The Role of PERK in Understanding Development of Neurodegenerative Diseases
Abstract
:1. Introduction
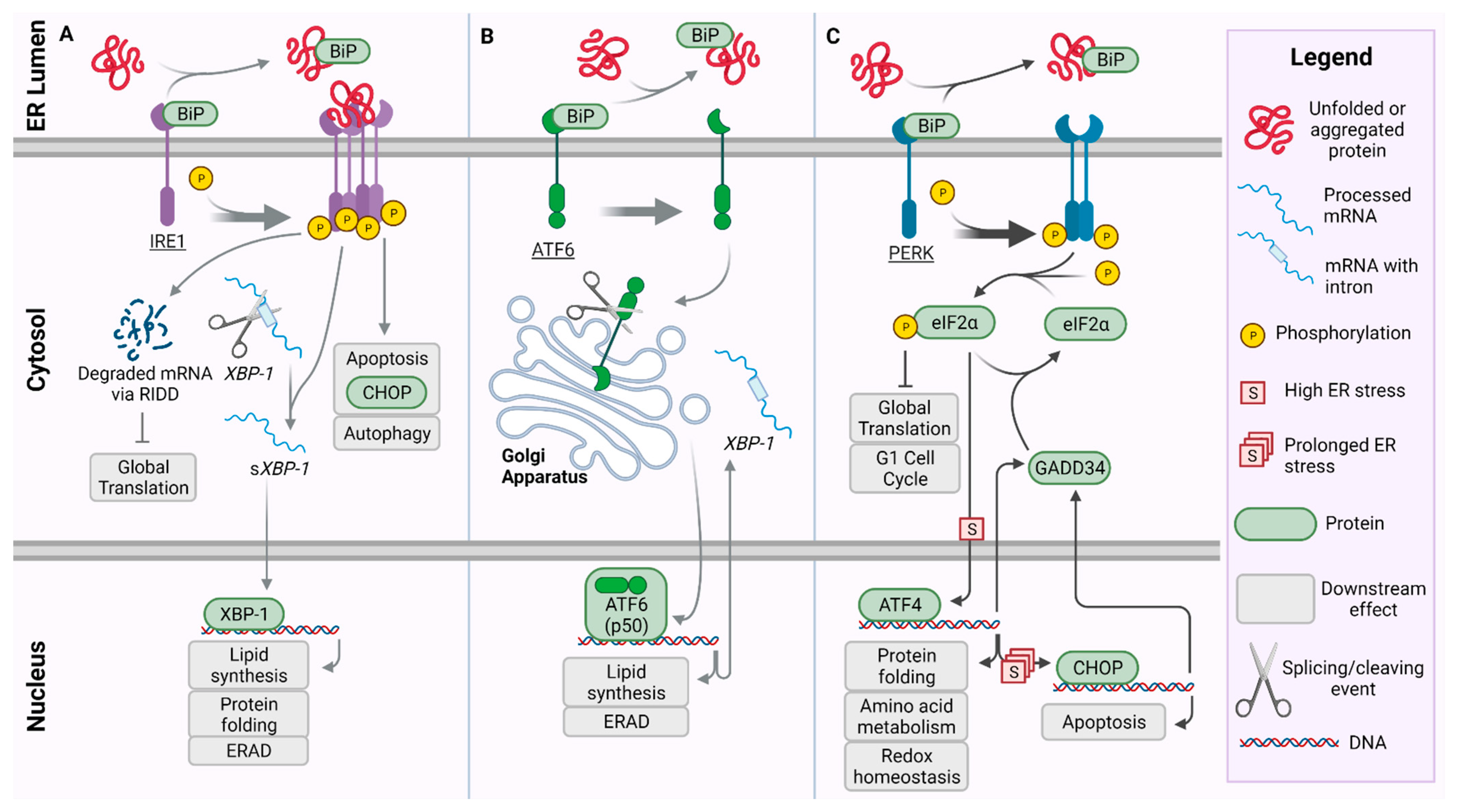
2. Unfolded Protein Response Pathway and PERK
3. Crosstalk between PERK and Pathways of the UPR
4. The Role of PERK in Neurodegeneration
5. Genetic Components of PERK Failure
6. Clinical Evidence of ER Stress Modulation as Treatment
7. Discussion/Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, R.C.; Lockwood, A.H.; Sonawane, B.R. Neurodegenerative diseases: An overview of environmental risk factors. Environ. Health Perspect. 2005, 113, 1250–1256. [Google Scholar] [CrossRef] [Green Version]
- Takalo, M.; Salminen, A.; Soininen, H.; Hiltunen, M.; Haapasalo, A. Protein aggregation and degradation mechanisms in neurodegenerative diseases. Am. J. Neurodegener. Dis. 2013, 2, 1–14. [Google Scholar] [PubMed]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10. [Google Scholar] [CrossRef]
- Halliday, M.; Hughes, D.; Mallucci, G.R. Fine-tuning PERK signaling for neuroprotection. J. Neurochem. 2017, 142, 812–826. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.; Ryten, M.; Simone, R.; Trabzuni, D.; Nicolaou, N.; Hondhamuni, G.; Ramasamy, A.; Vandrovcova, J.; UK Brain Expression Consortium; Weale, M.E.; et al. Assessment of common variability and expression quantitative trait loci for genome-wide associations for progressive supranuclear palsy. Neurobiol. Aging. 2014, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, Y.; Yang, X.; Lau, J.C.; Hung, C.H.; Wuwongse, S. Endoplasmic Reticulum Stress Induces Tau Pathology and Forms a Vicious Cycle: Implication in Alzheimer’s Disease Pathogenesis. J. Alzheimers Dis. 2014, 28, 839–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crary, J.F.; Trojanowski, J.Q.; Schneider, J.A.; Abisambra, J.F.; Abner, E.L.; Alafuzoff, I.; Arnold, S.E.; Attems, J.; Beach, T.G.; Bigio, E.H.; et al. Primary age-related tauopathy (PART): A common pathology associated with human aging. Acta Neuropathol. 2014, 128, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Ballatore, C.; Lee, V.M.Y.; Trojanowski, J.Q. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci. 2007, 8, 663–672. [Google Scholar] [CrossRef]
- Lee, V.M.Y.; Goedert, M.; Trojanowski, J.Q. Neurodegenerative Tauopathies. Genetics 2001, 24, 1121–1161. [Google Scholar]
- Rao, R.V.; Bredesen, D.E. Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr. Opin. Cell Biol. 2004, 16, 653–662. [Google Scholar] [CrossRef] [Green Version]
- Taylor, J.P.; Hardy, J.; Fischbeck, K.H. Toxic proteins in neurodegenerative disease. Science 2002, 296, 1991–1995. [Google Scholar] [CrossRef] [PubMed]
- Vonsattel, J.P.G.; DiFiglia, M. Huntington Disease. J. Neuropathol. Exp. Neurol. 1998, 57, 369–384. [Google Scholar] [CrossRef] [Green Version]
- Moreno, J.A.; Radford, H.; Peretti, D.; Steinert, J.R.; Verity, N.; Martin, M.G.; Halliday, M.; Morgan, J.; Disndale, D.; Ortori, C.A.; et al. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature 2012, 485, 507–511. [Google Scholar] [CrossRef] [Green Version]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef]
- Schröder, M.; Kaufman, R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005, 74, 739–789. [Google Scholar] [CrossRef] [PubMed]
- Tabara, K.; Iwata, Y.; Koizumi, N. The unfolded protein response. Methods Mol. Biol. 2018, 1691, 223–230. [Google Scholar] [CrossRef]
- Gardner, B.M.; Walter, P. Unfolded Proteins Are Ire1-Activating Ligands That Directly Induce the Unfolded Protein Response. Science 2011, 333, 1891–1895. [Google Scholar] [CrossRef] [Green Version]
