The Role of RNA Secondary Structure in Regulation of Gene Expression in Bacteria
Abstract
1. Introduction
2. Dynamics of RNA Structure as a Source of Regulatory Potential
2.1. RNA Free Energy Landscape
2.2. Free Energy Barriers
2.3. Types of RNA Conformation Dynamics
2.4. Co-Transcriptional Folding
2.5. Riboswitch Structure Dynamics
2.6. RNA Thermometer Dynamics
3. Regulation of Transcription by Changes in the RNA Secondary Structure
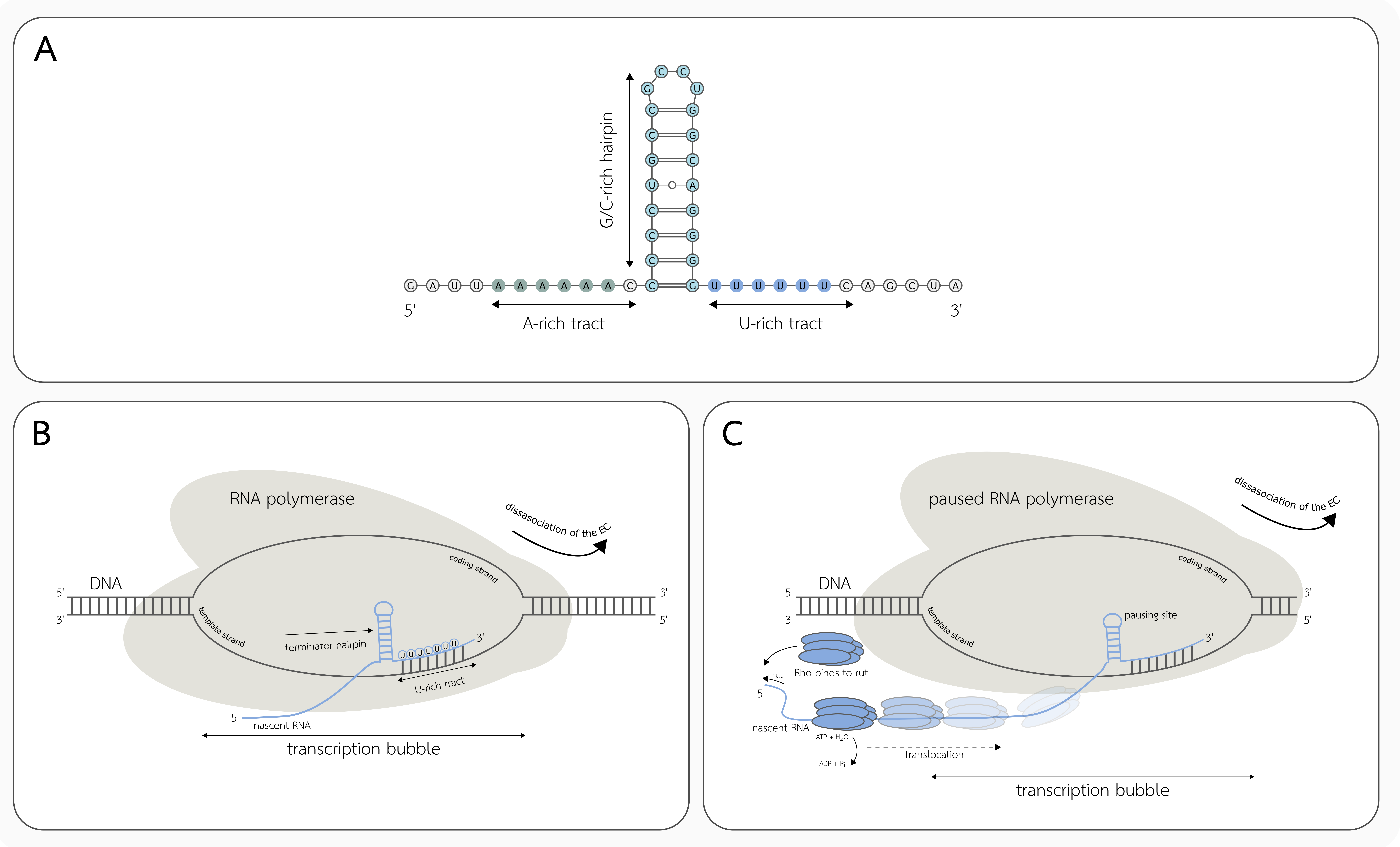
3.1. Intrinsic Transcription Termination
3.2. Factor-Dependent Transcription Termination
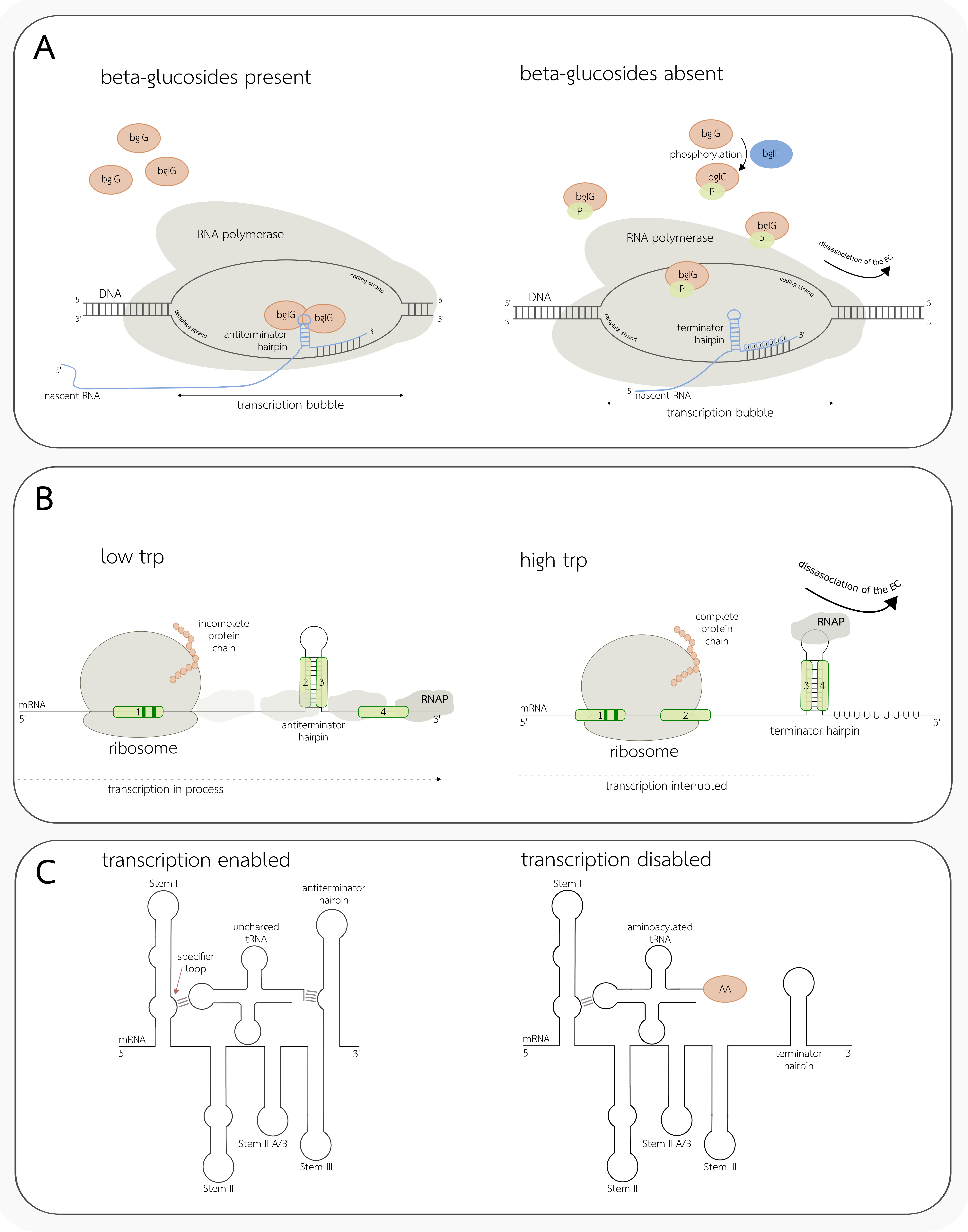
3.3. Conditional Terminators
4. Regulation of Translation by Changes in the RNA Secondary Structure
4.1. Cis-Acting Translation Regulators
4.2. Trans-Acting Translation Regulators
5. Future Challenges and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Bloomfield, V.A.; Crothers, D.M.; Tinoco, I., Jr. Nucleic Acids: Structures, Properties, and Functions; University Science Books: Sausalito, CA, USA, 2000. [Google Scholar]
- Leontis, N.B.; Westhof, E. Geometric nomenclature and classification of RNA base pairs. RNA 2001, 7, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Westhof, E.; Auffinger, P. RNA Tertiary Structure. In Encyclopedia of Analytical Chemistry; Meyer, R.A., Ed.; Wiley: New York, NY, USA, 2006; pp. 5222–5232. [Google Scholar]
- Wan, Y.; Kertesz, M.; Spitale, R.C.; Segal, E.; Chang, H.Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011, 12, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.; Tang, Y.; Assmann, S.M.; Bevilacqua, P.C. The RNA structurome: Transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 2015, 40, 221–232. [Google Scholar] [CrossRef]
- Merino, E.; Yanofsky, C. Transcription attenuation: A highly conserved regulatory strategy used by bacteria. Trends Genet. 2005, 21, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Babitzke, P.; Baker, C.S.; Romeo, T. Regulation of translation initiation by RNA binding proteins. Annu. Rev. Microbiol. 2009, 63, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E.; Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004, 29, 11–17. [Google Scholar] [CrossRef]
- Tucker, B.J.; Breaker, R.R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005, 15, 342–348. [Google Scholar] [CrossRef]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nature Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef]
