Application of Modified mRNA in Somatic Reprogramming to Pluripotency and Directed Conversion of Cell Fate
Abstract
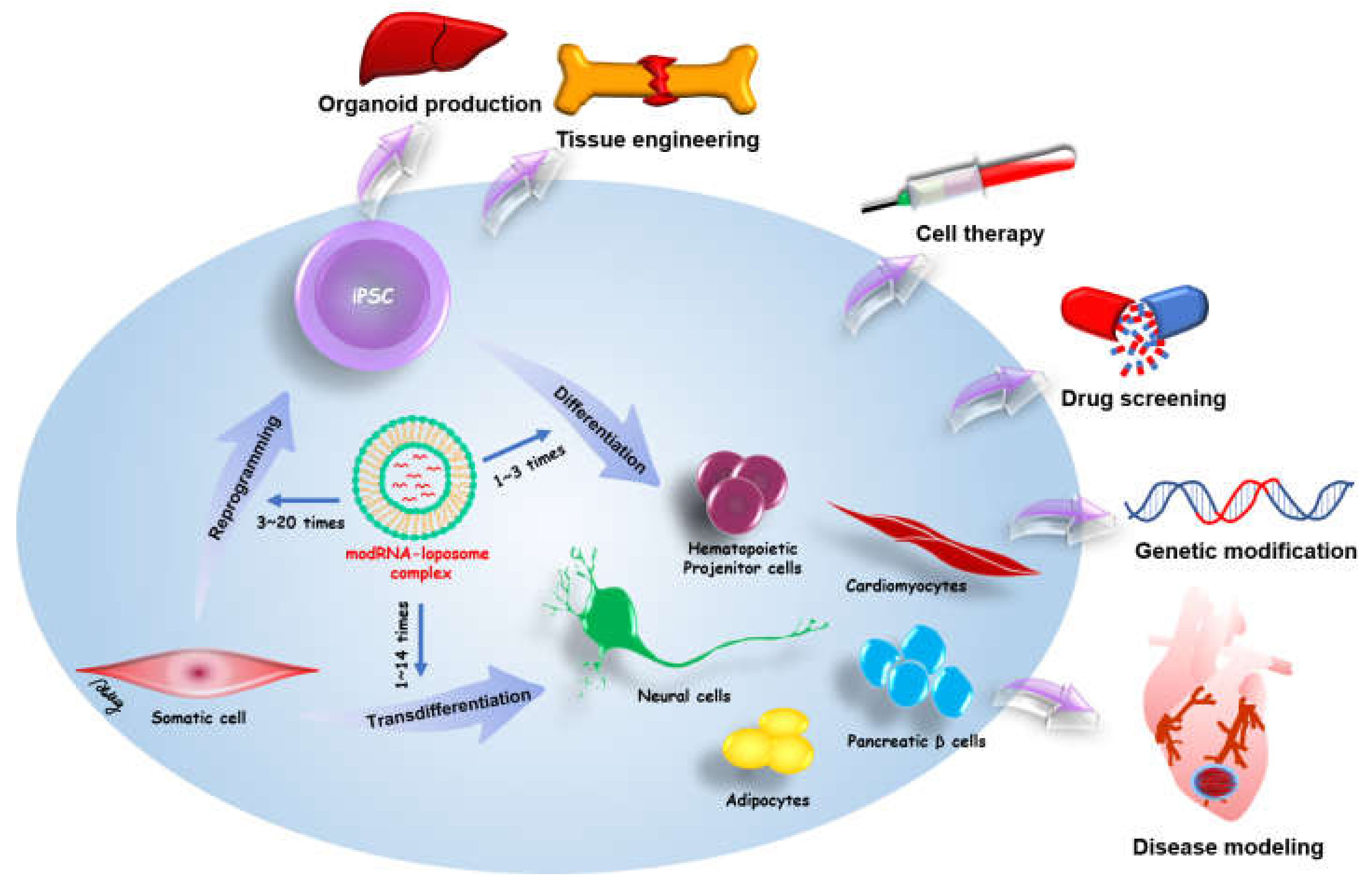
:1. Introduction
2. Messenger RNA
2.1. Natural mRNA
2.2. Modified mRNA
3. modRNA Delivery for Reprogramming
4. Modified mRNA-Based Reprogramming
5. modRNA Applications in Cell Differentiation
6. Safety
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amabile, G.; Meissner, A. Induced pluripotent stem cells: Current progress and potential for regenerative medicine. Trends Mol. Med. 2009, 15, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Narsinh, K.H.; Plews, J.; Wu, J.C. Comparison of human induced pluripotent and embryonic stem cells: Fraternal or identical twins? Mol. Ther. 2011, 19, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Y.L. Human induced pluripotent stem cell-derived exosomes as a new therapeutic strategy for various diseases. Int. J. Mol. Sci. 2021, 22, 1769. [Google Scholar] [CrossRef] [PubMed]
- Rowe, R.G.; Daley, G.Q. Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 2019, 20, 377–388. [Google Scholar] [CrossRef]
- Park, I.H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lerou, P.H.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 348–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ban, H.; Nishishita, N.; Fusaki, N.; Tabata, T.; Saeki, K.; Shikamura, M.; Takada, N.; Inoue, M.; Hasegawa, M.; Kawamata, S.; et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. USA 2011, 108, 14234–14239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, L.; Muench, M.O.; Fusaki, N.; Beyer, A.I.; Wang, J.; Qi, Z.; Yu, J.; Kan, Y.W. Blood cell-derived induced pluripotent stem cells free of reprogramming factors generated by Sendai viral vectors. Stem Cells Transl. Med. 2013, 2, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Y.L.; Loh, C.Y.Y. Episomal induced pluripotent stem cells: Functional and potential therapeutic applications. Cell Transplant. 2019, 28, 112S–131S. [Google Scholar] [CrossRef]
- Loh, C.Y.Y.; Wang, A.Y.L.; Kao, H.K.; Cardona, E.; Chuang, S.H.; Wei, F.C. Episomal induced pluripotent stem cells promote functional recovery of transected murine peripheral nerve. PLoS ONE 2016, 11, e0164696. [Google Scholar] [CrossRef] [Green Version]
- Cobb, M. Who discovered messenger RNA? Curr. Biol. 2015, 25, R526–R532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozak, M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol. Rev. 1983, 47, 1–45. [Google Scholar] [CrossRef]
- Lopez-Lastra, M.; Rivas, A.; Barria, M.I. Protein synthesis in eukaryotes: The growing biological relevance of cap-independent translation initiation. Biol. Res. 2005, 38, 121–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, S.E.; Hillner, P.E.; Vale, R.D.; Sachs, A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 1998, 2, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Wahle, E.; Keller, W. The biochemistry of polyadenylation. Trends Biochem. Sci. 1996, 21, 247–250. [Google Scholar] [CrossRef]
- Choi, Y.S.; Patena, W.; Leavitt, A.D.; McManus, M.T. Widespread RNA 3′-end oligouridylation in mammals. RNA 2012, 18, 394–401. [Google Scholar] [CrossRef] [Green Version]
- Korner, C.G.; Wahle, E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 1997, 272, 10448–10456. [Google Scholar] [CrossRef] [Green Version]
- Sachs, A.; Wahle, E. Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 1993, 268, 22955–22958. [Google Scholar] [CrossRef]
- Barrett, L.W.; Fletcher, S.; Wilton, S.D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol. Life Sci. 2012, 69, 3613–3634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, S.; Pal, J.K. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell 2009, 101, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Leppek, K.; Das, R.; Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, P.; Mebrahtu, A.; Barzadd, M.M.; Thalén, N.; Klanert, G.; Weinguny, M.; Sandegren, A.; Su, C.; Hatton, D.; Borth, N.; et al. Systematic use of synthetic 5′-UTR RNA structures to tune protein translation improves yield and quality of complex proteins in mammalian cell factories. Nucleic Acids Res. 2020, 48, e119. [Google Scholar] [CrossRef]
- Yergert, K.M.; Doll, C.A.; O’Rouke, R.; Hines, J.H.; Appel, B. Identification of 3′ UTR motifs required for mRNA localization to myelin sheaths in vivo. PLoS Biol. 2021, 19, e3001053. [Google Scholar] [CrossRef]
- Rasekhian, M.; Roohvand, F.; Habtemariam, S.; Marzbany, M.; Kazemimanesh, M. The role of 3′UTR of RNA viruses on mRNA stability and translation enhancement. Mini. Rev. Med. Chem. 2021. online ahead of print. [Google Scholar] [CrossRef]
