyon | - | responsive | [44] | ||
| - | 28 | B. rapa | - | BrSR3, BrRS2Z2, BrRSZ1, BrSCL2,3, 4, BrSR-like 3 | BrRS2Z1, BrSCL5, BrRS1,2 | [20] | |
| PUF | APUM9 | - | A. thaliana | T | - | - | [80] |
| GR-RBP | NtRGP-1a, 1b, 2,3 | - | N. tabacum | - | NtRGP-1a,1b,2,3 | - | [60] |
| DEAD-RH | OsRH58 | - | A. thaliana | - | OsRH58 | - | [19] |
| SlDEAD31 | - | S. lycopersicum | - | SlDEAD31 | - | [56] | |
| STRS1, 2 | - | A. thaliana | S | - | - | [39] | |
| SDP | At1g12800 | - | A. thaliana | S | - | - | [73] |
| KH | RCF3 | - | A. thaliana | T (M) | - | - | [81] |
| esr1-1, esr1-2 | - | A. thaliana | T (M) | - | - | [13] | |
| hnRNP | - | E. guineensis | - | EgRBP42 | - | [25] | |
| FCA | - | A. thaliana | T | - | - | [82] | |
| RBPs Class | Study | Total Genes | Species | Cold | Modulation | Ref. | |
|---|---|---|---|---|---|---|---|
| Upregulated | Downregulated | ||||||
| YTH | - | 39 | T. aestivum L | - | responsive | [39] | |
| - | 10 | C. sinensis | - | CitYTH1, 2, 3, 4, 5, 6, 7, 8, 9, 10 | - | [40] | |
| - | 5 | C. sativus | - | CsYTH1, 2, 4 | - | [38] | |
| - | 26 | M. domestica | - | MdYTP1, 2, 5, 11, 14 | MdYTP3, 4, 8, 15 | [42] | |
| - | 13 | A. thaliana | - | AtYTH05 | AtYTH10 | [18] | |
| - | 12 | O. sativa | - | OsYTH08 | OsYTH10, 5, 6, 7, 9 | [18] | |
| SR | - | 18 | B. distachyon | - | responsive | [44] | |
| - | 28 | B. rapa | - | BrRSZ3, BrSR1,2,4, BrRS2Z2-5, BrRSZ1, BrSCL4,5, BrSR-like 2 | BrSR3, BrSCL2, BrSR-like 3 | [20] | |
| GR-RBP | NtRGP-1a, 1b,2, 3 | - | N. tabacum | - | NtRGP-1a, NtRGP-1b, NtRGP-2, NtRGP-3 | - | [60] |
| HvGRRBP1 | - | H. vulgare L. | T | HvGRRBP1 | - | [31] | |
| OsGRP1, 4, 6 | - | Oryza sativa | T | - | - | [83] | |
| atRZ-1a | - | A. thaliana | T | - | - | [84] | |
| OsRZ2 | - | Oryza sativa | T | - | - | ||
| BrRZ1, Z2, Z3 | - | B. rapa | S | BrRZ1, BrRZ2, BrRZ3 | - | [21] | |
| atRZ-1a | - | A. thaliana | T | - | - | [84] | |
| TaRZ1 | - | T. aestivum | S | - | - | [49] | |
| CP31A, CP29A | - | Arabidopsis | T | - | - | [23] | |
| OsCSP1, 2 | - | O. sativa | T | OsCSP1, OsCSP2 | - | [85] | |
| CSDP1 | - | A. thaliana | T | - | - | [9] | |
| AtCSP2 | - | A. thaliana | S | - | - | [86] | |
| AtCSP3 | - | A. thaliana | - | AtCSP3 | - | [87] | |
| PPR | SOAR1 | - | A. thaliana | T | - | - | [52] |
| DEAD-RH | RH50 | - | A. thaliana | S (M) | - | - | [88] |
| AtRH9, AtRH25 | - | A. thaliana | - | AtRH9, AtRH25 | - | [68] | |
| AtRH7/PRH75 | - | A. thaliana | T | AtRH7/PRH75 | - | [89] | |
| TCD33 | - | O. sativa | S (M) | - | - | [90] | |
| KH | sh1 | A. thaliana | S (M) | - | - | [91] | |
| CRM | CFM4 | - | A. thaliana | S (M) | - | - | [77] |
3. Plant RBP Signatures/Domain Characteristics and Their Role in Abiotic Stress Responses
3.1. Glycine-Rich RBPs
3.1.1. Zinc Finger Containing Glycine-Rich RBPs/Zinc Finger RBPs
3.1.2. Cold Shock Domain Protein (CSDP)
3.2. Serine/Arginine-Rich (SR) Domain
3.3. PPR Proteins
3.4. YTH Domain
3.5. Pumilio/Fem-3 Binding Factors (PUF) RBPs
3.6. DEAD-Box RNA Helicases
3.7. KH Domain
3.8. S1 Domain Containing-Protein (SDP)
3.9. The Chloroplast RNA Splicing and Ribosome Maturation (CRM) Domain-Containing Proteins
3.10. Double-Stranded RNA-Binding Protein (DRBP)
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dedow, L.K.; Bailey-Serres, J. Searching for a Match: Structure, Function and Application of Sequence-Specific RNA-Binding Proteins. Plant Cell Physiol. 2019, 60, 1927–1938. [Google Scholar] [CrossRef] [PubMed]
- Muleya, V.; Marondedze, C. Functional Roles of RNA-Binding Proteins in Plant Signaling. Life 2020, 10, 288. [Google Scholar] [CrossRef]
- Marondedze, C. The increasing diversity and complexity of the RNA-binding protein repertoire in plants. Proc. R. Soc. B Biol. Sci. 2020, 287, 20201397. [Google Scholar] [CrossRef]
- Kim, M.K.; Jung, H.J.; Kim, D.H.; Kang, H. Characterization of glycine-rich RNA-binding proteins in Brassica napus under stress conditions. Physiol. Plant. 2012, 146, 297–307. [Google Scholar] [CrossRef]
- Prall, W.; Sharma, B.; Gregory, B.D. Transcription Is Just the Beginning of Gene Expression Regulation: The Functional Significance of RNA-Binding Proteins to Post-transcriptional Processes in Plants. Plant Cell Physiol. 2019, 60, 1939–1952. [Google Scholar] [CrossRef]