- Warf, M.B.; Berglund, J.A. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010, 35, 169–178. [Google Scholar] [CrossRef]
- Martin, K.C.; Ephrussi, A. mRNA localization: Gene expression in the spatial dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef]
- Kozak, M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 2005, 361, 13–37. [Google Scholar] [CrossRef]
- Breaker, R.R. Riboswitches and the RNA World. Cold Spring Harb. Perspect. Biol. 2012, 4, a003566. [Google Scholar] [CrossRef] [PubMed]
- Grosshans, H.; Filipowicz, W. Molecular biology: The expanding world of small RNAs. Nature 2008, 451, 414–416. [Google Scholar] [CrossRef] [PubMed]
- McCown, P.J.; Corbino, K.A.; Stav, S.; Sherlock, M.E.; Breaker, R.R. Riboswitch diversity and distribution. RNA 2017, 23, 995–1011. [Google Scholar] [CrossRef]
- Van Assche, E.; Van Puyvelde, S.; Vanderleyden, J.; Steenackers, H.P. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 2015, 6, 141. [Google Scholar]
- Crothers, D.M.; Cole, P.E.; Hilbers, C.W.; Shulman, R.G. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J. Mol. Biol. 1974, 87, 63–88. [Google Scholar] [CrossRef]
- Nowakowski, J.; Tinoco, I. RNA structure and stability. Sem. Virol. 1997, 8, 153–165. [Google Scholar] [CrossRef]
- Russell, R.; Zhuang, X.; Babcock, H.P.; Millett, I.S.; Doniach, S.; Chu, S.; Herschlag, D. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. USA 2002, 99, 155–160. [Google Scholar] [CrossRef]
- Dethoff, E.A.; Chugh, J.; Mustoe, A.M.; Al-Hashimi, H.M. Functional complexity and regulation through RNA dynamics. Nature 2012, 482, 322–330. [Google Scholar] [CrossRef]
- Fürtig, B.; Wenter, P.; Pitsch, S.; Schwalbe, H. Probing mechanism and transition state of RNA refolding. ACS Chem. Biol. 2010, 5, 753–765. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA website. Nucleic Acids Res. 2008, 1, W70–W74. [Google Scholar] [CrossRef]
- Mustoe, A.M.; Brooks, C.L.; Al-Hashimi, H.M. Hierarchy of RNA functional dynamics. Annu. Rev. Biochem. 2014, 83, 441–466. [Google Scholar] [CrossRef]
- Semrad, K.; Schroeder, R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 1998, 12, 1327–1337. [Google Scholar] [CrossRef][Green Version]
- Zemora, G.; Waldsich, C. RNA folding in living cells. RNA Biol. 2010, 7, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Scull, C.E.; Dandpat, S.S.; Romero, R.A.; Walter, N.G. Transcriptional Riboswitches Integrate Timescales for Bacterial Gene Expression Control. Front. Mol. Biosci. 2021, 7, 480. [Google Scholar] [CrossRef] [PubMed]
