Mechanisms of Viscous Media Effects on Elementary Steps of Bacterial Bioluminescent Reaction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Experimental Procedures
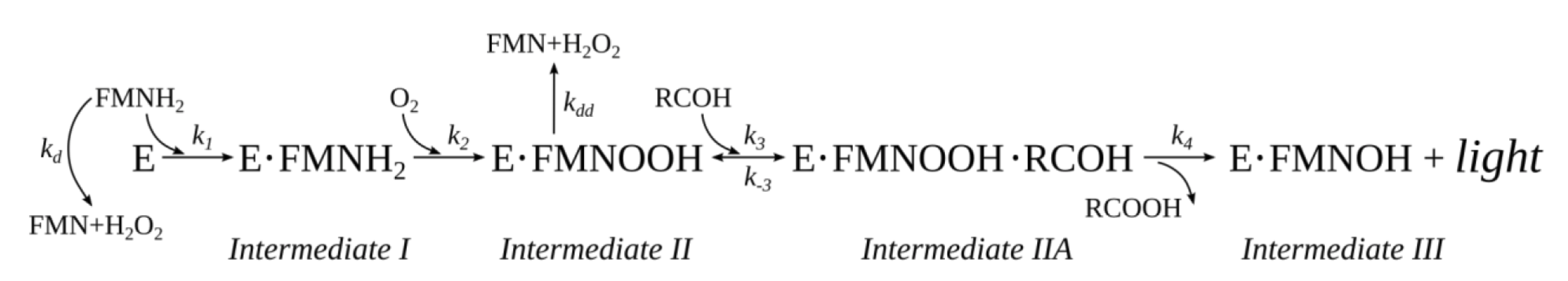