- Hollien, J.; Lin, J.H.; Li, H.; Stevens, N.; Walter, P.; Weissman, J.S. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009, 186, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.S.; Walter, P. A Novel Mechanism for Regulating Activity of a Transcription Factor That Controls the Unfolded Protein Response. Cell 1996, 87, 391–404. [Google Scholar] [CrossRef] [Green Version]
- Sidrauski, C.; Walter, P. The Transmembrane Kinase Ire1p Is a Site-Specific Endonuclease That Initiates mRNA Splicing in the Unfolded Protein Response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef] [Green Version]
- Cui, W.; Li, J. The structure of the PERK kinase domain suggests the mechanism for its activation research papers. Acta Cryst. 2011, 423–428. [Google Scholar] [CrossRef] [Green Version]
- Walter, P.; Ron, D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [Green Version]
- Schindler, A.J.; Schekman, R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc. Natl. Acad. Sci. USA 2009, 106, 17775–17780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Chen, X.; Hendershot, L.; Prywes, R. ER Stress Regulation of ATF6 Localization by Dissociation of BiP/GRP78 Binding and Unmasking of Golgi Localization Signals. Dev. Cell 2002, 3, 99–111. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, S.; Ng, D.T.W. Sensing ER Stress. Cell Biol. 2011, 333, 1830–1832. [Google Scholar] [CrossRef] [PubMed]
- Scheper, W.; Hoozemans, J.J.M. Endoplasmic Reticulum Protein Quality Control in Neurodegenerative Disease: The Good, the Bad and the Therapy. Curr. Med. Chem. 2009, 16, 615–626. [Google Scholar] [CrossRef]
- Lee, K.; Tirasophon, W.; Shen, X.; Michalak, M.; Prywes, R.; Okada, T.; Yoshida, H.; Mori, K.; Kaufman, R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002, 16, 452–466. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Lv, Y.; Zhao, N.; Guan, G.; Wang, J. Protein kinase R-like ER kinase and its role in endoplasmic reticulum stress-decided cell fate. Cell Death Dis. 2015, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Carrara, M.; Prischi, F.; Nowak, P.R.; Ali, M.M. Crystal structures reveal transient PERK luminal domain tetramerization in endoplasmic reticulum stress signaling. EMBO J. 2015, 34, 1589–1600. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Bertolotti, A.; Zeng, H.; Ron, D. Perk Is Essential for Translational Regulation and Cell Survival during the Unfolded Protein Response. Mol. Cell 2000, 5, 897–904. [Google Scholar] [CrossRef]
- Lu, P.D.; Harding, H.P.; Ron, D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004, 167, 27–33. [Google Scholar] [CrossRef]
- Yukawa, K.; Tanaka, T.; Tsuji, S.; Akira, S. Regulation of transcription factor C/ATF by the cAMP signal activation in hippocampal neurons, and molecular interaction of C/ATF with signal integrator CBP/p300. Mol. Brain Res. 1999, 69, 124–134. [Google Scholar] [CrossRef]
- Hetz, C.; Chevet, E.; Harding, H.P. Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 2013, 12, 703–719. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 2003, 11, 619–633. [Google Scholar] [CrossRef]
- Blais, J.D.; Filipenko, V.; Bi, M.; Harding, H.P.; Ron, D.; Koumenis, C.; Wouters, B.G.; Bell, J.C. Activating Transcription Factor 4 Is Translationally Regulated by Hypoxic Stress. Mol. Cell Biol. 2004, 24, 7469–7482. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Brewer, J.W.; Alan Diehl, J.; Hendershot, L.M. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 2002, 318, 1351–1365. [Google Scholar] [CrossRef]