- Kortmann, J.; Sczodrok, S.; Rinnenthal, J.; Schwalbe, H.; Narberhaus, F. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res. 2011, 39, 2855–2868. [Google Scholar] [CrossRef] [PubMed]
- Kramer, F.R.; Mills, D.R. Secondary structure formation during RNA synthesis. Nucleic Acids Res. 1981, 10, 5109–5124. [Google Scholar] [CrossRef] [PubMed]
- Boyle, J.; Robillard, G.; Kim, S. Sequential folding of transfer RNA. A nuclear magnetic resonance study of successively longer tRNA fragments with a common 5′ end. J. Mol. Biol. 1980, 139, 601–625. [Google Scholar] [CrossRef]
- Ganser, L.R.; Kelly, M.L.; Herschlag, D.; Al-Hashimi, H.M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019, 20, 474–489. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, R.; Grossberger, R.; Pichler, A.; Waldsich, C. RNA folding in vivo. Curr. Opp. Struct. Biol. 2002, 12, 296–300. [Google Scholar] [CrossRef]
- Lai, D.; Proctor, J.R.; Meyer, I.M. On the importance of cotranscriptional RNA structure formation. RNA 2013, 19, 1461–1473. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, Y.; Severinov, K.; Das, A.; Hanna, M.M. Role of Escherichia coli RNA polymerase alpha subunit in modulation of pausing, termination and anti-termination by the transcription elongation factor NusA. EMBO J. 1996, 15, 150–161. [Google Scholar] [CrossRef]
- Landick, R. RNA polymerase slides home: Pause and termination site recognition. Cell 1997, 88, 741–744. [Google Scholar] [CrossRef][Green Version]
- Mooney, R.A.; Artsimovitch, I.; Landick, R. Information processing by RNA polymerase: Recognition of regulatory signals during RNA chain elongation. J. Bacteriol. 1998, 180, 3265–3275. [Google Scholar] [CrossRef] [PubMed]
- Wickiser, J.K.; Winkler, W.C.; Breaker, R.R.; Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 2005, 18, 49–60. [Google Scholar] [CrossRef]
- Breaker, R.R. Riboswitches and Translational Control. Cold Spring Harbor Perspect. Biol. 2018, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lotz, T.S.; Suess, B. Small-Molecule-Binding Riboswitches. Microbiol. Spectr. 2018, 6, 26. [Google Scholar] [CrossRef]
- Waters, L.S.; Storz, G. Regulatory RNAs in Bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef]
- Serganov, A.; Nudler, E. A Decade of Riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef]
