Adaptor Molecules Epitranscriptome Reprograms Bacterial Pathogenicity
Abstract
:1. Introduction
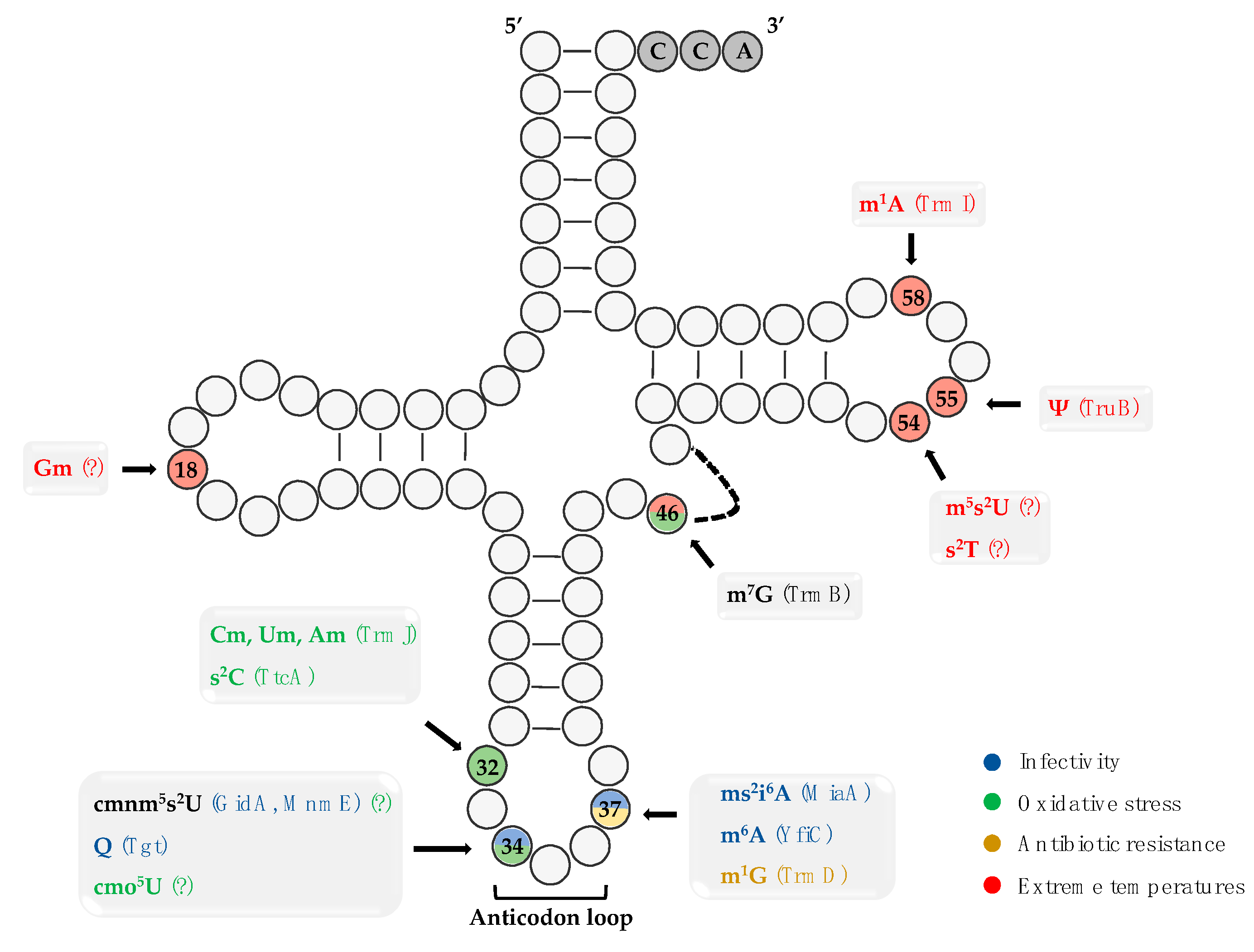
2. tRNA Epitranscriptome into the Bacterial Infectivity
2.1. tRNA Modification at the Wobble Position 34
2.2. ROS-Induced tRNA Modifications
2.3. Membrane-Associated tRNA Modifications
2.4. Temperature-Related tRNA Modifications
3. Methodologies for Detection of tRNA Modifications
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| VapC | Virulence associated protein C |
| TrmB | tRNA (guanine-N(7)-)-methyltransferase |
| TrmD | tRNA (guanine-N(1)-)-methyltransferase |
| TrmJ | tRNA (cytidine/uridine-2′-O-)-methyltransferase |
| IscS | iron-sulfur cluster assembly gene |
| MnmA | tRNA-specific 2-thiouridylase |
| TtcA | tRNA-cytidine(32) 2-sulfurtransferase |
| ThiI | tRNA sulfurtransferase |
| TtuA | RNA-5-methyluridine(54) 2-sulfurtransferase |
| TsaB, TsaE | tRNA threonylcarbamoyladenosine biosynthesis proteins B and E |
| TmcA | tRNA(Met) cytidine acetyltransferase |
| TadA | tRNA-specific adenosine deaminase |
| MiaA | tRNA dimethylallyltransferase |
| TrmL | tRNA (cytidine(34)-2′-O)-methyltransferase |
| TusA | Sulfur transfer protein |
| ALKB | alkane hydroxylase |
| ALKBH1 | AlkB homolog 1 |
| GidA | glucose-inhibited division protein A |
| MnmE | tRNA modification GTPase |
| SpeB | Streptococcus pyrogenic exotoxin B |
| VirF | virulence regulon transcriptional activator VirF |
| DosR | dormancy survival regulator |
| katA, katB | catalase A and B |
| DnaA | chromosomal replication initiator protein |
| LolB | lipoprotein localization protein |
| OmpA | outer membrane protein A |
References
- Pizarro-Cerdá, J.; Cossart, P. Bacterial Adhesion and Entry into Host Cells. Cell 2006, 124, 715–727. [Google Scholar] [CrossRef] [Green Version]
- Aujoulat, F.; Roger, F.; Bourdier, A.; Lotthé, A.; Lamy, B.; Marchandin, H.; Jumas-Bilak, E. From Environment to Man: Genome Evolution and Adaptation of Human Opportunistic Bacterial Pathogens. Genes 2012, 3, 191–232. [Google Scholar] [CrossRef] [Green Version]
- Tollerson, R.; Ibba, M. Translational Regulation of Environmental Adaptation in Bacteria. J. Biol. Chem. 2020, 295, 10434–10445. [Google Scholar] [CrossRef]
- Kaliatsi, E.G.; Giarimoglou, N.; Stathopoulos, C.; Stamatopoulou, V. Non-Coding RNA-Driven Regulation of RRNA Biogenesis. Int. J. Mol. Sci. 2020, 21, 9738. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Stamatopoulou, V.; Apostolidi, M.; Li, S.; Lamprinou, K.; Papakyriakou, A.; Zhang, J.; Stathopoulos, C. Direct Modulation of T-Box Riboswitch-Controlled Transcription by Protein Synthesis Inhibitors. Nucleic Acids Res. 2017, 45, 10242–10258. [Google Scholar] [CrossRef] [Green Version]
- Vandivier, L.E.; Gregory, B.D. Reading the Epitranscriptome. In The Enzymes; Elsevier: Amsterdam, The Netherlands, 2017; Volume 41, pp. 269–298. ISBN 978-0-12-811777-4. [Google Scholar]
- Christofi, T.; Zaravinos, A. RNA Editing in the Forefront of Epitranscriptomics and Human Health. J. Transl. Med. 2019, 17, 319. [Google Scholar] [CrossRef]