- Vattem, K.M.; Wek, R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11269–11274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harding, H.P.; Novoa, I.; Zhang, Y.; Zeng, H.; Wek, R.; Schapira, M.; Ron, D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 2000, 6, 1099–1108. [Google Scholar] [CrossRef]
- Novoa, I.; Zeng, H.; Harding, H.P.; Ron, D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 2001, 153, 1011–1021. [Google Scholar] [CrossRef] [Green Version]
- Marciniak, S.J.; Yun, C.Y.; Oyadomari, S.; Novoa, I.; Zhang, Y.; Jungreis, R.; Nagata, K.; Harding, H.P.; Ron, D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004, 18, 3066–3077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCullough, K.D.M.C.; Martindale, J.L.; Klotz, L.; Aw, T.; Holbrook, N.J. Gadd153 Sensitizes Cells to Endoplasmic Reticulum Stress by Down-Regulating Bcl2 and Perturbing the Cellular Redox State. Mol. Cell Biol. 2001, 21, 1249–1259. [Google Scholar] [CrossRef] [Green Version]
- Puthalakath, H.; Reilly, L.A.O.; Gunn, P.; Lee, L.; Kelly, P.N.; Huntington, N.D.; Hughes, P.D.; Michalak, E.M.; McKimm-Breschkin, J.; Motoyama, N.; et al. ER Stress Triggers Apoptosis by Activating BH3-Only Protein Bim. Cell 2007, 129, 1337–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danial, N.N.; Korsmeyer, S.J. Cell Death: Critical Control Points Review. Cell 2004, 116, 205–219. [Google Scholar] [CrossRef] [Green Version]
- Brewer, J.W.; Hendershot, L.M.; Sherr, C.J.; Diehl, J.A. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. USA 1999, 96, 8505–8510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brewer, J.W. Regulatory crosstalk within the mammalian unfolded protein response. Cell Mol. Life Sci. 2014, 71, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Scheuner, D.; Song, B.; Mcewen, E.; Liu, C.; Laybutt, R.; Gillespie, P.; Saunders, T.; Bonner-Weir, S.; Kaufman, R.J. Translational Control Is Required for the Unfolded Protein Response and In Vivo Glucose Homeostasis. Mol. Cell 2001, 7, 1165–1176. [Google Scholar] [CrossRef]
- Wu, J.; Rutkowski, D.T.; Dubois, M.; Swathirajan, J.; Saunders, T.; Wang, J.; Song, B.; Yau, G.D.-Y.; Kaufman, R.J. ATF6a Optimizes Long-Term Endoplasmic Reticulum Function to Protect Cells from Chronic Stress. Dev. Cell 2007, 351–364. [Google Scholar] [CrossRef] [Green Version]
- Adachi, Y.; Yamamoto, K.; Okada, T.; Yoshida, H. ATF6 is a Transcription Factor Specializing in the Regulation of Quality Control Proteins in the Endoplasmic Reticulum. Cell Struct. Funct. 2008, 33. [Google Scholar] [CrossRef] [Green Version]
- Teske, B.F.; Wek, S.A.; Bunpo, P.; Cundiff, J.K.; Mcclintick, J.N. The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol. Biol. Cell 2011, 6. [Google Scholar] [CrossRef]
- Calfon, M.; Zeng, H.; Urano, F.; Till, J.H.; Hubbard, S.R.; Harding, H.P.; Clark, S.G.; Ron, D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 2002, 415, 1–6. [Google Scholar] [CrossRef]
- Huang, C.C.; Li, Y.; Lopez, A.B.; Chiang, C.; Kaufman, R.J. Temporal regulation of Cat-1 (cationic amino acid transporter-1) gene transcription during endoplasmic reticulum stress. Biochem. J. 2010, 429, 215–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majumder, M.; Huang, C.; Snider, M.D.; Komar, A.A.; Tanaka, J.; Kaufman, R.J. A Novel Feedback Loop Regulates the Response to Endoplasmic Reticulum Stress via the Cooperation of Cytoplasmic Splicing and mRNA Translation. Mol. Cell Biol. 2012, 32, 992–1003. [Google Scholar] [CrossRef] [Green Version]