- Reichel, M.; Liao, Y.; Rettel, M.; Ragan, C.; Evers, M.; Alleaume, A.M.; Horos, R.; Hentze, M.W.; Preiss, T.; Millar, A.A. In planta determination of the mRNA-binding proteome of arabidopsis etiolated seedlings. Plant Cell 2016, 28, 2435–2452. [Google Scholar] [CrossRef] [Green Version]
- Bach-Pages, M.; Homma, F.; Kourelis, J.; Kaschani, F.; Mohammed, S.; Kaiser, M.; van der Hoorn, R.; Castello, A.; Preston, G. Discovering the RNA-Binding Proteome of Plant Leaves with an Improved RNA Interactome Capture Method. Biomolecules 2020, 10, 661. [Google Scholar] [CrossRef]
- Lou, L.; Ding, L.; Wang, T.; Xiang, Y. Emerging Roles of RNA-Binding Proteins in Seed Development and Performance. Int. J. Mol. Sci. 2020, 21, 6822. [Google Scholar] [CrossRef]
- Kim, J.S.; Park, S.J.; Kwak, K.J.; Kim, Y.O.; Kim, J.Y.; Song, J.; Jang, B.; Jung, C.H.; Kang, H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 2007, 35, 506–516. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Ogé, L.; Perez-Garcia, M.D.; Hamama, L.; Sakr, S. The PUF Protein Family: Overview on PUF RNA Targets, Biological Functions, and Post Transcriptional Regulation. Int. J. Mol. Sci. 2018, 19, 410. [Google Scholar] [CrossRef] [Green Version]
- Lurin, C.; Andreés, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyeère, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-Wide Analysis of Arabidopsis Pentatricopeptide Repeat Proteins Reveals Their Essential Role in Organelle Biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef] [Green Version]
- Clavel, M.; Pélissier, T.; Montavon, T.; Tschopp, M.A.; Pouch-Pélissier, M.N.; Descombin, J.; Jean, V.; Dunoyer, P.; Bousquet-Antonelli, C.; Deragon, J.M. Evolutionary history of double-stranded RNA binding proteins in plants: Identification of new cofactors involved in easiRNA biogenesis. Plant Mol. Biol. 2016, 91, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, L.F.; Kamphuis, L.G.; Hane, J.K.; Oñate-Sánchez, L.; Singh, K.B. The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [Green Version]
- Duque, P. A role for SR proteins in plant stress responses. Plant Signal. Behav. 2011, 6, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Kang, H. Roles of Organellar RNA-Binding Proteins in Plant Growth, Development, and Abiotic Stress Responses. Int. J. Mol. Sci. 2020, 21, 4548. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wu, J.; Liu, Y.; Gong, X.; Xu, J.; Lin, D.; Dong, Y. The Rice Pentatricopeptide Repeat Gene TCD10 is Needed for Chloroplast Development under Cold Stress. Rice 2016, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Burjoski, V.; Reddy, A.S.N. The Landscape of RNA-Protein Interactions in Plants: Approaches and Current Status. Int. J. Mol. Sci. 2021, 22, 2845. [Google Scholar] [CrossRef]
- Li, D.; Zhang, H.; Hong, Y.; Huang, L.; Li, X.; Zhang, Y.; Ouyang, Z.; Song, F. Genome-Wide Identification, Biochemical Characterization, and Expression Analyses of the YTH Domain-Containing RNA-Binding Protein Family in Arabidopsis and Rice. Plant Mol. Biol. Report. 2014, 32, 1169–1186. [Google Scholar] [CrossRef]
- Nawaz, G.; Kang, H. Chloroplast- or mitochondria-targeted DEAD-box RNA helicases play essential roles in organellar RNA metabolism and abiotic stress responses. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, E.K.; Krishnamurthy, P.; Kim, J.A.; Jeong, M.J.; Lee, S.I. Genome-wide Characterization of Brassica rapa Genes Encoding Serine/arginine-rich Proteins: Expression and Alternative Splicing Events by Abiotic Stresses. J. Plant Biol. 2018, 61, 198–209. [Google Scholar] [CrossRef]