- Stamatopoulou, V.; Zaravinos, A. Epitranscriptomics markers regulate the infection by RNA viruses. In Epitranscriptomics; RNA Technologies; Springer Nature Series; Springer: Chem, Switzerland, 2021; Volume 11, ISBN 978-3-030-71612-7. [Google Scholar]
- Saletore, Y.; Meyer, K.; Korlach, J.; Vilfan, I.D.; Jaffrey, S.; Mason, C.E. The Birth of the Epitranscriptome: Deciphering the Function of RNA Modifications. Genome Biol. 2012, 13, 175. [Google Scholar] [CrossRef] [Green Version]
- Helm, M.; Motorin, Y. Detecting RNA Modifications in the Epitranscriptome: Predict and Validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [Green Version]
- Yi, C.; Pan, T. Cellular Dynamics of RNA Modification. Acc. Chem. Res. 2011, 44, 1380–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaefer, M.; Kapoor, U.; Jantsch, M.F. Understanding RNA Modifications: The Promises and Technological Bottlenecks of the ‘Epitranscriptome’. Open Biol. 2017, 7, 170077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz, A.; Elgamal, S.; Rajkovic, A.; Ibba, M. Non-Canonical Roles of TRNAs and TRNA Mimics in Bacterial Cell Biology: Non-Canonical Roles of TRNAs and TRNA Mimics. Mol. Microbiol. 2016, 101, 545–558. [Google Scholar] [CrossRef] [Green Version]
- Schimmel, P. The Emerging Complexity of the TRNA World: Mammalian TRNAs beyond Protein Synthesis. Nat. Rev. Mol. Cell Biol. 2018, 19, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.S.; Sarin, L.P. Transfer RNA Modification and Infection—Implications for Pathogenicity and Host Responses. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P.; Bagiński, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A Database of RNA Modification Pathways. 2017 Update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Zhao, B.S.; He, C. Pseudouridine in a New Era of RNA Modifications. Cell Res. 2015, 25, 153–154. [Google Scholar] [CrossRef] [Green Version]
- Tuorto, F.; Lyko, F. Genome Recoding by TRNA Modifications. Open Biol. 2016, 6, 160287. [Google Scholar] [CrossRef] [Green Version]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA Modifications: Nature’s Combinatorial Chemistry Playground: Transfer RNA Modifications. WIREs RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Edwards, A.M.; Addo, M.A.; Dos Santos, P.C. Extracurricular Functions of TRNA Modifications in Microorganisms. Genes 2020, 11, 907. [Google Scholar] [CrossRef]
- Chen, Z.; Qi, M.; Shen, B.; Luo, G.; Wu, Y.; Li, J.; Lu, Z.; Zheng, Z.; Dai, Q.; Wang, H. Transfer RNA Demethylase ALKBH3 Promotes Cancer Progression via Induction of TRNA-Derived Small RNAs. Nucleic Acids Res. 2019, 47, 2533–2545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motorin, Y.; Helm, M. TRNA Stabilization by Modified Nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef]
- Torres, A.G.; Reina, O.; Stephan-Otto Attolini, C.; Ribas de Pouplana, L. Differential Expression of Human TRNA Genes Drives the Abundance of TRNA-Derived Fragments. Proc. Natl. Acad. Sci. USA 2019, 116, 8451–8456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sørensen, M.A.; Fehler, A.O.; Lo Svenningsen, S. Transfer RNA Instability as a Stress Response in Escherichia coli: Rapid Dynamics of the TRNA Pool as a Function of Demand. RNA Biol. 2018, 15, 586–593. [Google Scholar] [CrossRef] [Green Version]