- Bell, M.C.; Meier, S.E.; Ingram, A.L.; Abisambra, J.F. PERK-Opathies: An Endoplasmic Reticulum Stress Mechanism Underlying Neurodegeneration. Curr. Alzheimers Res. 2016, 13, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Nijholt, D.A.T.; Haastert, E.S.; Van Rozemuller, A.J.M.; Scheper, W.; Jm, J. The unfolded protein response is associated with early tau pathology in the hippocampus of tauopathies. J. Pathol. 2012, 226, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Klann, E. PERK: A novel therapeutic target for neurodegenerative diseases? Alzheimers Res. Ther. 2014, 6, 4–6. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.H.; Hiramatsu, N.; Liu, Q.; Sun, X.V.; Lenh, D.; Chan, P.; Chiang, K.; Koo, E.H.; Kao, A.W.; Litvan, I.; et al. Tauopathy-associated PERK alleles are functional hypomorphs that increase neuronal vulnerability to ER stress. Hum. Mol. Genet. 2018, 27, 3951–3963. [Google Scholar] [CrossRef]
- Kouri, N.; Ross, O.A.; Dombroski, B.; Younkin, C.S.; Serie, D.J.; Soto-Ortolaza, A.; Baker, M.; Finch, N.C.A.; Yoon, H.; Kim, J.; et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat. Commun. 2015, 6, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkwood, T.B.L. The most pressing problem of our age. Br. Med. J. 2003, 326, 1297–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaFerla, F.M.; Green, K.N.; Oddo, S. Intracellular amyloid-β in Alzheimer’s disease. Nat. Rev. Neurosci. 2007, 8, 499–509. [Google Scholar] [CrossRef]
- Ray, I.; Chauhan, A.; Wisniewski, H.M.; Wegiel, J.; Kim, K.S.; Chauhan, V.P.S. Binding of amyloid beta-protein to intracellular brain proteins in rat and human. Neurochem. Res. 1998, 23, 1277–1282. [Google Scholar] [CrossRef]
- Muralidar, S.; Visaga, S.; Sekaran, S.; Thirumalai, D.; Palaniappan, B. Role of tau protein in Alzheimer ’ s disease: The prime pathological player. Int. J. Biol. Macromol. 2020, 163, 1599–1617. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Imaizumi, K.; Honda, A.; Yoneda, T.; Kudo, T.; Takeda, M.; Moi, K.; Rozmahel, R.; Fraser, P.; George-Hyslop, P.S.; et al. Disturbed Activation of Endoplasmic Reticulum Stress Transducers by Familial Alzheimer’s Disease-linked Presenilin-1 Mutations. J. Biol. Chem. 2001, 276, 43446–43454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katayama, T.; Imaizumi, K.; Sato, N.; Miyoshi, K.; Kudo, T.; Morihara, T.; Yoneda, T.; Gomi, F.; Mori, Y.; Nakano, Y.; et al. Presenilin-1 mutations downregulate the signalling pathway of the unfolded- protein response. Nat. Cell Biol. 1999, 1, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Hamos, J.E.; Oblas, B.; Pulaski-Salo, D.; Welch, W.J.; Bole, D.G.; Drachman, D.A. Expression of heat shock proteins in Alzheimer’s disease. Neurology 1991, 41, 345–350. [Google Scholar] [CrossRef]
- Hoozemans, J.J.M.; Veerhuis, R.; Van Haastert, E.S.; Rozemuller, J.M.; Baas, F.; Eikelenboom, P.; Scheper, W. The unfolded protein response is activated in Alzheimer’s disease. Acta Neuropathol. 2005, 110, 165–172. [Google Scholar] [CrossRef]
- Yu, M.; Suen, K.; Kwok, N.; So, K.; Hugon, J.; Chuen-chung, R. Beta-amyloid peptides induces neuronal apoptosis via a mechanism independent of unfolded protein responses. Apoptosis 2006, 11, 687–700. [Google Scholar] [CrossRef]
- Hoozemans, J.J.M.; Van Haastert, E.S.; Nijholt, D.A.T.; Rozemuller, A.J.M.; Eikelenboom, P.; Scheper, W. The unfolded protein response is activated in pretangle neurons in alzheimer’s disease hippocampus. Am. J. Pathol. 2009, 174, 1241–1251. [Google Scholar] [CrossRef] [Green Version]