- Shabalina, S.A.; Ogurtsov, A.Y.; Spiridonov, N.A. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 2006, 34, 2428–2437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz, L.; Burge, C.B. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003, 13, 2042–2051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasquinelli, A.E.; Dahlberg, J.E.; Lund, E. Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA 1995, 1, 957–967. [Google Scholar]
- Stepinski, J.; Waddell, C.; Stolarski, R.; Darzynkiewicz, E.; Rhoads, R.E. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA 2001, 7, 1486–1495. [Google Scholar] [PubMed]
- Mockey, M.; Gonçalves, C.; Dupuy, F.P.; Lemoine, F.M.; Pichon, C.; Midoux, P. mRNA transfection of dendritic cells: Synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem. Biophys. Res. Commun. 2006, 340, 1062–1068. [Google Scholar] [CrossRef]
- Kormann, M.S.D.; Hasenpusch, G.; Aneja, M.K.; Nica, G.; Flemmer, A.W.; Herber-Jonat, S.; Huppmann, M.; Mays, L.E.; Illenyi, M.; Schams, A.; et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011, 29, 154–157. [Google Scholar] [CrossRef]
- Sahin, U.; Kariko, K.; Tureci, O. mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Hadas, Y.; Katz, M.G.; Bridges, C.R.; Zangi, L. Modified mRNA as a therapeutic tool to induce cardiac regeneration in ischemic heart disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Quabius, E.S.; Krupp, G. Synthetic mRNAs for manipulating cellular phenotypes: An overview. New Biotechnol. 2015, 32, 229–235. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.; Athirasala, A.; Menezes, P.P.; Ashwanikumar, N.; Zou, T.; Sahay, G.; Bertassoni, L.E. Messenger RNA delivery for tissue engineering and regenerative medicine applications. Tissue Eng. Part A 2019, 25, 91–112. [Google Scholar] [CrossRef]
- Isaacs, A.; Cox, R.A.; Rotem, Z. Foreign nucleic acids as the stimulus to make interferon. Lancet 1963, 2, 113–116. [Google Scholar] [CrossRef]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Sousa, C.R. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar] [CrossRef] [Green Version]
- Machnicka, M.A.; Milanowska, K.; Oglou, O.O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef]
- Haque, A.K.M.A.; Dewerth, A.; Antony, J.S.; Riethmüller, J.; Schweizer, G.R.; Weinmann, P.; Latifi, N.; Yasar, H.; Pedemonte, N.; Sondo, E.; et al. Chemically modified hCFTR mRNAs recuperate lung function in a mouse model of cystic fibrosis. Sci. Rep. 2018, 8, 16776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Durbin, A.F.; Wang, C.; Marcotrigiano, J.; Gehrke, L. RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 2016, 7, e00833. [Google Scholar] [CrossRef] [Green Version]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [Green Version]
- Andries, O.; Cafferty, S.M.; Smedt, S.C.D.; Weiss, R.; Sanders, N.N.; Kitada, T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 2015, 217, 337–344. [Google Scholar] [CrossRef]
- Li, B.; Luo, X.; Dong, Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjug. Chem. 2016, 27, 849–853. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef] [Green Version]
- Kariko, K. In vitro-Transcribed mRNA Therapeutics: Out of the Shadows and Into the Spotlight. Mol. Ther. 2019, 27, 691–692. [Google Scholar] [CrossRef] [Green Version]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magadum, A.; Kaur, K.; Zangi, L. mRNA-based protein replacement therapy for the heart. Mol. Ther. 2019, 27, 785–793. [Google Scholar] [CrossRef] [Green Version]
- Yakubov, E.; Rechavi, G.; Rozenblatt, S.; Givol, D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem. Biophys. Res. Commun. 2010, 394, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Plews, J.R.; Li, J.L.; Jones, M.; Moore, H.D.; Mason, C.; Andrews, P.W.; Na, J. Activation of pluripotency genes in human fibroblast cells by a novel mRNA based approach. PLoS ONE 2010, 5, e14397. [Google Scholar] [CrossRef]
- Antje Arnold, Y.M.N.; Fabian, C.; Wirth, H.; Hans Binder, G.N.; Armstrong, L.; Stolzing, A. Reprogramming of human Huntington fibroblasts using mRNA. Int. Sch. Res. Netw. 2011, 2012, 124878. [Google Scholar] [CrossRef] [Green Version]
- Tavernier, G.; Wolfrum, K.; Demeester, J.; Smedt, S.C.D.; Adjaye, J.; Rejman, J. Activation of pluripotency-associated genes in mouse embryonic fibroblasts by non-viral transfection with in vitro-derived mRNAs encoding Oct4, Sox2, Klf4 and cMyc. Biomaterials 2012, 33, 412–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, L.; Ni, Y.; Wang, J.; Guo, X. Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Sci. Rep. 2012, 2, 657. [Google Scholar] [CrossRef]
- Heng, B.C.; Heinimann, K.; Miny, P.; Iezzi, G.; Glatz, K.; Scherberich, A.; Zulewski, H.; Fussenegger, M. mRNA transfection-based, feeder-free, induced pluripotent stem cells derived from adipose tissue of a 50-year-old patient. Metab. Eng. 2013, 18, 9–24. [Google Scholar] [CrossRef]
- Mandal, P.K.; Rossi, D.J. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat. Protoc. 2013, 8, 568–582. [Google Scholar] [CrossRef]