- Chionh, Y.H.; McBee, M.; Babu, I.R.; Hia, F.; Lin, W.; Zhao, W.; Cao, J.; Dziergowska, A.; Malkiewicz, A.; Begley, T.J.; et al. TRNA-Mediated Codon-Biased Translation in Mycobacterial Hypoxic Persistence. Nat. Commun. 2016, 7, 13302. [Google Scholar] [CrossRef] [Green Version]
- Cruz, J.W.; Sharp, J.D.; Hoffer, E.D.; Maehigashi, T.; Vvedenskaya, I.O.; Konkimalla, A.; Husson, R.N.; Nickels, B.E.; Dunham, C.M.; Woychik, N.A. Growth-Regulating Mycobacterium Tuberculosis VapC-Mt4 Toxin Is an Isoacceptor-Specific TRNase. Nat. Commun. 2015, 6, 7480. [Google Scholar] [CrossRef] [PubMed]
- de Crécy-Lagard, V.; Jaroch, M. Functions of Bacterial TRNA Modifications: From Ubiquity to Diversity. Trends Microbiol. 2021, 29, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.; Leonardi, A.; Dedon, P.; Begley, T. The Versatile Roles of the TRNA Epitranscriptome during Cellular Responses to Toxic Exposures and Environmental Stress. Toxics 2019, 7, 17. [Google Scholar] [CrossRef] [Green Version]
- Gebetsberger, J.; Wyss, L.; Mleczko, A.M.; Reuther, J.; Polacek, N. A TRNA-Derived Fragment Competes with MRNA for Ribosome Binding and Regulates Translation during Stress. RNA Biol. 2017, 14, 1364–1373. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; He, F.; Luo, J.; Dou, S.; Wang, Y.; Guo, A.; Lu, J. Drosophila TsRNAs Preferentially Suppress General Translation Machinery via Antisense Pairing and Participate in Cellular Starvation Response. Nucleic Acids Res. 2018, 46, 5250–5268. [Google Scholar] [CrossRef] [Green Version]
- Ren, B.; Wang, X.; Duan, J.; Ma, J. Rhizobial TRNA-Derived Small RNAs Are Signal Molecules Regulating Plant Nodulation. Science 2019, 365, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.M.; Zhou, C.; Huang, R.H. Reconstituting Bacterial RNA Repair and Modification in Vitro. Science 2009, 326, 247. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, X.; Shi, J.; Yan, M.; Zhou, T. Origins and Evolving Functionalities of TRNA-Derived Small RNAs. Trends Biochem. Sci. 2021, in press. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, Y.; Tan, D.; Zhang, X.; Yan, M.; Zhang, Y.; Franklin, R.; Shahbazi, M.; Mackinlay, K.; Liu, S.; et al. PANDORA-Seq Expands the Repertoire of Regulatory Small RNAs by Overcoming RNA Modifications. Nat. Cell Biol. 2021, 23, 424–436. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Rao, Y.S.P.; Cherayil, J.D. Studies on Chemical Modification of Thionucleosides in the Transfer Ribonucleic Acid of Escherichia Coli. Biochem. J. 1974, 143, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Chock, P.B. Oxidative Modifications of RNA and Its Potential Roles in Biosystem. Front. Mol. Biosci. 2021, 8, 685331. [Google Scholar] [CrossRef] [PubMed]
- Golovina, A.Y.; Sergiev, P.V.; Golovin, A.V.; Serebryakova, M.V.; Demina, I.; Govorun, V.M.; Dontsova, O.A. The YfiC Gene of E. Coli Encodes an Adenine-N6 Methyltransferase That Specifically Modifies A37 of TRNA1Val(Cmo5UAC). RNA 2009, 15, 1134–1141. [Google Scholar] [CrossRef] [Green Version]
- Pollo-Oliveira, L.; de Crécy-Lagard, V. Can Protein Expression Be Regulated by Modulation of TRNA Modification Profiles? Biochemistry 2019, 58, 355–362. [Google Scholar] [CrossRef]