- Chang, R.Y.K.; Nouwens, A.S.; Dodd, P.R.; Etheridge, N. The synaptic proteome in Alzheimer’s disease. Alzheimers Dement 2013, 9, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Abisambra, J.F.; Jinwal, U.K.; Blair, L.J.; Leary, J.C.O.; Li, Q.; Brady, S.; Wang, L.; Guidi, C.E.; Zhang, B.; Nordhues, B.A.; et al. Tau Accumulation Activates the Unfolded Protein Response by Impairing Endoplasmic Reticulum-Associated Degradation. Neurobiol. Dis. 2013, 33, 9498–9507. [Google Scholar] [CrossRef]
- Devi, L.; Ohno, M. PERK mediates eIF2α phosphorylation responsible for BACE1 elevation, CREB dysfunction and neurodegeneration in a mouse model of Alzheimer’s disease. Neurobiol. Aging 2014, 35, 2272–2281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braakman, I.; Bulleid, N.J. Protein Folding and Modification in the Mammalian Endoplasmic Reticulum. Annu. Rev. Biochem. 2011, 80, 71–99. [Google Scholar] [CrossRef]
- Scheper, W.; Hoozemans, J.J.M. The unfolded protein response in neurodegenerative diseases: A neuropathological perspective. Acta Neuropathol. 2015, 130, 315–331. [Google Scholar] [CrossRef] [Green Version]
- Unterberger, U.; Ho, R.; Gelpi, E.; Flicker, H.; Budka, H.; Voigtla, T. Endoplasmic Reticulum Stress Features Are Prominent in Alzheimer Disease but Not in Prion Diseases In Vivo. J. Neuropathol. Exp. Neurol. 2006, 65, 348–358. [Google Scholar] [CrossRef] [Green Version]
- Stutzbach, L.D.; Xie, S.X.; Naj, A.C.; Albin, R.; Gilman, S.; PSP Genetics Study Group; Lee, V.M.Y.; Trojanowski, J.Q.; Devlin, B.; Schellenberg, G.D. The unfolded protein response is activated in disease-affected brain regions in progressive supranuclear palsy and Alzheimer’s disease. Acta Neuropathol. Commun. 2013, 1, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruch, J.; Xu, H.; Rösler, T.W.; De Andrade, A.; Kuhn, P.; Lichtenthaler, S.F.; Arzberger, T.; Winklhofer, K.F.; Muller, U.; Hoglinger, G.U. PERK activation mitigates tau pathology in vitro and in vivo. EMBO Mol. Med. 2017, 9, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Foltynie, T.; Brayne, C.E.G.; Robbins, T.W.; Barker, R.A. The cognitive ability of an incident cohort of Parkinson’s patients in the, U.K. The CamPaIGN study. Brain 2004, 127, 550–560. [Google Scholar] [CrossRef] [Green Version]
- Hindle, J.V. Ageing, neurodegeneration and Parkinson’s disease. Age Ageing 2010, 39, 156–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoozemans, J.J.M.; Rozemuller, J.M.; Scheper, W. Activation of the unfolded protein response in Parkinson’s disease. Biochem. Biophys. Res. Commun. 2007, 354, 707–711. [Google Scholar] [CrossRef]
- Ryu, E.J.; Harding, H.P.; Angelastro, J.M.; Vitolo, O.V.; Ron, D.; Greene, L.A. Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson’s disease. J. Neurosci. 2002, 22, 10690–10698. [Google Scholar] [CrossRef] [Green Version]
- Colla, E.; Coune, P.; Liu, Y.; Pletnikova, O.; Troncoso, J.C.; Iwatsubo, T.; Schneider, B.L.; Lee, M.K. Endoplasmic reticulum stress is important for the manifestations of α-synucleinopathy in vivo. J. Neurosci. 2012, 32, 3306–3320. [Google Scholar] [CrossRef]
- Carnemolla, A.; Fossale, E.; Agostoni, E.; Michelazzi, S.; Calligaris, R.; De Maso, L.; Del Sal, G.; MacDonald, M.E.; Persichetti, F. Rrs1 is involved in endoplasmic reticulum stress response in huntington disease. J. Biol. Chem. 2009, 284, 18167–18173. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Noh, J.Y.; Oh, Y.; Kim, Y.; Chang, J.W.; Chung, C.W.; Lee, S.T.; Kin, M.; Ryu, H.; Jung, Y.K. IRE1 plays an essential role in ER stress-mediated aggregation of mutant huntingtin via the inhibition of autophagy flux. Hum. Mol. Genet. 2012, 21, 101–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hetz, C.; Russelakis-Carneiro, M.; Maundrell, K.; Castilla, J.; Soto, C. Caspase-12 and endoplasmic reticulum stress mediate neurotoxicity of pathological prion protein. EMBO J. 2003, 22, 5435–5445. [Google Scholar] [CrossRef] [Green Version]