- Park, Y.R.; Choi, M.J.; Park, S.J.; Kang, H. Three zinc-finger RNA-binding proteins in cabbage (Brassica rapa) play diverse roles in seed germination and plant growth under normal and abiotic stress conditions. Physiol. Plant. 2017, 159, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Cui, P.; Xiong, L. The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2015, 43, 8283–8298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kupsch, C.; Ruwe, H.; Gusewski, S.; Tillich, M.; Small, I.; Schmitz-Linneweber, C. Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell 2012, 24, 4266–4280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Chen, Y.; Qian, L.; Mu, R.; Yuan, X.; Fang, H.; Huang, X.; Xu, E.; Zhang, H.; Huang, J. A novel RNA-binding protein involves ABA signaling by post-transcriptionally repressing ABI2. Front. Plant Sci. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeap, W.C.; Ooi, T.E.K.; Namasivayam, P.; Kulaveerasingam, H.; Ho, C.L. EgRBP42 encoding an hnRNP-like RNA-binding protein from Elaeis guineensis Jacq. is responsive to abiotic stresses. Plant Cell Rep. 2012, 31, 1829–1843. [Google Scholar] [CrossRef]
- Bazin, J.; Romero, N.; Rigo, R.; Charon, C.; Blein, T.; Ariel, F.; Crespi, M. Nuclear Speckle RNA Binding Proteins Remodel Alternative Splicing and the Non-coding Arabidopsis Transcriptome to Regulate a Cross-Talk Between Auxin and Immune Responses. Front. Plant Sci. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Doroshenk, K.A.; Zhang, L.; Fukuda, M.; Washida, H.; Kumamaru, T.; Okita, T. Zipcode RNA-binding proteins and membrane trafficking proteins cooperate to transport glutelin mRNAs in rice endosperm. Plant Cell 2020, 32, 2566–2581. [Google Scholar] [CrossRef]
- Jung, H.J.; Kim, M.K.; Kang, H. An ABA-regulated putative RNA-binding protein affects seed germination of Arabidopsis under ABA or abiotic stress conditions. J. Plant Physiol. 2013, 170, 179–184. [Google Scholar] [CrossRef]
- Pieczynski, M.; Kruszka, K.; Bielewicz, D.; Dolata, J.; Szczesniak, M.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. A role of U12 intron in proper pre-mRNA splicing of plant cap binding protein 20 genes. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Zinc finger-containing glycine-rich RNA-binding protein in Oryza sativa has an RNA chaperone activity under cold stress conditions. Plant Cell Environ. 2010, 33, 759–768. [Google Scholar] [CrossRef]
- Tripet, B.P.; Mason, K.E.; Eilers, B.J.; Burns, J.; Powell, P.; Fischer, A.M.; Copié, V. Structural and biochemical analysis of the hordeum vulgare L. Hv GR-RBP1 protein, a glycine-rich RNA-binding protein involved in the regulation of barley plant development and stress response. Biochemistry 2014, 53, 7945–7960. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, G.; Kang, H. Rice OsRH58, a chloroplast DEAD-box RNA helicase, improves salt or drought stress tolerance in Arabidopsis by affecting chloroplast translation. BMC Plant Biol. 2019, 19, 1–11. [Google Scholar] [CrossRef]
- Martín, G.; Márquez, Y.; Mantica, F.; Duque, P.; Irimia, M. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 2021, 22, 1–26. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Q.; Liu, P.; Huang, J.; Zhang, S.; Yang, G.; Wu, C.; Zheng, C.; Yan, K. Dual roles of the serine/arginine-rich splicing factor SR45a in promoting and interacting with nuclear cap-binding complex to modulate the salt-stress response in Arabidopsis. New Phytol. 2021, 230, 641–655. [Google Scholar] [CrossRef]
- Melo, J.P.; Kalyna, M.; Duque, P. Current Challenges in Studying Alternative Splicing in Plants: The Case of Physcomitrella patens SR Proteins. Front. Plant Sci. 2020, 11, 286. [Google Scholar] [CrossRef] [Green Version]
- Rosenkranz, R.R.E.; Bachiri, S.; Vraggalas, S.; Keller, M.; Simm, S.; Schleiff, E.; Fragkostefanakis, S. Identification and Regulation of Tomato Serine/Arginine-Rich Proteins Under High Temperatures. Front. Plant Sci. 2021, 12, 645689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chen, M.X.; Zhu, F.Y.; Zhang, J.; Liu, Y.G. Emerging Functions of Plant Serine/Arginine-Rich (SR) Proteins: Lessons from Animals. CRC. Crit. Rev. Plant Sci. 2020, 39, 173–194. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.; Jiang, L.; Liu, S. Genome-wide identification and expression analysis of YTH domain-containing RNA-binding protein family in cucumber (Cucumis sativus). Genes Genom. 2018, 40, 579–589. [Google Scholar] [CrossRef]