- Pavlova, N.; Kaloudas, D.; Penchovsky, R. Riboswitch distribution, structure, and function in bacteria. Gene 2019, 708, 38–48. [Google Scholar] [CrossRef]
- Gilbert, S.D.; Stoddard, C.D.; Wise, S.J.; Batey, R.T. Thermodynamic and Kinetic Characterization of Ligand Binding to the Purine Riboswitch Aptamer Domain. J. Mol. Biol. 2006, 359, 754–768. [Google Scholar] [CrossRef] [PubMed]
- Kortmann, J.; Narberhaus, F. Bacterial RNA thermometers: Molecular zippers and switches. Nat. Rev. Microbiol. 2012, 10, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Ragaz, C.; Kreuger, E.; Narberhaus, F. Temperature-controlled Structural Alterations of an RNA Thermometer. J. Biol. Chem. 2003, 278, 47915–47921. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Maris, C.; Allain, F.H.; Narberhaus, F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006, 25, 2487–2497. [Google Scholar] [CrossRef]
- Narberhaus, F.; Waldminghaus, T.; Chowdhury, S. RNA thermometers. FEMS Microbiol. Rev. 2006, 30, 3–16. [Google Scholar] [CrossRef]
- Soper, T.J.; Woodson, S.A. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 2008, 14, 1907–1917. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.W. Mechanisms of Bacterial Transcription Termination. J. Mol. Biol. 2019, 431, 4030–4039. [Google Scholar] [CrossRef]
- Ray-Soni, A.; Bellecourt, M.J.; Landick, R. Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annu. Rev. Biochem. 2016, 85, 319–347. [Google Scholar] [CrossRef]
- Henkin, T.M. Transcription termination control in bacteria. Curr. Opp. Microbiol. 2000, 2, 149–153. [Google Scholar] [CrossRef]
- Peters, J.M.; Vangeloff, A.D.; Landick, R. Bacterial Transcription Terminators: The RNA 3′-End Chronicles. J. Mol. Biol. 2011, 412, 793–813. [Google Scholar] [CrossRef]
- Ahmad, E.; Hegde, S.R.; Nagaraja, V. Revisiting intrinsic transcription termination in mycobacteria: U-tract downstream of secondary structure is dispensable for termination. Biochem. Biophys. Res. Com. 2020, 522, 226–232. [Google Scholar] [CrossRef]
- Mondal, S.; Yakhnin, A.V.; Sebastian, A.; Albert, I.; Babitzke, P. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat. Microbiol. 2015, 1, 15007. [Google Scholar] [CrossRef]
- Strauss, M.; Vitiello, C.; Schwieger, K.; Gottesman, M.; Rosch, P.; Knauer, S.H. Transcription is regulated by NusA:NusG interaction. Nucleic Acid Res. 2016, 44, 5971–5982. [Google Scholar] [CrossRef] [PubMed]
- Artsimovitch, I.; Landick, R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. USA 2000, 97, 7090–7095. [Google Scholar] [CrossRef] [PubMed]