- Ito, Y.; Yamada, M.; Tanaka, H.; Aida, K.; Tsuruma, K.; Shimazawa, M.; Hozumi, I.; Inuzuka, T.; Takahashi, H.; Hara, H. Involvement of CH.O.P.; an ER-stress apoptotic mediator, in both human sporadic ALS and ALS model mice. Neurobiol. Dis. 2009, 36, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Thielen, P.; Matus, S.; Nassif, M.; Court, F.; Kiffin, R.; Martinez, G.; Cuervo, A.M.; Brown, R.H.; Glimcher, L.H. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009, 23, 2294–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, S.; Cabuy, E.; Caroni, P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 2009, 12, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zeng, H.; Zhang, Y.; Jungries, R.; Chung, P.; Plesken, H.; Sabatini, D.D.; Ron, D. Diabetes mellitus and exocrine pancreatic dysfunction in Perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell 2001, 7, 1153–1163. [Google Scholar] [CrossRef]
- Senee, V.; Vattem, K.M.; Delepine, M.; Rainbow, L.A.; Haton, C.; Lecoq, A.; Shaw, N.J.; Robert, J.J.; Rooman, R.; Diatloff-Zito, D.; et al. Wolcott-Rallison Syndrome Clinical, Genetic, and Functional Study of EIF2AK3 Mutations and Suggestion of Genetic Heterogeneity. Diabetes 2004, 53, 1876–1883. [Google Scholar] [CrossRef] [Green Version]
- Julier, C.; Nicolino, M. Wolcott-Rallison syndrome. Orphanet. J. Rare Dis. 2010, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Bruch, J.; Kurz, C.; Vasiljevic, A.; Nicolino, M.; Arzberger, T.; Höglinger, G.U. Early Neurodegeneration in the Brain of a Child Without Functional PKR-like Endoplasmic Reticulum Kinase. J. Neuropathol. Exp. Neurol. 2015, 74, 850–857. [Google Scholar] [CrossRef] [Green Version]
- Höglinger, G.U.; Melhem, N.M.; Dickson, D.W.; Sleiman, P.M.A.; Wang, L.; Klei, L.; Rademakers, R.; de Silva, R.; Litvan, I.; Riley, D.E.; et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat. Publ. Gr. 2011, 43. [Google Scholar] [CrossRef]
- Sanchez-Contreras, M.Y.; Kouri, N.; Cook, C.N.; Serie, D.J.; Heckman, M.G.; Finch, N.A.; Caselli, R.J.; Uitti, R.J.; Wszolek, Z.K.; Graff-Radford, N.; et al. Replication of progressive supranuclear palsy genome-wide association study identifies SLCO1A2 and DUSP10 as new susceptibility loci. Mol. Neurodegener. 2018, 13, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Yu, J.; Miao, D.; Ma, X.; Wang, H.; Wang, W.; Tan, L. An exploratory study on STX6, MO.B.P.; MA.P.T.; and EIF2AK3 and late-onset Alzheimer’s disease. Neurobiol. Aging 2013, 34, 1519.e13–1519.e17. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hoppman, N.; Connell, J.R.O.; Wang, H.; Streeten, E.A.; Mclenithan, J.C.; Mitchell, B.D.; Shuldiner, A.R. A Functional Haplotype in EIF2AK3, an ER Stress Sensor, Is Associated With Lower Bone Mineral Density. J. Bone Miner. Res. 2012, 27, 331–341. [Google Scholar] [CrossRef] [Green Version]
- Khatoon, S.; Grundke-Iqbal, I.; Iqbal, K. Brain Levels of Microtubule-Associated Protein τ Are Elevated in Alzheimer’s Disease: A Radioimmuno-Slot-Blot Assay for Nanograms of the Protein. J. Neurochem. 1992, 59, 750–753. [Google Scholar] [CrossRef]