- Sun, J.; Bie, X.M.; Wang, N.; Zhang, X.S.; Gao, X.-Q. Genome-wide identification and expression analysis of YTH domain-containing RNA-binding protein family in common wheat. BMC Plant Biol. 2020, 20, 351. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Z.; Duan, H.; Mi, L.; Hu, W.; Chen, J.; Li, X.; Zhong, B. Genome-wide identification and expression analysis of the YTH domain-containing RNA-binding protein family in Citrus Sinensis. J. Am. Soc. Hortic. Sci. 2019, 144, 79–91. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Wang, N.; Xue, Y.; Guan, Q.; van Nocker, S.; Liu, C.; Ma, F. Overexpression of the RNA binding protein MhYTP1 in transgenic apple enhances drought tolerance and WUE by improving ABA level under drought condition. Plant Sci. 2019, 280, 397–407. [Google Scholar] [CrossRef]
- Wang, N.; Yue, Z.; Liang, D.; Ma, F. Genome-wide identification of members in the YTH domain-containing RNA-binding protein family in apple and expression analysis of their responsiveness to senescence and abiotic stresses. Gene 2014, 538, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Ma, S.; Zhang, Y.; Wang, D.; Cao, S.; Wang, Z.-Y. Genome-Wide Identification of Cassava Serine/Arginine-Rich Proteins: Insights into Alternative Splicing of Pre-mRNAs and Response to Abiotic Stress. Plant Cell Physiol. 2020, 61, 178–191. [Google Scholar] [CrossRef]
- Chen, S.; Li, J.; Liu, Y.; Li, H. Genome-Wide Analysis of Serine/Arginine-Rich Protein Family in Wheat and Brachypodium distachyon. Plants 2019, 8, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthusamy, M.; Yoon, E.K.; Kim, J.A.; Jeong, M.-J.; Lee, S.I. Brassica Rapa SR45a Regulates Drought Tolerance via the Alternative Splicing of Target Genes. Genes 2020, 11, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; Jung, H.J.; Lee, H.J.; Kim, K.A.; Goh, C.H.; Woo, Y.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 2008, 55, 455–466. [Google Scholar] [CrossRef]
- Yang, W.; Yu, M.; Zou, C.; Lu, C.; Yu, D.; Cheng, H.; Jiang, P.; Feng, X.; Zhang, Y.; Wang, Q.; et al. Genome-wide comparative analysis of RNA-binding Glycine-rich protein family genes between Gossypium arboreum and Gossypium raimondii. PLoS ONE 2018, 14, 1–22. [Google Scholar] [CrossRef]
- Shim, J.S.; Park, S.H.; Lee, D.K.; Kim, Y.S.; Park, S.C.; Redillas, M.C.F.R.; Seo, J.S.; Kim, J.K. The Rice GLYCINE-RICH PROTEIN 3 Confers Drought Tolerance by Regulating mRNA Stability of ROS Scavenging-Related Genes. Rice 2021, 14. [Google Scholar] [CrossRef]
- Xu, T.; Gu, L.; Choi, M.J.; Kim, R.J.; Suh, M.C.; Kang, H. Comparative functional analysis of wheat (Triticum aestivum) zinc finger-containing glycine-rich RNA-binding proteins in response to abiotic stresses. PLoS ONE 2014, 9, e96877. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J.; Kwak, K.J.; Oh, T.R.; Kim, Y.O.; Kang, H. Cold Shock Domain Proteins Affect Seed Germination and Growth of Arabidopsis thaliana Under Abiotic Stress Conditions. Plant Cell Physiol. 2009, 50, 869–878. [Google Scholar] [CrossRef]
- Yu, T.F.; Xu, Z.S.; Guo, J.K.; Wang, Y.X.; Abernathy, B.; Fu, J.D.; Chen, X.; Zhou, Y.B.; Chen, M.; Ye, X.G.; et al. Improved drought tolerance in wheat plants overexpressing a synthetic bacterial cold shock protein gene SeCspA. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Mei, C.; Liang, S.; Yu, Y.T.; Lu, K.; Wu, Z.; Wang, X.F.; Zhang, D.P. Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Mol. Biol. 2015, 88, 369–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Zou, Y.; Hu, J.; Ding, Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genom. 2018, 19, 1–14. [Google Scholar] [CrossRef]
- Su, H.G.; Li, B.; Song, X.Y.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Min, D.H.; Xu, Z.S.; Ma, Y.Z. Genome-wide analysis of the DYW subgroup PPR gene family and identification of gmPPR4 responses to drought stress. Int. J. Mol. Sci. 2019, 20, 5667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emami, H.; Kumar, A.; Kempken, F. Transcriptomic analysis of poco1, a mitochondrial pentatricopeptide repeat protein mutant in Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Chen, G.; Dong, T.; Wang, L.; Zhang, J.; Zhao, Z.; Hu, Z. SlDEAD31, a putative DEAD-Box RNA helicase gene, regulates salt and drought tolerance and stress-related genes in tomato. PLoS ONE 2015, 10, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Macovei, A.; Vaid, N.; Tula, S.; Tuteja, N. A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signal. Behav. 2012, 7, 1138–1143. [Google Scholar] [CrossRef] [Green Version]