- Sjogren, A.K.M.; Liljevald, M.; Glinghammar, B.; Sagemark, J.; Li, X.Q.; Jonebring, A.; Cotgreave, I.; Brolén, G.; Andersson, T.B. Critical differences in toxicity mechanisms in induced pluripotent stem cell-derived hepatocytes, hepatic cell lines and primary hepatocytes. Arch. Toxicol. 2014, 88, 1427–1437. [Google Scholar] [CrossRef]
- Varela, I.; Karagiannidou, A.; Oikonomakis, V.; Tzetis, M.; Tzanoudaki, M.; Siapati, E.K.; Vassilopoulos, G.; Graphakos, S.; Kanavakis, E.; Goussetis, E. Generation of human beta-thalassemia induced pluripotent cell lines by reprogramming of bone marrow-derived mesenchymal stromal cells using modified mRNA. Cell. Reprogram. 2014, 16, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Durruthy-Durruthy, J.; Briggs, S.F.; Awe, J.; Ramathal, C.Y.; Karumbayaram, S.; Lee, P.C.; Heidmann, J.D.; Clark, A.; Karakikes, I.; Loh, K.M.; et al. Rapid and efficient conversion of integration-free human induced pluripotent stem cells to GMP-grade culture conditions. PLoS ONE 2014, 9, e94231. [Google Scholar] [CrossRef]
- Ramakrishnan, V.M.; Yang, J.Y.; Tien, K.T.; McKinley, T.R.; Bocard, B.R.; Maijub, J.G.; Burchell, P.O.; Williams, S.K.; Morris, M.E.; Hoying, J.B.; et al. Restoration of physiologically responsive low-density lipoprotein receptor-mediated endocytosis in genetically deficient induced pluripotent stem cells. Sci. Rep. 2015, 5, 13231. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.I.; Lee, S.Y.; Hwang, S.Y. Extracellular matrix-dependent generation of integration- and xeno-free iPS cells using a modified mRNA transfection method. Stem Cells Int. 2016, 2016, 6853081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preskey, D.; Allison, T.F.; Jones, M.; Mamchaoui, K.; Unger, C. Synthetically modified mRNA for efficient and fast human iPS cell generation and direct transdifferentiation to myoblasts. Biochem. Biophys. Res. Commun. 2016, 473, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.Y.; Lee, T.J.; Yang, G.M.; Oh, J.; Won, J.; Han, J.; Jeong, G.J.; Kim, J.; Kim, J.H.; Kim, B.S.; et al. Efficient mRNA delivery with graphene oxide-polyethylenimine for generation of footprint-free human induced pluripotent stem cells. J. Control. Release 2016, 235, 222–235. [Google Scholar] [CrossRef]
- Velasquez-Mao, A.J.; Tsao, C.J.M.; Monroe, M.N.; Legras, X.; Bissig-Choisat, B.; Bissig, K.D.; Ruano, R.; Jacot, J.G. Differentiation of spontaneously contracting cardiomyocytes from non-virally reprogrammed human amniotic fluid stem cells. PLoS ONE 2017, 12, e0177824. [Google Scholar]
- Chen, H.; Zuo, Q.; Wang, Y.; Song, J.; Yang, H.; Zhang, Y.; Li, B. Inducing goat pluripotent stem cells with four transcription factor mRNAs that activate endogenous promoters. BMC Biotechnol. 2017, 17, 11. [Google Scholar] [CrossRef] [Green Version]
- Kogut, I.; McCarthy, S.M.; Pavlova, M.; Astling, D.P.; Chen, X.; Jakimenko, A.; Jones, K.L.; Getahun, A.; Cambier, J.C.C.; Pasmooij, A.M.G.; et al. High-efficiency RNA-based reprogramming of human primary fibroblasts. Nat. Commun. 2018, 9, 745. [Google Scholar] [CrossRef] [Green Version]
- McGrath, P.S.; McGarvey, S.S.; Kogut, I.; Bilousova, G. Efficient RNA-based reprogramming of disease-associated primary human fibroblasts into induced pluripotent stem cells. Methods Mol. Biol. 2020, 2117, 271–284. [Google Scholar]
- Kaczmarek, J.C.; Kowalski, P.S.; Anderson, D.G. Advances in the delivery of RNA therapeutics: From concept to clinical reality. Genome Med. 2017, 9, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef] [Green Version]
- Guevara, M.L.; Persano, F.; Persano, S. Advances in Lipid Nanoparticles for mRNA-based cancer immunotherapy. Front. Chem. 2020, 8, 589959. [Google Scholar] [CrossRef]
- Trepotec, Z.; Lichtenegger, E.; Plank, C.; Aneja, M.K.; Rudolph, C. Delivery of mRNA therapeutics for the treatment of hepatic diseases. Mol. Ther. 2019, 27, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef]
- Balazs, D.A.; Godbey, W. Liposomes for use in gene delivery. J. Drug Deliv. 2011, 2011, 326497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, H.Y.; Guo, P.; Wen, W.C.; Wong, H.L. Lipid-based nanocarriers for RNA delivery. Curr. Pharm. Des. 2015, 21, 3140–3147. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.H.; Yu, B.; Wang, X.; Lu, Y.; Schmidt, C.R.; Lee, R.J.; Lee, L.J.; Jacob, S.T.; Ghoshal, K. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomedicine 2013, 9, 1169–1180. [Google Scholar] [CrossRef] [Green Version]
- Simberg, D.; Weisman, S.; Talmon, Y.; Barenholz, Y. DOTAP (and other cationic lipids): Chemistry, biophysics, and transfection. Crit. Rev. Ther. Drug. Carrier. Syst. 2004, 21, 257–317. [Google Scholar] [CrossRef]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Heyes, J.; Palmer, L.; Bremner, K.; MacLachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release 2005, 107, 276–287. [Google Scholar] [CrossRef]