- Hoover, B.R.; Reed, M.N.; Su, J.; Penrod, R.D.; Kotilinek, L.A.; Grant, M.K.; Pitstick, R.; Carlson, G.A.; Lanier, L.M.; Yuan, L.L.; et al. Tau Mislocalization to Dendritic Spines Mediates Synaptic Dysfunction Independently of Neurodegeneration. Neuron 2010, 68, 1067–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thies, E.; Mandelkow, E.M. Missorting of tau in neurons causes degeneration of synapses that can be rescued by the kinase MARK2/Par-1. J. Neurosci. 2007, 27, 2896–2907. [Google Scholar] [CrossRef] [Green Version]
- Alonso, A.D.C.; Zaidi, T.; Novak, M.; Grundke-Iqbal, I.; Iqbal, K. Hyperphosphorylation induces self-assembly of τ into tangles of paired helical filaments/straight filaments. Proc. Natl. Acad. Sci. USA 2001, 98, 6923–6928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.P.; Biernat, J.; Pickhardt, M.; Mandelkow, E.; Mandelkow, E.M. Stepwise proteolysis liberates tau fragments that nucleate the Alzheimer-like aggregation of full-length tau in a neuronal cell model. Proc. Natl. Acad. Sci. USA 2007, 104, 10252–10257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eftekharzadeh, B.; Daigle, J.G.; Kapinos, L.E.; Coyne, A.; Schiantarelli, J.; Carlomagno, Y.; Cook, C.; Miller, S.J.; Dujardin, S.; Amaral, A.S.; et al. Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron 2018, 99, 925–940.e7. [Google Scholar] [CrossRef] [Green Version]
- Lasagna-Reeves, C.A.; Castillo-Carranza, D.L.; Sengupta, U.; Sarmiento, J.; Troncoso, J.; Jackson, G.R.; Kayed, R. Identification of oligomers at early stages of tau aggregation in Alzheimer’s disease. FASEB J. 2012, 26, 1946–1959. [Google Scholar] [CrossRef] [Green Version]
- Wong, T.H.; van der Lee, S.J.; van Rooij, J.G.J.; Meeter, L.H.H.; Frick, P.; Melhem, S.; Harro, S.; Ikram, M.A.; Rozemuller, A.J.; Holstege, H.; et al. EIF2AK3 variants in Dutch patients with Alzheimer’s disease. Neurobiol. Aging 2019, 73, 229.e11–229.e18. [Google Scholar] [CrossRef]
- Urano, F. Wolfram Syndrome: Diagnosis, Management, and Treatment. Curr. Diab. Rep. 2016, 16, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, S.G.; Fukuma, M.; Lipson, K.L.; Nguyen, L.X.; Allen, J.R.; Oka, Y.; Urano, F. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pacreatic β-cells. J. Biol. Chem. 2005, 280, 39609–39615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bugiani, M.; Boor, I.; Powers, J.M.; Scheper, G.C.; Van Der Knaap, M.S. Leukoencephalopathy with vanishing white matter: A review. J. Neuropathol. Exp. Neurol. 2010, 69, 987–996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Wang, X.; van der Knaap, M.S.; Proud, C.G. Mutations Linked to Leukoencephalopathy with Vanishing White Matter Impair the Function of the Eukaryotic Initiation Factor 2B Complex in Diverse Ways. Mol. Cell Biol. 2004, 24, 3295–3306. [Google Scholar] [CrossRef] [Green Version]
- Matsukawa, T.; Wang, X.; Liu, R.; Wortham, N.C.; Onuki, Y.; Kubota, A.; Hida, A.; Kowa, H.; Fukuda, Y.; Ishiura, H.; et al. Adult-onset leukoencephalopathies with vanishing white matter with novel missense mutations in EIF2B2, EIF2B3, and EIF2B5. Neurogenetics 2011, 12, 259–261. [Google Scholar] [CrossRef]
- Leng, X.; Wu, Y.; Wang, X.; Pan, Y.; Wang, J.; Li, J.; Du, L.; Dai, L.; Wu, X.; Proud, C.G.; et al. Functional analysis of recently identified mutations in eukaryotic translation initiation factor 2Bε (eIF2Bε) identified in Chinese patients with vanishing white matter disease. J. Hum. Genet. 2011, 56, 300–305. [Google Scholar] [CrossRef]