- Huh, S.U.; Paek, K.H. APUM5, encoding a Pumilio RNA binding protein, negatively regulates abiotic stress responsive gene expression. BMC Plant Biol. 2014, 14, 75. [Google Scholar] [CrossRef] [Green Version]
- Albaqami, M.; Laluk, K.; Reddy, A.S.N. The Arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner. Plant Mol. Biol. 2019, 100, 379–390. [Google Scholar] [CrossRef]
- Chen, X.; Zeng, Q.; Lu, X.; Yu, D.; Li, W. Characterization and Expression Analysis of Four Glycine-Rich RNA-Binding Proteins Involved in Osmotic Response in Tobacco (Nicotiana tabacum cv. Xanthi). Agric. Sci. China 2010, 9, 1577–1587. [Google Scholar] [CrossRef]
- Ortega-Amaro, M.A.; Aída, A.; Rodríguez-Hernández, M.R.-K.; Hernández-Lucero, S.R.-M.E.; Ibáñez-Salazar, A.; Delgado-Sánchez, P.; Jiménez-Bremont, J.F. Overexpression of AtGRDP2, a novel glycine-rich domain protein, accelerates plant growth and improves stress tolerance. Front. Plant Sci. 2015, 5, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.; Wang, M.; Yuan, F.; Sui, N.; Song, J.; Wang, B. The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Mol. Biol. 2014, 86, 237–253. [Google Scholar] [CrossRef]
- Tan, J.; Tan, Z.; Wu, F.; Sheng, P.; Heng, Y.; Wang, X.; Ren, Y.; Wang, J.; Guo, X.; Zhang, X.; et al. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant 2014, 7, 1329–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laluk, K.; AbuQamar, S.; Mengiste, T. The Arabidopsis Mitochondria-Localized Pentatricopeptide Repeat Protein PGN Functions in Defense against Necrotrophic Fungi and Abiotic Stress Tolerance. Plant Physiol. 2011, 156, 2053–2068. [Google Scholar] [CrossRef] [Green Version]
- Zsigmond, L.; Szepesi, Á.; Tari, I.; Rigó, G.; Király, A.; Szabados, L. Overexpression of the mitochondrial PPR40 gene improves salt tolerance in Arabidopsis. Plant Sci. 2012, 182, 87–93. [Google Scholar] [CrossRef]
- Liu, J.M.; Zhao, J.Y.; Lu, P.P.; Chen, M.; Guo, C.H.; Xu, Z.S.; Ma, Y.Z. The E-subgroup pentatricopeptide repeat protein family in Arabidopsis thaliana and confirmation of the responsiveness PPR96 to abiotic stresses. Front. Plant Sci. 2016, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Xing, H.; Fu, X.; Yang, C.; Tang, X.; Guo, L.; Li, C.; Xu, C.; Luo, K. Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.V.; Seok, H.Y.; Woo, D.H.; Lee, S.Y.; Moon, Y.H. Overexpression of the DEAD-Box RNA helicase gene AtRH17 confers tolerance to salt stress in arabidopsis. Int. J. Mol. Sci. 2018, 19, 3777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; Kim, K.A.; Oh, T.R.; Park, C.M.; Kang, H. Functional characterization of DEAD-box RNA helicases in Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol. 2008, 49, 1563–1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Garbelli, A.; Grossi, S.; Florentin, A.; Batelli, G.; Acuna, T.; Zolla, G.; Kaye, Y.; Paul, L.K.; Zhu, J.K.; et al. The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing. Plant J. 2014, 79, 28–43. [Google Scholar] [CrossRef]
- Tuteja, N.; Sahoo, R.K.; Garg, B.; Tuteja, R. OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64). Plant J. 2013, 76, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.C.; Lin, W.C.; Cheng, W.H. Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 2018, 18, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Dinh, S.N.; Park, S.J.; Han, J.H.; Kang, H. A Chloroplast-targeted S1 RNA-binding Domain Protein Plays a Role in Arabidopsis Response to Diverse Abiotic Stresses. J. Plant Biol. 2019, 62, 74–81. [Google Scholar] [CrossRef]