- Torrent, M.; Chalancon, G.; de Groot, N.S.; Wuster, A.; Madan Babu, M. Cells Alter Their TRNA Abundance to Selectively Regulate Protein Synthesis during Stress Conditions. Sci. Signal. 2018, 11, eaat6409. [Google Scholar] [CrossRef] [Green Version]
- Björk, G.R.; Hagervall, T.G. Transfer RNA Modification: Presence, Synthesis, and Function. EcoSal Plus 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.R.; Keffer-Wilkes, L.C.; Dobing, S.R.; Kothe, U. Pre-Steady-State Kinetic Analysis of the Three Escherichia Coli Pseudouridine Synthases TruB, TruA, and RluA Reveals Uniformly Slow Catalysis. RNA 2011, 17, 2074–2084. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.-M.; Matsubara, R.; Takase, R.; Masuda, I.; Sulkowska, J.I. TrmD. In The Enzymes; Elsevier: Amsterdam, The Netherlands, 2017; Volume 41, pp. 89–115. ISBN 978-0-12-811777-4. [Google Scholar]
- Jaroensuk, J.; Atichartpongkul, S.; Chionh, Y.H.; Wong, Y.H.; Liew, C.W.; McBee, M.E.; Thongdee, N.; Prestwich, E.G.; DeMott, M.S.; Mongkolsuk, S.; et al. Methylation at Position 32 of TRNA Catalyzed by TrmJ Alters Oxidative Stress Response in Pseudomonas aeruginosa. Nucleic Acids Res. 2016, 44, 10834–10848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thongdee, N.; Jaroensuk, J.; Atichartpongkul, S.; Chittrakanwong, J.; Chooyoung, K.; Srimahaeak, T.; Chaiyen, P.; Vattanaviboon, P.; Mongkolsuk, S.; Fuangthong, M. TrmB, a TRNA M7G46 Methyltransferase, Plays a Role in Hydrogen Peroxide Resistance and Positively Modulates the Translation of KatA and KatB MRNAs in Pseudomonas Aeruginosa. Nucleic Acids Res. 2019, 47, 9271–9281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Čavužić, M.; Liu, Y. Biosynthesis of Sulfur-Containing TRNA Modifications: A Comparison of Bacterial, Archaeal, and Eukaryotic Pathways. Biomolecules 2017, 7, 27. [Google Scholar] [CrossRef] [Green Version]
- Bou-Nader, C.; Montémont, H.; Guérineau, V.; Jean-Jean, O.; Brégeon, D.; Hamdane, D. Unveiling Structural and Functional Divergences of Bacterial TRNA Dihydrouridine Synthases: Perspectives on the Evolution Scenario. Nucleic Acids Res. 2018, 46, 1386–1394. [Google Scholar] [CrossRef] [Green Version]
- Thiaville, P.C.; El Yacoubi, B.; Köhrer, C.; Thiaville, J.J.; Deutsch, C.; Iwata-Reuyl, D.; Bacusmo, J.M.; Armengaud, J.; Bessho, Y.; Wetzel, C.; et al. Essentiality of Threonylcarbamoyladenosine (t6A), a Universal TRNA Modification, in Bacteria: T6A Essentiality. Mol. Microbiol. 2015, 98, 1199–1221. [Google Scholar] [CrossRef] [Green Version]
- Grosjean, H.; Gaspin, C.; Marck, C.; Decatur, W.A.; de Crécy-Lagard, V. RNomics and Modomics in the Halophilic Archaea Haloferax Volcanii: Identification of RNA Modification Genes. BMC Genom. 2008, 9, 470. [Google Scholar] [CrossRef] [Green Version]
- Ikeuchi, Y.; Kitahara, K.; Suzuki, T. The RNA Acetyltransferase Driven by ATP Hydrolysis Synthesizes N4-Acetylcytidine of TRNA Anticodon. EMBO J. 2008, 27, 2194–2203. [Google Scholar] [CrossRef] [Green Version]
- Matuszek, Z.; Pan, T. Quantification of Queuosine Modification Levels in TRNA from Human Cells Using APB Gel and Northern Blot. Bio-Protoc. 2019, 9, e3191. [Google Scholar] [CrossRef]