- Mitra, P.; Ghosh, G.; Hafeezunnisa, M.; Sen, R. Rho Protein: Roles and Mechanisms, Ann. Rev. Microbiol. 2017, 71, 687–709. [Google Scholar] [CrossRef]
- Washburn, R.S.; Gottesman, M.E. Regulation of Transcription Elongation and Termination. Biomolecules 2015, 5, 1063–1078. [Google Scholar] [CrossRef]
- Kriner, M.A.; Sevostyanova, A.; Groisman, E.A. Learning from the Leaders: Gene Regulation by the Transcription Termination Factor Rho. Trends Biochem. Sci. 2016, 41, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Hollands, K.; Proshkin, S.; Sklyarova, S.; Epshtein, V.; Mironov, A.; Nudler, E.; Groisman, E.A. Riboswitch control of Rho-dependent transcription termination. Proc. Natl. Acad. Sci. USA 2012, 109, 5376–5381. [Google Scholar] [CrossRef]
- Cossart, P.; Mellin, J.R. Unexpected versatility in bacterial riboswitches. Trends Genet. 2015, 31, 3. [Google Scholar]
- Turnbough, C.L., Jr. Regulation of Bacterial Gene Expression by Transcription Attenuation. Microbiol. Mol. Biol. Rev. 2019, 83, e00019-19. [Google Scholar] [CrossRef]
- Zhang, J. Unboxing the T-box riboswitches—A glimpse into multivalent and multimodal RNA-RNA interactions. Wiley Interdiscip. Rev. RNA 2020, 11, e1600. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ferré-D’Amaré, A.R. Structure and mechanism of the T-box riboswitches. Wiley Interdiscip. Rev. RNA 2015, 6, 419–433. [Google Scholar] [CrossRef]
- Kreuzer, K.D.; Henkin, T.M. The T-Box Riboswitch: tRNA as an Effector to Modulate Gene Regulation. Microbiol. Spectr. 2018, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Samatova, E.; Daberger, J.; Liutkute, M.; Rodnina, M.V. Translational control by ribosome pausing in bacteria: How a non-uniform pace of translation affects protein production and folding. Front. Microbiol. 2021, 11, 619430. [Google Scholar] [CrossRef] [PubMed]
- De Smith, M.H.; van Duin, J. Translational Standby Sites: How Ribosomes May Deal with the Rapid Folding Kinetics of mRNA. JMB 2003, 4, 737–743. [Google Scholar] [CrossRef]
- Schlax, P.J.; Worhunsky, D.J. Translational repression mechanisms in prokaryotes. Mol. Microbiol. 2003, 48, 1157–1169. [Google Scholar] [CrossRef]
- Wieland, M.; Hartig, J.S. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007, 14, 757–763. [Google Scholar] [CrossRef]
- Geissmann, T.; Marzi, S.; Romby, P. The role of mRNA structure in translational control in bacteria. RNA Biol. 2009, 6, 153–160. [Google Scholar] [CrossRef]