- Sharma, S.; Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. OsSRO1a interacts with RNA binding domain-containing protein (OsRBD1) and functions in abiotic stress tolerance in yeast. Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, L.; Lee, H.; Ishitani, M.; Tanaka, Y.; Stevenson, B.; Koiwa, H.; Bressan, R.A.; Hasegawa, P.M.; Zhu, J.K. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10899–10904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawano, H.; Matsuzaki, T.; Usui, T.; Tabara, M.; Fukudome, A.; Kanaya, A.; Tanoue, D.; Hiraguri, A.; Horiguchi, G.; Ohtani, M.; et al. Double-stranded RNA-binding protein DRB3 negatively regulates anthocyanin biosynthesis by modulating PAP1 expression in Arabidopsis thaliana. J. Plant Res. 2017, 130, 45–55. [Google Scholar] [CrossRef]
- Lee, K.; Lee, H.J.; Kim, D.H.; Jeon, Y.; Pai, H.S.; Kang, H. A nuclear-encoded chloroplast protein harboring a single CRM domain plays an important role in the Arabidopsis growth and stress response. BMC Plant Biol. 2014, 14, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Shen, L.; Ren, D.; Hu, J.; Zhu, L.; Gao, Z.; Zhang, G.; Guo, L.; Zeng, D.; Qian, Q. Characterization of the CRM gene family and elucidating the function of OsCFM2 in rice. Biomolecules 2020, 10, 327. [Google Scholar] [CrossRef] [Green Version]
- Dit Frey, N.F.; Muller, P.; Jammes, F.; Kizis, D.; Leung, J.; Perrot-Rechenmann, C.; Bianchi, M.W. The RNA Binding Protein Tudor-SN Is Essential for Stress Tolerance and Stabilizes Levels of Stress-Responsive mRNAs Encoding Secreted Proteins in Arabidopsis. Plant Cell 2010, 22, 1575–1591. [Google Scholar] [CrossRef] [Green Version]
- Nyikó, T.; Auber, A.; Bucher, E. Functional and molecular characterization of the conserved Arabidopsis PUMILIO protein, APUM9. Plant Mol. Biol. 2019, 100, 199–214. [Google Scholar] [CrossRef] [Green Version]
- Guan, Q.; Wen, C.; Zeng, H.; Zhu, J. A KH domain-containing putative RNA-binding protein is critical for heat stress-responsive gene regulation and thermotolerance in arabidopsis. Mol. Plant 2013, 6, 386–395. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Lee, H.J.; Jung, J.H.; Park, C.M. The Arabidopsis thaliana RNA-binding protein FCA regulates thermotolerance by modulating the detoxification of reactive oxygen species. New Phytol. 2015, 205, 555–569. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. J. Exp. Bot. 2010, 61, 2317–2325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.O.; Pan, S.O.; Jung, C.H.; Kang, H. A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol. 2007, 48, 1170–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaikam, V.; Karlson, D. Functional characterization of two cold shock domain proteins from Oryza sativa. Plant Cell Environ. 2008, 31, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Kim, M.H.; Imai, R. Arabidopsis COLD SHOCK DOMAIN PROTEIN 2 is a negative regulator of cold acclimation. New Phytol. 2013, 198, 95–102. [Google Scholar] [CrossRef]
- Kim, M.H.; Sasaki, K.; Imai, R. Cold shock domain protein 3 regulates freezing tolerance in Arabidopsis thaliana. J. Biol. Chem. 2009, 284, 23454–23460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paieri, F.; Tadini, L.; Manavski, N.; Kleine, T.; Ferrari, R.; Morandini, P.; Pesaresi, P.; Meurer, J.; Leister, D. The DEAD-box RNA helicase RH50 is a 23S-4.5S rRNA maturation factor that functionally overlaps with the plastid signaling factor GUN1. Plant Physiol. 2018, 176, 634–648. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.K.; Shen, Y.L.; Huang, L.F.; Wu, S.J.; Yeh, C.H.; Lu, C.A. The DEAD-box RNA helicase AtRH7/PRH75 participates in pre-rRNA processing, plant development and cold tolerance in arabidopsis. Plant Cell Physiol. 2016, 57, 174–191. [Google Scholar] [CrossRef] [Green Version]
- Xiaomei, W.; Rongrong, K.; Ting, Z.; Yuanyuan, G.; Jianlong, X.; Zhongze, P.; Gangseob, L.; Dongzhi, L.; Yanjun, D. A DEAD-box RNA helicase TCD33 that confers chloroplast development in rice at seedling stage under cold stress. J. Plant Physiol. 2020, 248. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, B.; Shen, Y.; Wang, H.; Feng, Q.; Shi, H. The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression. PLoS Genet. 2013, 9, e1003625. [Google Scholar] [CrossRef] [Green Version]