- Nie, W.; Wang, S.; He, R.; Xu, Q.; Wang, P.; Wu, Y.; Tian, F.; Yuan, J.; Zhu, B.; Chen, G. A-to-I RNA Editing in Bacteria Increases Pathogenicity and Tolerance to Oxidative Stress. PLoS Pathog. 2020, 16, e1008740. [Google Scholar] [CrossRef] [PubMed]
- Takakura, M.; Ishiguro, K.; Akichika, S.; Miyauchi, K.; Suzuki, T. Biogenesis and Functions of Aminocarboxypropyluridine in TRNA. Nat. Commun. 2019, 10, 5542. [Google Scholar] [CrossRef] [Green Version]
- Reichle, V.F.; Petrov, D.P.; Weber, V.; Jung, K.; Kellner, S. NAIL-MS Reveals the Repair of 2-Methylthiocytidine by AlkB in E. Coli. Nat. Commun. 2019, 10, 5600. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated TRNA Demethylation Regulates Translation. Cell 2016, 167, 816–828.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, K.; Jäger, G.; Björk, G.R. An Unmodified Wobble Uridine in TRNAs Specific for Glutamine, Lysine, and Glutamic Acid from Salmonella Enterica Serovar Typhimurium Results in Nonviability—Due to Increased Missense Errors? PLoS ONE 2017, 12, e0175092. [Google Scholar] [CrossRef] [Green Version]
- Shippy, D.C.; Fadl, A.A. TRNA Modification Enzymes GidA and MnmE: Potential Role in Virulence of Bacterial Pathogens. Int. J. Mol. Sci. 2014, 15, 18267–18280. [Google Scholar] [CrossRef] [Green Version]
- Rehl, J.M.; Shippy, D.C.; Eakley, N.M.; Brevik, M.D.; Sand, J.M.; Cook, M.E.; Fadl, A.A. GidA Expression in Salmonella Is Modulated under Certain Environmental Conditions. Curr. Microbiol. 2013, 67, 279–285. [Google Scholar] [CrossRef]
- Shippy, D.C.; Eakley, N.M.; Bochsler, P.N.; Fadl, A.A. Biological and Virulence Characteristics of Salmonella Enterica Serovar Typhimurium Following Deletion of Glucose-Inhibited Division (GidA) Gene. Microb. Pathog. 2011, 50, 303–313. [Google Scholar] [CrossRef]
- Shippy, D.C.; Eakley, N.M.; Lauhon, C.T.; Bochsler, P.N.; Fadl, A.A. Virulence Characteristics of Salmonella Following Deletion of Genes Encoding the TRNA Modification Enzymes GidA and MnmE. Microb. Pathog. 2013, 57, 1–9. [Google Scholar] [CrossRef]
- Yu, H.; Kim, K.S. MRNA Context Dependent Regulation of Cytotoxic Necrotizing Factor 1 Translation by GidA, a TRNA Modification Enzyme in Escherichia Coli. Gene 2012, 491, 116–122. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Shibata, Y.; Takeshita, T.; Yamashita, Y. A Novel Gene Involved in the Survival of Streptococcus Mutans under Stress Conditions. Appl. Environ. Microbiol. 2014, 80, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.H.; Caparon, M.G. TRNA Modification by GidA/MnmE Is Necessary for Streptococcus Pyogenes Virulence: A New Strategy to Make Live Attenuated Strains. Infect. Immun. 2008, 76, 3176–3186. [Google Scholar] [CrossRef] [Green Version]
- Sha, J.; Kozlova, E.V.; Fadl, A.A.; Olano, J.P.; Houston, C.W.; Peterson, J.W.; Chopra, A.K. Molecular Characterization of a Glucose-Inhibited Division Gene, GidA, That Regulates Cytotoxic Enterotoxin of Aeromonas Hydrophila. Infect. Immun. 2004, 72, 1084–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durand, J.M.; Björk, G.R.; Kuwae, A.; Yoshikawa, M.; Sasakawa, C. The Modified Nucleoside 2-Methylthio-N6-Isopentenyladenosine in TRNA of Shigella Flexneri Is Required for Expression of Virulence Genes. J. Bacteriol. 1997, 179, 5777–5782. [Google Scholar] [CrossRef] [Green Version]