- Kauffman, K.J.; Webber, M.J.; Anderson, D.G. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Release 2016, 240, 227–234. [Google Scholar] [CrossRef]
- Shigekawa, K.; Dower, W.J. Electroporation of eukaryotes and prokaryotes: A general approach to the introduction of macromolecules into cells. Biotechniques 1988, 6, 742–751. [Google Scholar]
- Zhao, Y.; Zheng, Z.; Cohen, C.J.; Gattinoni, L.; Palmer, D.C.; Restifo, N.P.; Rosenberg, S.A.; Morgan, R.A. High-efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol. Ther. 2006, 13, 151–159. [Google Scholar] [CrossRef]
- Buganim, Y.; Faddah, D.A.; Jaenisch, R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013, 14, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Anokye-Danso, F.; Trivedi, C.M.; Juhr, D.; Gupta, M.; Cui, Z.; Tian, Y.; Zhang, Y.; Yang, W.; Gruber, P.J.; Epstein, J.A.; et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011, 8, 376–388. [Google Scholar] [CrossRef] [Green Version]
- Papapetrou, E.P.; Lee, G.; Malani, N.; Setty, M.; Riviere, I.; Tirunagari, L.M.S.; Kadota, K.; Roth, S.L.; Giardina, P.; Viale, A.; et al. Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zheng, C.G.; Jiang, Y.; Zhang, J.; Chen, J.; Yao, C.; Zhao, Q.; Liu, S.; Chen, K.; Du, J.; et al. Genetic correction of beta-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice. Cell Res. 2012, 22, 637–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lui, K.O.; Zangi, L.; Silva, E.A.; Bu, L.; Sahara, M.; Li, R.A.; Mooney, D.J.; Chien, K.R. Driving vascular endothelial cell fate of human multipotent Isl1+ heart progenitors with VEGF modified mRNA. Cell Res. 2013, 23, 1172–1186. [Google Scholar] [CrossRef] [Green Version]
- Elcheva, I.; Brok-Volchanskaya, V.; Kumar, A.; Liu, P.; Lee, J.H.; Tong, L.; Vodyanik, M.; Swanson, S.; Stewart, R.; Kyba, M.; et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat. Commun. 2014, 5, 4372. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Lin, R.Z.; Hong, X.; Ng, A.H.; Lee, C.N.; Neumeyer, J.; Wang, G.; Wang, X.; Ma, M.; Pu, W.T.; et al. Robust differentiation of human pluripotent stem cells into endothelial cells via temporal modulation of ETV2 with modified mRNA. Sci. Adv. 2020, 6, eaba7606. [Google Scholar] [CrossRef]
- Brok-Volchanskaya, V.S.; Bennin, D.A.; Suknuntha, K.; Klemm, L.C.; Huttenlocher, A.; Slukvin, I. Effective and rapid generation of functional neutrophils from induced pluripotent stem cells using ETV2-modified mRNA. Stem Cell Rep. 2019, 13, 1099–1110. [Google Scholar] [CrossRef] [Green Version]
- Majumder, A.; Suknuntha, K.; Bennin, D.; Klemm, L.; Brok-Volchanskaya, V.S.; Huttenlocher, A.; Slukvin, I. Generation of human neutrophils from induced pluripotent stem cells in chemically defined conditions using ETV2 modified mRNA. STAR Protoc. 2020, 1, 100075. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Yu, L.; Ding, Y.; Guo, X.R.; Yuan, Y.H.; Li, D.S. Gene manipulation of human embryonic stem cells by in vitro-synthesized mRNA for gene therapy. Curr. Gene Ther. 2015, 15, 428–435. [Google Scholar] [CrossRef]
- Guo, X.R.; Wang, X.L.; Li, M.C.; Yuan, Y.H.; Chen, Y.; Zou, D.D.; Bian, L.J.; Li, D.S. PDX-1 mRNA-induced reprogramming of mouse pancreas-derived mesenchymal stem cells into insulin-producing cells in vitro. Clin. Exp. Med. 2015, 15, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Goparaju, S.K.; Kohda, K.; Ibata, K.; Soma, A.; Nakatake, Y.; Akiyama, T.; Wakabayashi, S.; Matsushita, M.; Sakota, M.; Kimura, H.; et al. Rapid differentiation of human pluripotent stem cells into functional neurons by mRNAs encoding transcription factors. Sci. Rep. 2017, 7, 42367. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Zhao, M.; Mundy, G.R. Bone morphogenetic proteins. Growth Factors 2004, 22, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Marie, P.J.; Debiais, F.; Hay, E. Regulation of human cranial osteoblast phenotype by FGF-2, FGFR-2 and BMP-2 signaling. Histol. Histopathol. 2002, 17, 877–885. [Google Scholar] [PubMed]
- Balmayor, E.R.; Geiger, J.P.; Koch, C.; Aneja, M.K.; Griensven, M.V.; Rudolph, C.; Plank, C. Modified mRNA for BMP-2 in combination with biomaterials serves as a transcript-activated matrix for effectively inducing osteogenic pathways in stem cells. Stem Cells Dev. 2017, 26, 25–34. [Google Scholar] [CrossRef]
- White, H.D.; Chew, D.P. Acute myocardial infarction. Lancet 2008, 372, 570–584. [Google Scholar] [CrossRef] [Green Version]