- Czolpinska, M.; Rurek, M. Plant glycine-rich proteins in stress response: An emerging, still prospective story. Front. Plant Sci. 2018, 9, 302. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.O.; Kang, H. The role of a zinc finger-containing glycine-rich RNA-binding protein during the cold adaptation process in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 793–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pi, B.; He, X.; Ruan, Y.; Jang, J.C.; Huang, Y. Genome-wide analysis and stress-responsive expression of CCCH zinc finger family genes in Brassica rapa. BMC Plant Biol. 2018, 18, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Kang, H. Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol. Cells 2016, 39, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Kim, H.R.; Hwang, B.H.; Yi, H.; Hur, Y. Natural variation in glycine-rich region of Brassica oleracea cold shock domain protein 5 (BoCSDP5) is associated with low temperature tolerance. Genes Genom. 2020, 42, 1407–1417. [Google Scholar] [CrossRef]
- Rausin, G.; Tillemans, V.; Stankovic, N.; Hanikenne, M.; Motte, P. Dynamic Nucleocytoplasmic Shuttling of an Arabidopsis SR Splicing Factor: Role of the RNA-Binding Domains. Plant Physiol. 2010, 153, 273–284. [Google Scholar] [CrossRef] [Green Version]
- Barta, A.; Kalyna, M.; Reddy, A.S.N. Implementing a Rational and Consistent Nomenclature for Serine/Arginine-Rich Protein Splicing Factors (SR Proteins) in Plants. Plant Cell 2010, 22, 2926–2929. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.N.; Shad Ali, G. Plant serine/arginine-rich proteins: Roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip. Rev. RNA 2011, 2, 875–889. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Carvalho, S.D.; Duque, P. The Plant-Specific SR45 Protein Negatively Regulates Glucose and ABA Signaling during Early Seedling Development in Arabidopsis. Plant Physiol. 2010, 154, 772–783. [Google Scholar] [CrossRef] [Green Version]
- Reddy, A.S.N.; Day, I.S.; Göhring, J.; Barta, A. Localization and dynamics of nuclear speckles in plants. Plant Physiol. 2012, 158, 67–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelisch, F.; Gerez, J.; Druker, J.; Schor, I.E.; Muñoz, M.J.; Risso, G.; Petrillo, E.; Westman, B.J.; Lamond, A.I.; Arzt, E.; et al. The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proc. Natl. Acad. Sci. USA 2010, 107, 16119–16124. [Google Scholar] [CrossRef] [Green Version]
- Butt, H.; Piatek, A.; Li, L.; Reddy, A.S.N.; Mahfouz, M.M. Multiplex CRISPR mutagenesis of the serine/arginine-rich (SR) gene family in rice. Genes 2019, 10, 596. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; An, Y.; Xu, P.; Xiao, J. Functioning of PPR Proteins in Organelle RNA Metabolism and Chloroplast Biogenesis. Front. Plant Sci. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Sun, T. Expanded Function of the P-Type Pentatricopeptide Repeat Protein ATP4 in RNA Editing. Plant Physiol. 2020, 184, 1625–1626. [Google Scholar] [CrossRef]
- Park, Y.J.; Lee, H.J.; Kwak, K.J.; Lee, K.; Hong, S.W.; Kang, H. MicroRNA400-guided cleavage of pentatricopeptide repeat protein mRNAs renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol. 2014, 55, 1660–1668. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Dugardeyn, J.; Zhang, C.; Mühlenbock, P.; Eastmond, P.J.; Valcke, R.; De Coninck, B.; Öden, S.; Karampelias, M.; Cammue, B.P.A.; et al. The arabidopsis thaliana RNA editing factor SLO2, which affects the mitochondrial electron transport chain, participates in multiple stress and hormone responses. Mol. Plant 2014, 7, 290–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Guo, T.; Wang, P.; Sun, X.; Shao, Y.; Jia, X.; Liang, B.; Gong, X.; Ma, F. MhYTP1 and MhYTP2 from apple confer tolerance to multiple abiotic stresses in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1367. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Guo, T.; Sun, X.; Jia, X.; Wang, P.; Shao, Y.; Liang, B.; Gong, X.; Ma, F. Functions of two Malus hupehensis (Pamp.) Rehd. YTPs (MhYTP1 and MhYTP2) in biotic- and abiotic-stress responses. Plant Sci. 2017, 261, 18–27. [Google Scholar] [CrossRef]