- Loh, E.; Dussurget, O.; Gripenland, J.; Vaitkevicius, K.; Tiensuu, T.; Mandin, P.; Johansson, J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 2009, 139, 770–779. [Google Scholar] [CrossRef]
- Urban, J.H.; Vogel, J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008, 6, e64. [Google Scholar] [CrossRef]
- Collins, J.A.; Irnov, I.; Baker, S.; Winkler, W.C. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007, 21, 3356–3368. [Google Scholar] [CrossRef]
- Caron, M.P.; Bastet, L.; Lussier, A.; Simoneau-Roy, M.; Massé, E.; Lafontaine, D.A. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc. Natl. Acad. Sci. USA 2012, 109, E3444–E3453. [Google Scholar] [CrossRef]
- Shao, X.; Zhang, W.; Umar, M.I.; Wong, H.Y.; Seng, Z.; Xie, Y.; Deng, X. RNA G-Quadruplex structures mediate gene regulation in bacteria. MBio 2020, 11, e02926-19. [Google Scholar] [CrossRef] [PubMed]
- Heidrich, N.; Moll, I.; Brantl, S. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 2007, 35, 4331–4346. [Google Scholar] [CrossRef] [PubMed]
- Babitzke, P.; Romeo, T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opp. Microbiol. 2007, 10, 156–163. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chełkowska-Pauszek, A.; Kosiński, J.G.; Marciniak, K.; Wysocka, M.; Bąkowska-Żywicka, K.; Żywicki, M. The Role of RNA Secondary Structure in Regulation of Gene Expression in Bacteria. Int. J. Mol. Sci. 2021, 22, 7845. https://doi.org/10.3390/ijms22157845
Chełkowska-Pauszek A, Kosiński JG, Marciniak K, Wysocka M, Bąkowska-Żywicka K, Żywicki M. The Role of RNA Secondary Structure in Regulation of Gene Expression in Bacteria. International Journal of Molecular Sciences. 2021; 22(15):7845. https://doi.org/10.3390/ijms22157845
Chicago/Turabian StyleChełkowska-Pauszek, Agnieszka, Jan Grzegorz Kosiński, Klementyna Marciniak, Marta Wysocka, Kamilla Bąkowska-Żywicka, and Marek Żywicki. 2021. "The Role of RNA Secondary Structure in Regulation of Gene Expression in Bacteria" International Journal of Molecular Sciences 22, no. 15: 7845. https://doi.org/10.3390/ijms22157845
APA StyleChełkowska-Pauszek, A., Kosiński, J. G., Marciniak, K., Wysocka, M., Bąkowska-Żywicka, K., & Żywicki, M. (2021). The Role of RNA Secondary Structure in Regulation of Gene Expression in Bacteria. International Journal of Molecular Sciences, 22(15), 7845. https://doi.org/10.3390/ijms22157845