- Zangi, L.; Lui, K.O.; Gise, A.V.; Ma, Q.; Ebina, W.; Ptaszek, L.M.; Später, D.; Xu, H.; Tabebordbar, M.; Gorbatov, R.; et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013, 31, 898–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zangi, L.; Oliveira, M.S.; Ye, L.Y.; Ma, Q.; Sultana, N.; Hadas, Y.; Chepurko, E.; Später, D.; Zhou, B.; Chew, W.L.; et al. Insulin-like growth factor 1 receptor-dependent pathway drives epicardial adipose tissue formation after myocardial injury. Circulation 2017, 135, 59–72. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Yamada, S.; Shi, A.; Singh, R.D.; Rolland, T.J.; Jeon, R.; Lopez, N.; Shelerud, L.; Terzic, A.; Behfar, A. Brachyury engineers cardiac repair competent stem cells. Stem Cells Transl. Med. 2021, 10, 385–397. [Google Scholar] [CrossRef]
- Singh, R.D.; Hillestad, M.L.; Livia, C.; Li, M.; Alekseev, A.E.; Witt, T.A.; Stalboerger, P.G.; Yamada, S.; Terzic, A.; Behfar, A. M(3)RNA drives targeted gene delivery in acute myocardial infarction. Tissue Eng. Part A 2019, 25, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Elangovan, S.; Khorsand, B.; Do, A.V.; Hong, L.; Dewerth, A.; Kormann, M.; Ross, R.D.; Sumner, D.R.; Allamargot, C.; Salem, A.K. Chemically modified RNA activated matrices enhance bone regeneration. J. Control. Release 2015, 218, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Balmayor, E.R.; Geiger, J.P.; Aneja, M.K.; Berezhanskyy, T.; Utzinger, M.; Mykhaylyk, O.; Rudolph, C.; Plank, C. Chemically modified RNA induces osteogenesis of stem cells and human tissue explants as well as accelerates bone healing in rats. Biomaterials 2016, 87, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Badieyan, Z.S.; Berezhanskyy, T.; Utzinger, M.; Aneja, M.K.; Emrich, D.; Erben, R.; Schüler, C.; Altpeter, P.; Ferizi, M.; Hasenpusch, G.; et al. Transcript-activated collagen matrix as sustained mRNA delivery system for bone regeneration. J. Control. Release 2016, 239, 137–148. [Google Scholar] [CrossRef]
- Khorsand, B.; Elangovan, S.; Hong, L.; Dewerth, A.; Kormann, M.S.D.; Salem, A.K. A comparative study of the bone regenerative effect of chemically modified RNA encoding BMP-2 or BMP-9. AAPS J. 2017, 19, 438–446. [Google Scholar] [CrossRef]
- Hausburg, F.; Na, S.; Voronina, N.; Skorska, A.; Müller, P.; Steinhoff, G.; David, R. Defining optimized properties of modified mRNA to enhance virus- and DNA- independent protein expression in adult stem cells and fibroblasts. Cell Physiol. Biochem. 2015, 35, 1360–1371. [Google Scholar] [CrossRef]
- Lee, K.; Yu, P.; Lingampalli, N.; Kim, H.J.; Tang, R.; Murthy, N. Peptide-enhanced mRNA transfection in cultured mouse cardiac fibroblasts and direct reprogramming towards cardiomyocyte-like cells. Int. J. Nanomed. 2015, 10, 1841–1854. [Google Scholar]
- Koblas, T.; Leontovyc, I.; Loukotova, S.; Kosinova, L.; Saudek, F. Reprogramming of pancreatic exocrine cells AR42J into insulin-producing cells using mRNAs for Pdx1, Ngn3, and MafA transcription factors. Mol. Ther. Nucleic Acids 2016, 5, e320. [Google Scholar] [CrossRef] [Green Version]
- Connor, B.; Firmin, E.; McCaughey-Chapman, A.; Monk, R.; Lee, K.; Liot, S.; Geiger, J.; Rudolph, C.; Jones, K. Conversion of adult human fibroblasts into neural precursor cells using chemically modified mRNA. Heliyon 2018, 4, e00918. [Google Scholar] [CrossRef] [Green Version]
- Pham, P.V.; Vu, N.B.; Dao, T.T.T.; Le, H.T.N.; Phi, L.T.; Phan, N.K. Production of endothelial progenitor cells from skin fibroblasts by direct reprogramming for clinical usages. In Vitro Cell. Dev. Biol. Anim. 2017, 53, 207–216. [Google Scholar] [CrossRef]
- Corritore, E.; Lee, Y.S.; Pasquale, V.; Liberati, D.; Hsu, M.J.; Lombard, C.A.; Smissen, P.V.D.; Vetere, A.; Bonner-Weir, S.; Piemonti, L.; et al. V-Maf musculoaponeurotic fibrosarcoma oncogene homolog A synthetic modified mRNA drives reprogramming of human pancreatic duct-derived cells into insulin-secreting cells. Stem Cells Transl. Med. 2016, 5, 1525–1537. [Google Scholar] [CrossRef] [Green Version]
- Angel, M.; Yanik, M.F. Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS ONE 2010, 5, e11756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conry, R.M.; LoBuglio, A.F.; Wright, M.; Sumerel, L.; Pike, M.J.; Johanning, F.; Benjamin, R.; Lu, D.; Curiel, D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995, 55, 1397–1400. [Google Scholar] [PubMed]
- Crozat, K.; Beutler, B. TLR7: A new sensor of viral infection. Proc. Natl. Acad. Sci. USA 2004, 101, 6835–6836. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Karikó, K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.R.; Muramatsu, H.; Jha, B.K.; Silverman, R.H.; Weissman, D.; Karikó, K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011, 39, 9329–9338. [Google Scholar] [CrossRef] [Green Version]