- Song, P.; Yang, J.; Wang, C.; Lu, Q.; Shi, L.; Tayier, S.; Jia, G. Arabidopsis N6-methyladenosine reader CPSF30-L recognizes FUE signals to control polyadenylation site choice in liquid-like nuclear bodies. Mol. Plant 2021, 14, 571–587. [Google Scholar] [CrossRef]
- Park, S.H.; Kim, H.S.; Kalita, P.J.; Choi, S.B. Structural and functional similarities and differences in nucleolar Pumilio RNA-binding proteins between Arabidopsis and the charophyte Chara corallina. BMC Plant Biol. 2020, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, N.; Kim, H.B.; Park, N.I.L.; Kim, H.S.; Kim, Y.K.; Park, Y.I.L.; Choi, S.B. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010, 64, 960–976. [Google Scholar] [CrossRef]
- Huh, S.U.; Kim, M.J.; Paek, K.H. Arabidopsis Pumilio protein APUM5 suppresses Cucumber mosaic virus infection via direct binding of viral RNAs. Proc. Natl. Acad. Sci. USA 2013, 110, 779–784. [Google Scholar] [CrossRef] [Green Version]
- Raj, S.; Bagchi, D.; Orero, J.V.; Banroques, J.; Kyle Tanner, N.; Croquette, V. Mechanistic characterization of the DEAD-box RNA helicase Ded1 from yeast as revealed by a novel technique using single-molecule magnetic tweezers. Nucleic Acids Res. 2019, 47, 3699–3710. [Google Scholar] [CrossRef]
- Sahoo, R.K.; Ansari, M.W.; Tuteja, R.; Tuteja, N. OsSUV3 transgenic rice maintains higher endogenous levels of plant hormones that mitigates adverse effects of salinity and sustains crop productivity. Rice 2014, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorković, Z.J.; Barta, A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002, 30, 623–635. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Cui, P.; Chen, H.; Ali, S.; Zhang, S.; Xiong, L. A KH-Domain RNA-Binding Protein Interacts with FIERY2/CTD Phosphatase-Like 1 and Splicing Factors and Is Important for Pre-mRNA Splicing in Arabidopsis. PLoS Genet. 2013, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Yuan, H.; Yang, Y.; Fish, T.; Lyi, S.M.; Thannhauser, T.W.; Zhang, L.; Li, L. Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J. Exp. Bot. 2016, 67, 2731–2744. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Park, S.J.; Park, Y.I.L.; Kang, H. CFM9, a Mitochondrial CRM Protein, Is Crucial for Mitochondrial Intron Splicing, Mitochondria Function and Arabidopsis Growth and Stress Responses. Plant Cell Physiol. 2019, 60, 2538–2548. [Google Scholar] [CrossRef]
- Montavon, T.; Kwon, Y.; Zimmermann, A.; Michel, F.; Dunoyer, P. New DRB complexes for new DRB functions in plants. RNA Biol. 2017, 14, 1637–1641. [Google Scholar] [CrossRef] [Green Version]
- Hirata, R.; Mishiba, K.I.; Koizumi, N.; Iwata, Y. Deficiency in the double-stranded RNA binding protein HYPONASTIC LEAVES1 increases sensitivity to the endoplasmic reticulum stress inducer tunicamycin in Arabidopsis. BMC Res. Notes 2019, 12, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Fedoroff, N. A Mutation in the Arabidopsis HYL1 Gene Encoding a dsRNA Binding Protein Affects Responses to Abscisic Acid, Auxin, and Cytokinin. Plant Cell 2000, 12, 2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muthusamy, M.; Kim, J.-H.; Kim, J.A.; Lee, S.-I. Plant RNA Binding Proteins as Critical Modulators in Drought, High Salinity, Heat, and Cold Stress Responses: An Updated Overview. Int. J. Mol. Sci. 2021, 22, 6731. https://doi.org/10.3390/ijms22136731
Muthusamy M, Kim J-H, Kim JA, Lee S-I. Plant RNA Binding Proteins as Critical Modulators in Drought, High Salinity, Heat, and Cold Stress Responses: An Updated Overview. International Journal of Molecular Sciences. 2021; 22(13):6731. https://doi.org/10.3390/ijms22136731
Chicago/Turabian StyleMuthusamy, Muthusamy, Jong-Hee Kim, Jin A Kim, and Soo-In Lee. 2021. "Plant RNA Binding Proteins as Critical Modulators in Drought, High Salinity, Heat, and Cold Stress Responses: An Updated Overview" International Journal of Molecular Sciences 22, no. 13: 6731. https://doi.org/10.3390/ijms22136731
APA StyleMuthusamy, M., Kim, J.-H., Kim, J. A., & Lee, S.-I. (2021). Plant RNA Binding Proteins as Critical Modulators in Drought, High Salinity, Heat, and Cold Stress Responses: An Updated Overview. International Journal of Molecular Sciences, 22(13), 6731. https://doi.org/10.3390/ijms22136731