- Zhou, L.; Li, P.; Chen, N.; Dai, L.F.; Gao, K.; Liu, Y.N.; Shen, L.; Wang, J.M.; Jiang, Y.W.; Wu, Y. Modeling vanishing white matter disease with patient-derived induced pluripotent stem cells reveals astrocytic dysfunction. CNS Neurosci. Ther. 2019, 25, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Van der Knaap, M.S.; Pronk, J.C.; Scheper, G.C. Vanishing White Matter Disease. Lancet Neurol. 2006, 5, 413–423. [Google Scholar] [CrossRef]
- Wong, Y.L.; Lebon, L.; Edalji, R.; Lim, H.B.; Sun, C.; Sidrauski, C. The small molecule ISRIB rescues the stability and activity of vanishing white matter disease eIF2B mutant complexes. eLife 2018, 7, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Abbink, T.E.M.; Wisse, L.E.; Jaku, E.; Thiecke, M.J.; Voltolini-González, D.; Fritsen, H.; Bobeldijk, S.; ter Braak, T.J.; Polder, E.; Postma, N.L.; et al. Vanishing white matter: Deregulated integrated stress response as therapy target. Ann. Clin. Transl. Neurol. 2019, 6, 1407–1422. [Google Scholar] [CrossRef] [Green Version]
- Brauer, R.; Lau, W.C.Y.; Hayes, J.F.; Man, K.K.C.; Osborn, D.P.J.; Howard, R.; Kim, J.; Wong, I.C.K. Trazodone use and risk of dementia: A population-based cohort study. PLoS Med. 2019, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- La, A.L.; Walsh, C.M.; Neylan, T.C.; Vossel, K.A.; Yaffe, K.; Krystal, A.D.; Miller, B.L.; Karageorgiou, E. Long-Term Trazodone Use and Cognition: A Potential Therapeutic Role for Slow-Wave Sleep Enhancers. J. Alzheimers Dis. 2019, 67, 911–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paganoni, S.; Hendrix, S.; Dickson, S.P.; Knowlton, N.; Macklin, E.A.; Berry, J.D.; Elliott, M.A.; Maiser, S.; Karam, C.; Caress, J.B.; et al. Long-term survival of participants in the CENTAUR trial of sodium phenylbutyrate-taurursodiol in amyotrophic lateral sclerosis. Muscle Nerve 2021, 63, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Paganoni, S.; Macklin, E.A.; Hendrix, S.; Berry, J.D.; Elliott, M.A.; Maiser, S.; Karam, C.; Caress, J.B.; Owegi, M.A.; Quick, A.; et al. Trial of Sodium Phenylbutyrate–Taurursodiol for Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2020, 383, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Bhat, A.; Chakraborty, R.; Adlakha, K.; Sengupta, S.; Roy, S.; Chakraborty, K. Proteomic profile of 4-PBA treated human neuronal cells during ER stress. Mol. Omi. 2018, 14, 53–63. [Google Scholar] [CrossRef]
- Suaud, L.; Miller, K.; Panichelli, A.E.; Randell, R.L.; Marando, C.M.; Rubenstein, R.C. 4-Phenylbutyrate stimulates Hsp70 expression through the Elp2 component of elongator and STAT-3 in cystic fibrosis epithelial cells. J. Biol. Chem. 2011, 286, 45083–45092. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, C.M.P.; Solá, S.; Sharpe, J.C.; Moura, J.J.G.; Steer, C.J. Tauroursodeoxycholic acid prevents Bax-induced membrane perturbation and cytochrome c release in isolated mitochondria. Biochemistry 2003, 42, 3070–3080. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smedley, G.D.; Walker, K.E.; Yuan, S.H. The Role of PERK in Understanding Development of Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 8146. https://doi.org/10.3390/ijms22158146
Smedley GD, Walker KE, Yuan SH. The Role of PERK in Understanding Development of Neurodegenerative Diseases. International Journal of Molecular Sciences. 2021; 22(15):8146. https://doi.org/10.3390/ijms22158146
Chicago/Turabian StyleSmedley, Garrett Dalton, Keenan E. Walker, and Shauna H. Yuan. 2021. "The Role of PERK in Understanding Development of Neurodegenerative Diseases" International Journal of Molecular Sciences 22, no. 15: 8146. https://doi.org/10.3390/ijms22158146