- Warren, L.; Lin, C. mRNA-Based Genetic Reprogramming. Mol. Ther. 2019, 27, 729–734. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Sayed, N.; Hunter, A.; Au, K.F.; Wong, W.H.; Mocarski, E.S.; Pera, R.R.; Yakubov, E.; Cooke, J.P. Activation of innate immunity is required for efficient nuclear reprogramming. Cell 2012, 151, 547–558. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Xu, L.; Gibson, T.M.; Gersbach, C.A.; Sullenger, B.A. Differential effects of toll-like receptor stimulation on mRNA-driven myogenic conversion of human and mouse fibroblasts. Biochem. Biophys. Res. Commun. 2016, 478, 1484–1490. [Google Scholar] [CrossRef]
- Zhou, G.; Meng, S.; Li, Y.; Ghebre, Y.T.; Cooke, J.P. Optimal ROS signaling is critical for nuclear reprogramming. Cell Rep. 2016, 15, 919–925. [Google Scholar] [CrossRef] [Green Version]
- Brady, J.J.; Li, M.; Suthram, S.; Jiang, H.; Wong, W.H.; Blau, H.M. Early role for IL-6 signalling during generation of induced pluripotent stem cells revealed by heterokaryon RNA-Seq. Nat. Cell Biol. 2013, 15, 1244–1252. [Google Scholar] [CrossRef] [Green Version]
- Xue, H.Y.; Liu, S.; Wong, H.L. Nanotoxicity: A key obstacle to clinical translation of siRNA-based nanomedicine. Nanomedicine 2014, 9, 295–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Lizarzaburu, M.E.; Kurth, M.J.; Liu, L.; Wege, H.; Zern, M.A.; Nantz, M.H. Cationic lipid polymerization as a novel approach for constructing new DNA delivery agents. Bioconjug. Chem. 2001, 12, 251–257. [Google Scholar] [CrossRef]
- Aberle, A.M.; Tablin, F.; Zhu, J.; Walker, N.J.; Gruenert, D.C.; Nantz, M.H. A novel tetraester construct that reduces cationic lipid-associated cytotoxicity. Implications for the onset of cytotoxicity. Biochemistry 1998, 37, 6533–6540. [Google Scholar] [CrossRef]
- Rudin, C.M.; Marshall, J.L.; Huang, C.H.; Kindler, H.L.; Zhang, C.; Kumar, D.; Gokhale, P.C.; Steinberg, J.; Wanaski, S.; Kasid, U.N.; et al. Delivery of a liposomal c-raf-1 antisense oligonucleotide by weekly bolus dosing in patients with advanced solid tumors: A phase I study. Clin. Cancer Res. 2004, 10, 7244–7251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coelho, T.; Adams, D.; Silva, A.; Lozeron, P.; Hawkins, P.N.; Mant, T.; Perez, J.; Chiesa, J.; Warrington, S.; Tranter, E.; et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013, 369, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Pollard, C.; Rejman, J.; Haes, W.D.; Verrier, B.; Gulck, E.V.; Naessens, T.; Smedt, S.D.; Bogaert, P.; Grooten, J.; Vanham, G.; et al. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 2013, 21, 251–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, N.; Magadum, A.; Hadas, Y.; Kondrat, J.; Singh, N.; Youssef, E.; Calderon, D.; Chepurko, E.; Dubois, N.; Hajjar, R.J.; et al. Optimizing cardiac delivery of modified mRNA. Mol. Ther. 2017, 25, 1306–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlsson, L.; Clarke, J.C.; Yen, C.; Gregoire, F.; Albery, T.; Billger, M.; Egnell, A.C.; Gan, L.M.; Jennbacken, K.; Johansson, E.; et al. Biocompatible, purified VEGF-A mRNA improves cardiac function after intracardiac injection 1 week post-myocardial infarction in swine. Mol. Ther. Methods Clin. Dev. 2018, 9, 330–346. [Google Scholar] [CrossRef] [Green Version]

| Cell Sources | modRNA | Transfection Methods | Transfection Numbers | Total modRNA | Modifications | Differentiation to Three Germ Layers | Further Differentiation | References |
|---|---|---|---|---|---|---|---|---|
| BJ human neonatal foreskin fibroblasts, MRC-5 human fetal lung fibroblasts, Detroit 551 human fetal skin fibroblasts, dH1f fibroblasts, and skin cells of a cystic fibrosis patient | KLF4, c-MYC, OCT4, SOX2, LIN28 | Cationic lipid | 17 | 20 μg in 6-well plate; 136 μg in 10 cm dish | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | Yes | Myogenic cells | [47] |
| Human foreskin fibroblasts | OCT4, LIN28, SOX2, NANOG | Cationic lipid | 5 | 20 μg | Poly-A tail, ARCA, IRES sequence | No | N/A | [54] |
| Human fetal skin fibroblasts (HuF1), human embryonic lung fibroblasts (MRC5), human foreskin fibroblasts (HFF) | OCT4, SOX2, c-MYC, KLF4, SV40 large T (LT) | Electroporation | 1 | 43 μg | Poly-A tail, ARCA, 5′ and 3′ UTRs of Xenopus b-globin | No | N/A | [55] |
| Human foreskin, adult Huntington fibroblasts, and adult skin fibroblasts of healthy donors | OCT4, NANOG, KLF4, c-MYC, SOX2, hTERT | Electroporation and Lipofectamine 2000 | 4 | 12 μg | Poly-A tail, Cap | Yes | N/A | [56] |
| Mouse embryonic fibroblasts (MEF) | OCT4, SOX2, KLF4, c-MYC | Cationic lipid | 3 | 12 μg | Poly-A tail, ARCA | No | N/A | [57] |
| BJ neonatal fibroblasts, HDF-f fetal fibroblasts, HDF-n neonatal fibroblasts, HDF-a adult fibroblasts, and XFF xeno-free neonatal fibroblasts | OCT4, SOX2, KLF4, c-MYC-T58A, LIN28, NANOG | RNAiMAX | 9 | 9 μg | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | Yes | Cardiomyocytes | [58] |
| Adipose-derived mesenchymal stem cells of a 50-year-old patient | OCT4, KLF4, SOX2, LIN28, c-MYC | RNAiMAX | 18 | 9.6 μg | 5mC, ψU, Cap | Yes | N/A | [59] |