- Durand, J.M.; Dagberg, B.; Uhlin, B.E.; Björk, G.R. Transfer RNA Modification, Temperature and DNA Superhelicity Have a Common Target in the Regulatory Network of the Virulence of Shigella Flexneri: The Expression of the VirF Gene. Mol. Microbiol. 2000, 35, 924–935. [Google Scholar] [CrossRef] [Green Version]
- Romsang, A.; Duang-nkern, J.; Khemsom, K.; Wongsaroj, L.; Saninjuk, K.; Fuangthong, M.; Vattanaviboon, P.; Mongkolsuk, S. Pseudomonas Aeruginosa TtcA Encoding TRNA-Thiolating Protein Requires an Iron-Sulfur Cluster to Participate in Hydrogen Peroxide-Mediated Stress Protection and Pathogenicity. Sci. Rep. 2018, 8, 11882. [Google Scholar] [CrossRef] [PubMed]
- Nakayashiki, T.; Saito, N.; Takeuchi, R.; Kadokura, H.; Nakahigashi, K.; Wanner, B.L.; Mori, H. The TRNA Thiolation Pathway Modulates the Intracellular Redox State in Escherichia Coli. J. Bacteriol. 2013, 195, 2039–2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamper, H.B.; Masuda, I.; Frenkel-Morgenstern, M.; Hou, Y.-M. Maintenance of Protein Synthesis Reading Frame by EF-P and M1G37-TRNA. Nat. Commun. 2015, 6, 7226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamper, H.; Masuda, I.; Frenkel-Morgenstern, M.; Hou, Y.-M. The UGG Isoacceptor of TRNAPro Is Naturally Prone to Frameshifts. Int. J. Mol. Sci. 2015, 16, 14866–14883. [Google Scholar] [CrossRef] [PubMed]
- Masuda, I.; Matsubara, R.; Christian, T.; Rojas, E.R.; Yadavalli, S.S.; Zhang, L.; Goulian, M.; Foster, L.J.; Huang, K.C.; Hou, Y.-M. TRNA Methylation Is a Global Determinant of Bacterial Multi-Drug Resistance. Cell Syst. 2019, 8, 302–314.e8. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.; Masuda, I.; Foster, L.J. tRNA Methylation: An Unexpected Link to Bacterial Resistance and Persistence to Antibiotics and Beyond. WIREs RNA 2020, 11, e1609. [Google Scholar] [CrossRef]
- Kinghorn, S.M.; O’Byrne, C.P.; Booth, I.R.; Stansfield, I. Physiological Analysis of the Role of TruB in Escherichia Coli: A Role for TRNA Modification in Extreme Temperature Resistance. Microbiology 2002, 148, 3511–3520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, K.; Kunibayashi, T.; Tomikawa, C.; Ochi, A.; Kanai, T.; Hirata, A.; Iwashita, C.; Hori, H. Pseudouridine at Position 55 in TRNA Controls the Contents of Other Modified Nucleotides for Low-Temperature Adaptation in the Extreme-Thermophilic Eubacterium Thermus Thermophilus. Nucleic Acids Res. 2011, 39, 2304–2318. [Google Scholar] [CrossRef] [Green Version]
- Droogmans, L. Cloning and Characterization of TRNA (M1A58) Methyltransferase (TrmI) from Thermus Thermophilus HB27, a Protein Required for Cell Growth at Extreme Temperatures. Nucleic Acids Res. 2003, 31, 2148–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomikawa, C.; Yokogawa, T.; Kanai, T.; Hori, H. N7-Methylguanine at Position 46 (M7G46) in TRNA from Thermus Thermophilus Is Required for Cell Viability at High Temperatures through a TRNA Modification Network. Nucleic Acids Res. 2010, 38, 942–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigi, N.; Sakaguchi, Y.; Suzuki, T.; Watanabe, K. Identification of Two TRNA Thiolation Genes Required for Cell Growth at Extremely High Temperatures. J. Biol. Chem. 2006, 281, 14296–14306. [Google Scholar] [CrossRef] [Green Version]