| Primary human fibroblasts | KLF4, c-MYC, OCT4, SOX2, LIN28, NDG | Cationic lipid | 14 | 14 μg | 5mC, ψU, 5′ UTR, 3′ UTR, Poly-A tail, ARCA | No | N/A | [60] |
| Human newborn foreskin fibroblasts | OCT4, KLF4, SOX2, LIN28, c-MYC | RNAiMAX | 17 | 74.8 μg (Stemgent) | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | Yes | Hepatocytes | [61] |
| Human bone marrow-derived mesenchymal stromal cells from a patient with β-thalassemia | OCT4, KLF4, SOX2, c-MYC, LIN28 | RNAiMAX | 18 | 21.6 μg | 5mC, ψU, Poly-A tail, ARCA | Yes | Hematopoietic progenitors | [62] |
| Human adult dermal fibroblasts (HUF1 and HUF58), GM13325 fibroblasts from a 9-day-old patient with DiGeorge Syndrome, BJ human fibroblasts | OCT4, KLF4, SOX2, c-MYC, LIN28 | RNAiMAX | 12 | 14.4 μg | 5mC, ψU, ARCA | Yes | Cardiomyocytes | [63] |
| Skin fibroblasts from a patient with low-density lipoprotein receptor (LDLR) deficiency, familial hypercholesterolemia (FH) | OCT4, SOX2, KLF4, c-MYC, LIN28 | RNAiMAX | 20 | 23.5 μg | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | Yes | Hepatocytes, mesenchymal cells | [64] |
| Human adult dermal fibroblasts | OCT4, SOX2, KLF4, c-MYC, LIN28, miR302a-d, miR367 | Stemfect RNA Transfection reagent | 11 | 11 μg | 5mC, ψU, Cap | Yes | N/A | [65] |
| Human BJ fibroblasts | OCT4, SOX2, KLF4, c-MYC, NANOG, LIN28, | Stemfect RNA Transfection reagent | 9 | 2.2 μg | 5mC, ψU, 5′ UTR containing Kozak sequence, 3′ UTR, Poly-A tail, ARCA | Yes | N/A | [66] |
| Human adipose-derived fibroblasts (ADFs), rat ADFs, mouse embryonic fibroblasts (MEF) | OCT4, SOX2, KLF4, c-MYC, or mRNA extracted from cells overexpressing OSKM | Graphene oxide-polyethylenimine (GO-PEI) | 3 | 6 μg | 5′ UTR, 3′ UTR, Poly-A tail, Cap | Yes | N/A | [67] |
| Human amniotic fluid stem Cells (AFSC) | OCT4, SOX2, KLF4, c-MYC, LIN28 | RNAiMAX | 18 | 79.2 μg | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | Yes | Cardiomyocytes | [68] |
| Goat embryonic fibroblasts (GEF) | OCT4, SOX2, KLF4, c-MYC | Lipofectamine 2000 | 5 | 5 μg | Poly-A tail, ARCA | Yes | N/A | [69] |
| Human primary fibroblasts from two healthy donors and a patient with Down syndrome | OCT4, SOX2, KLF4, c-MYC, LIN28A, NANOG, mWasabi miR367/302s | RNAiMAX | 7 | 4.4 μg | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | Yes | N/A | [70] |
| Human primary fibroblasts | OCT4, SOX2, KLF4, c-MYC, LIN28A, NANOG, mWasabi miR367/302s | RNAiMAX | 7 | 7 μg | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | No | N/A | [71] |
| Cell Sources | modRNA | Modifications | Transfection Methods | Transfection Numbers | Total modRNA | Differentiated Cell Types | Animal Models | Therapeutic Effects | References |
|---|---|---|---|---|---|---|---|---|---|
| modRNA-induced hiPSCs | MYOD | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | RNAiMAX | 3 | 3.6 μg | Myogenic cells | N/A | N/A | [47] |
| Human foreskin fibroblasts | MYOD | 5mC, ψU, 5′ UTR containing Kozak sequence, 3′ UTR, Poly-A tail, ARCA | Stemfect RNA transfection reagent | 4 | 1.2 μg | Myoblasts | N/A | MYOD1 modRNA can directly transdifferentiate human fibroblasts into myoblasts without a transgene footprint | [66] |
| Mouse fibroblasts and hMSCs | MYOD | 5mC, ψU, ARCA | Lipofectamine 2000 | 3 | 0.75 μg | Skeletal myoblasts | N/A | Defining optimized properties of modRNA-based protein expression in adult stem cells and fibroblasts | [110] |
| hESC-derived ISL1+ heart progenitors | VEGF-A | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | RNAiMAX | In vitro-2 In vivo-1 | In vitro-2 μg In vivo-5 μg | Human Isl1+ vascular endothelial cells | N/A | VEGF-A modRNA promotes not only the endothelial specification but also engraftment, proliferation, and survival (reduced apoptosis) of the human Isl1+ progenitors in vivo | [90] |
| Heart WT1+ epicardial progenitors | VEGF-A | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | RNAiMAX | In vitro-1 In vivo-1 | In vitro-3 μg In vivo-100 μg/heart | Endothelial cells and cardiovascular cells | Mouse myocardial infarction model | Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction | [102] |
| Endogenous heart epicardial progenitors | IGF1 | 5mC, ψU, ARCA | N/A | 1 | 100 μg/heart | Epicardial adipose tissues | Mouse myocardial injury | An IGF1R modRNA-induced pathway drives epicardial adipose tissue formation after myocardial injury | [103] |
| Human ADSCs | Brachyury | 5mC, Poly-A tail, ARCA | Microencapsulated-modified-mRNA (M3RNA) technique | 1 | 1.75 μg | Cardiopoietic stem cells | Mouse myocardial infarction | Intramyocardial delivery of Brachyury modRNA-induced cardiopoietic stem cells can improve cardiac performance and protect against decompensated heart failure | [104] |
| Cardiac fibroblasts | Gata4, Mef2c, Tbx5 | 5mC, ψU, Poly-A tail, ARCA | C-Lipo (polyarginine-fused heart-targeting peptide and lipofectamine complex) | 14 | 16.8 μg | Cardiomyocytes | N/A | C-Lipo can enhance modRNA transfection and results in the direct reprogramming of fibroblasts into cardiomyocytes | [111] |