- Grosjean, H.; Droogmans, L.; Roovers, M.; Keith, G. Detection of Enzymatic Activity of Transfer RNA Modification Enzymes Using Radiolabeled tRNA Substrates. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2007; Volume 425, pp. 55–101. ISBN 978-0-12-374155-4. [Google Scholar]
- Motorin, Y.; Marchand, V. Analysis of RNA Modifications by Second- and Third-Generation Deep Sequencing: 2020 Update. Genes 2021, 12, 278. [Google Scholar] [CrossRef]
- Yan, T.-M.; Pan, Y.; Yu, M.-L.; Hu, K.; Cao, K.-Y.; Jiang, Z.-H. Full-Range Profiling of TRNA Modifications Using LC–MS/MS at Single-Base Resolution through a Site-Specific Cleavage Strategy. Anal. Chem. 2021, 93, 1423–1432. [Google Scholar] [CrossRef]
- Schwartz, M.H.; Wang, H.; Pan, J.N.; Clark, W.C.; Cui, S.; Eckwahl, M.J.; Pan, D.W.; Parisien, M.; Owens, S.M.; Cheng, B.L.; et al. Microbiome Characterization by High-Throughput Transfer RNA Sequencing and Modification Analysis. Nat. Commun. 2018, 9, 5353. [Google Scholar] [CrossRef]
- Gogakos, T.; Brown, M.; Garzia, A.; Meyer, C.; Hafner, M.; Tuschl, T. Characterizing Expression and Processing of Precursor and Mature Human TRNAs by Hydro-TRNAseq and PAR-CLIP. Cell Rep. 2017, 20, 1463–1475. [Google Scholar] [CrossRef] [Green Version]
- Behrens, A.; Rodschinka, G.; Nedialkova, D.D. High-Resolution Quantitative Profiling of TRNA Abundance and Modification Status in Eukaryotes by Mim-TRNAseq. Mol. Cell 2021, 81, 1802–1815.e7. [Google Scholar] [CrossRef] [PubMed]
- Thomas, N.; Poodari, V.; Jain, M.; Olsen, H.; Akeson, M.; Abu-Shumays, R. Direct Nanopore Sequencing of Individual Full Length TRNA Strands. BioRxiv 2021. [Google Scholar] [CrossRef]
- Su, D.; Chan, C.T.Y.; Gu, C.; Lim, K.S.; Chionh, Y.H.; McBee, M.E.; Russell, B.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Quantitative Analysis of Ribonucleoside Modifications in TRNA by HPLC-Coupled Mass Spectrometry. Nat. Protoc. 2014, 9, 828–841. [Google Scholar] [CrossRef]
- Meyer, B.; Immer, C.; Kaiser, S.; Sharma, S.; Yang, J.; Watzinger, P.; Weiß, L.; Kotter, A.; Helm, M.; Seitz, H.-M.; et al. Identification of the 3-Amino-3-Carboxypropyl (Acp) Transferase Enzyme Responsible for Acp3U Formation at Position 47 in Escherichia Coli TRNAs. Nucleic Acids Res. 2020, 48, 1435–1450. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Dedon, P.C.; Waldor, M.K. Comparative TRNA Sequencing and RNA Mass Spectrometry for Surveying TRNA Modifications. Nat. Chem. Biol. 2020, 16, 964–972. [Google Scholar] [CrossRef]
- Antoine, L.; Wolff, P.; Westhof, E.; Romby, P.; Marzi, S. Mapping Post-Transcriptional Modifications in Staphylococcus Aureus TRNAs by NanoLC/MSMS. Biochimie 2019, 164, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Su, Y.; Zhang, X.; Turkel, S.J.; Shi, S.; Wang, X.; Choi, E.-J.; Wu, W.; Liu, H.; Viner, R.; et al. MLC Seq: De Novo Sequencing of Full-Length TRNA Isoforms by Mass Ladder Complementation. BioRxiv 2021. [Google Scholar] [CrossRef]
- Goto-Ito, S.; Ito, T.; Yokoyama, S. Trm5 and TrmD: Two Enzymes from Distinct Origins Catalyze the Identical TRNA Modification, M1G37. Biomolecules 2017, 7, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, W.; Pasunooti, K.K.; Balamkundu, S.; Wong, Y.H.; Nah, Q.; Gadi, V.; Gnanakalai, S.; Chionh, Y.H.; McBee, M.E.; Gopal, P.; et al. Thienopyrimidinone Derivatives That Inhibit Bacterial TRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism. J. Med. Chem. 2019, 62, 7788–7805. [Google Scholar] [CrossRef] [Green Version]
- Zhong, W.; Koay, A.; Ngo, A.; Li, Y.; Nah, Q.; Wong, Y.H.; Chionh, Y.H.; Ng, H.Q.; Koh-Stenta, X.; Poulsen, A.; et al. Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel TRNA-(N1 G37) Methyltransferase (TrmD) Inhibitors. ACS Infect. Dis. 2019, 5, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; Iwata-Reuyl, D.; de Crécy-Lagard, V. Diversity of the Biosynthesis Pathway for Threonylcarbamoyladenosine (t6A), a Universal Modification of TRNA. RNA Biol. 2014, 11, 1529–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Modification | tRNA | Writer | Organism | Function | Reference |
|---|---|---|---|---|---|
| (c)mnm5s2U34 | tRNAGln/tRNALys/tRNAGlu | GidA/MnmE | S. enterica |
| [58,59,60,61] |
| E. coli |
| [63] | |||
| S. mutans |
| [64] | |||
| tRNALys/tRNALeu/tRNAArg | S. pyogenes |
| [65] | ||
| A. hydrophila |
| [66] | |||
| cmo5U34 | tRNAThr(UGU) | CmoB | M. bovis BCG |
| [27] |
| Q34 | tRNATyr/tRNAHis/tRNAAsp/tRNAAsn | Tgt | S. flexneri |
| [67] |
| ms2i6A37 | tRNAAxx | MiaA | S. flexneri |
| [67,68] |
| m6A37 | tRNA1Val | YfiC | E. coli |
| [40] |
| Cm32 Um32 Am32 | tRNAMet tRNATrp tRNAGln/tRNAPro/tRNAHis tRNAGln/tRNAPro | TrmJ | P. aeruginosa |
| [46] |
| m7G46 | tRNAPhe/tRNAAsp | TrmB | P. aeruginosa |
| [47] |
| s2C32 | TtcA | P. aeruginosa |
| [69] | |
| m1G37 | tRNAPro | TrmD | E. coli/S. enterica |
| [70,71,72,73,74] |
| Ψ55 | Universal | TruB | E. coli |
| [75] |
| Universal | T. thermophilus |
| [76] | ||
| m1A58 | tRNAAsp | TrmI | T. thermophilus |
| [77] |
| m7G46 | tRNAPhe/tRNAIle | TrmB | T. thermophilus |
| [78] |
| s2T54 | TtuA/TtuB | T. thermophilus |
| [79] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kouvela, A.; Zaravinos, A.; Stamatopoulou, V. Adaptor Molecules Epitranscriptome Reprograms Bacterial Pathogenicity. Int. J. Mol. Sci. 2021, 22, 8409. https://doi.org/10.3390/ijms22168409
Kouvela A, Zaravinos A, Stamatopoulou V. Adaptor Molecules Epitranscriptome Reprograms Bacterial Pathogenicity. International Journal of Molecular Sciences. 2021; 22(16):8409. https://doi.org/10.3390/ijms22168409
Chicago/Turabian StyleKouvela, Adamantia, Apostolos Zaravinos, and Vassiliki Stamatopoulou. 2021. "Adaptor Molecules Epitranscriptome Reprograms Bacterial Pathogenicity" International Journal of Molecular Sciences 22, no. 16: 8409. https://doi.org/10.3390/ijms22168409
APA StyleKouvela, A., Zaravinos, A., & Stamatopoulou, V. (2021). Adaptor Molecules Epitranscriptome Reprograms Bacterial Pathogenicity. International Journal of Molecular Sciences, 22(16), 8409. https://doi.org/10.3390/ijms22168409