| hESCs | ETV2, GATA2 | 5mC, ψU, Poly-A tail, ARCA | Electroporation | 2 | 7 μg | CD43+ hematopoietic cells | N/A | Transient expression of ETV2 and GATA2 is indeed sufficient to commit the hPSCs to blood fate | [91] |
| Human skin fibroblasts | ETV2 | Poly-A tail, Cap | Electroporation | 1 | 3 μg | Endothelial progenitor cells | Hindlimb ischemia model | ETV2 modRNA combined with hypoxia can produce functional EPCs from fibroblasts and improve mouse ischemia | [114] |
| Human iPSCs | ETV2 | ψU, Poly-A tail, ARCA | TransIT-mRNA | 1 | 0.2 μg | Hemogenic endothelium | N/A | ETV2 modRNA-induced hematoendothelial progenitors can differentiate into functional neutrophils in the presence of G-CSF and Am580 | [93,94] |
| Human iPSCs | ETV2 | 5′ UTR, 3′ UTR, Poly-A tail, Cap | Electroporation or RNAiMax | 1 | 0.6 μg | Endothelial cells | N/A | Direct differentiation of human iPSCs into endothelial cells via transient modulation of ETV2 modRNA | [92] |
| hESCs | PDX1 | 5mC, ψU, Poly-A tail, ARCA | Electroporation | 1 | N/A | Insulin-producing cells | N/A | PDX1 modRNA can directly induce the transdifferentiation of insulin-producing cells | [95] |
| Mouse pancreas-derived MSCs | PDX1 | 5mC, ψU, Poly-A tail, ARCA | TransIT-mRNA | 1 | N/A | Insulin-producing cells | N/A | Mouse pMSCs can be transdifferentiated into functional glucose-responsive insulin-producing cells through transfecting PDX-1 modRNA | [96] |
| Pancreatic exocrine cells AR42J | PDX1, Ngn3, MafA | 5mC, ψU, Poly-A tail, ARCA | Lipofectamine MessengerMAX | 10 | 15 μg | Insulin-producing cells | N/A | Reprogramming of pancreatic exocrine cells into insulin-producing cells through modRNAs, represents a promising approach for cell-based diabetes therapy | [112] |
| Human pancreatic duct-derived cells | MafA | 5mC, ψU, 5′ UTR containing Kozak sequence, α-Globin 3′ UTR, Poly-A tail, ARCA | jetPEI | 7 | 8.4 μg | Insulin-producing cells | Diabetic SCID-beige mice | MafA modRNA can drive the reprogramming of human pancreatic duct-derived cells into functional insulin-secreting cells, and reverse diabetes | [115] |
| Human pluripotent stem cells | NEUROG1, NEUROG2, NEUROG3, NEUROD1, and NEUROD2 | 5mC, ψU, 5′ UTR, 3′ UTR, Poly-A tail, ARCA | Lipofectamine MessengerMAX | 2 | 2 μg | Neurons | N/A | The modRNA cocktail can differentiate hPSCs into motor neurons | [97] |
| Human adult fibroblasts | SOX2, PAX6 | 5′ UTR, 3′ UTR, Poly-A tail, Cap | Lipofectamine RNAiMAX | 4 | 8 μg | Neural precursor cells | N/A | Direct conversion of human fibroblasts into neural precursor cells using modRNA | [113] |
| HumanBMSCs | BMP-2 | 5mC, ψU, Poly-A tail, ARCA | Branched PEI | 1 | 25 μg | Bone regeneration | Rat calvarial bone defect model | Scaffolds loaded with BMP-2 modRNA can enhance bone regeneration | [106] |
| Rat mesenchymal stem cells | BMP-2 | 5mC, 2TU, Poly-A tail, ARCA | C12-EPE | 1 | 2.5 μg | Bone regeneration | Rat femur defect model | Delivering hBMP-2 modRNA to a femur defect can result in new bone tissue formation | [107] |
| Rat mesenchymal stem cells | BMP-2 | 5mC, 2TU, Poly-A tail, Cap | Proprietary lipid | 1 | 2.5 μg | Bone regeneration | Rat femur defect model | BMP-2 modRNA-loaded collagen sponges can induce bone regeneration | [108] |
| HumanBMSCs | BMP-9, BMP-2 | 5mC, ψU, Poly-A tail, ARCA | PEI | 1 | 50 μg | Bone regeneration | Rat calvarial bone defect model | BMP-9 modRNA can induce increased connectivity density of the regenerated bone compared with BMP-2 modRNA | [109] |
| Rat BMSCs | BMP-2 | 5mC, 2TU, Poly-A tail, ARCA | DF-gold | 1 | 1 μg | Osteogenesis | N/A | The micro-macro biphasic calcium phosphate (MBCP) granules synergistically enhance the hBMP-2 modRNA-induced osteogenic pathway | [100] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.Y.L. Application of Modified mRNA in Somatic Reprogramming to Pluripotency and Directed Conversion of Cell Fate. Int. J. Mol. Sci. 2021, 22, 8148. https://doi.org/10.3390/ijms22158148
Wang AYL. Application of Modified mRNA in Somatic Reprogramming to Pluripotency and Directed Conversion of Cell Fate. International Journal of Molecular Sciences. 2021; 22(15):8148. https://doi.org/10.3390/ijms22158148
Chicago/Turabian StyleWang, Aline Yen Ling. 2021. "Application of Modified mRNA in Somatic Reprogramming to Pluripotency and Directed Conversion of Cell Fate" International Journal of Molecular Sciences 22, no. 15: 8148. https://doi.org/10.3390/ijms22158148
APA StyleWang, A. Y. L. (2021). Application of Modified mRNA in Somatic Reprogramming to Pluripotency and Directed Conversion of Cell Fate. International Journal of Molecular Sciences, 22(15), 8148. https://doi.org/10.3390/ijms22158148