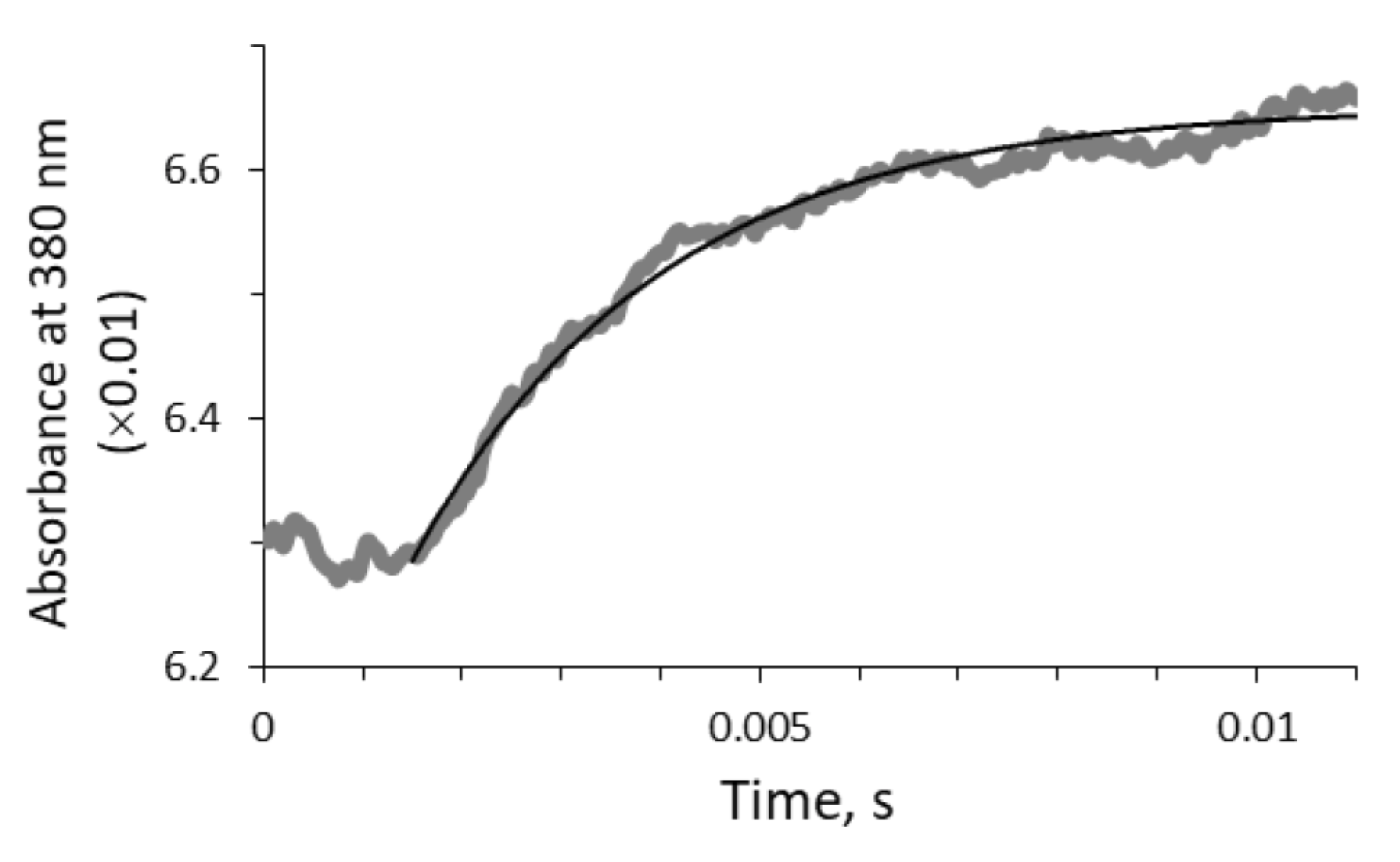
2.3. Mathematical Modelling of the Reaction Kinetics
2.4. Data Analysis
2.5. Computational Analysis of Bacterial Luciferase—Cosolvents Interactions
3. Results
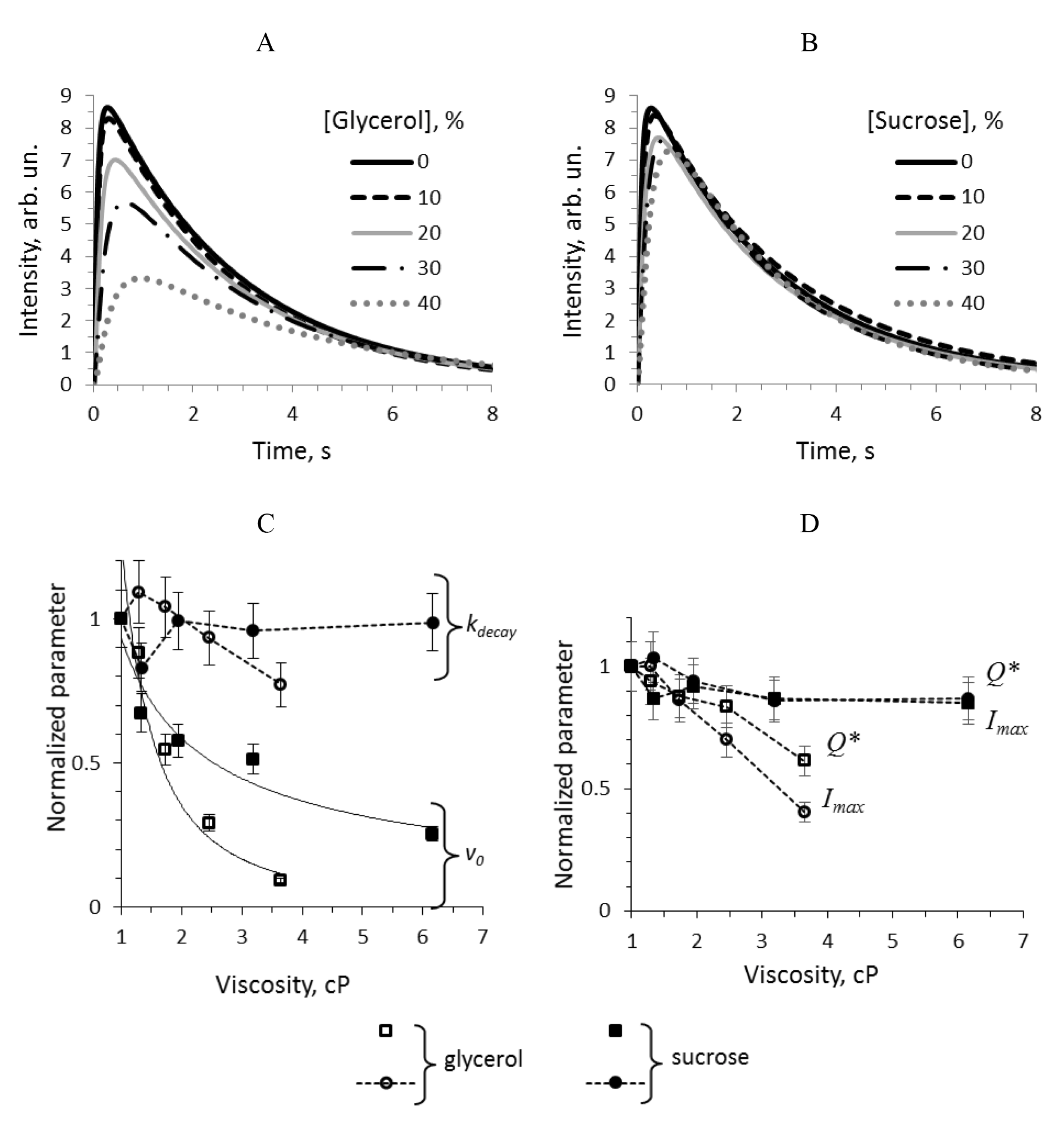
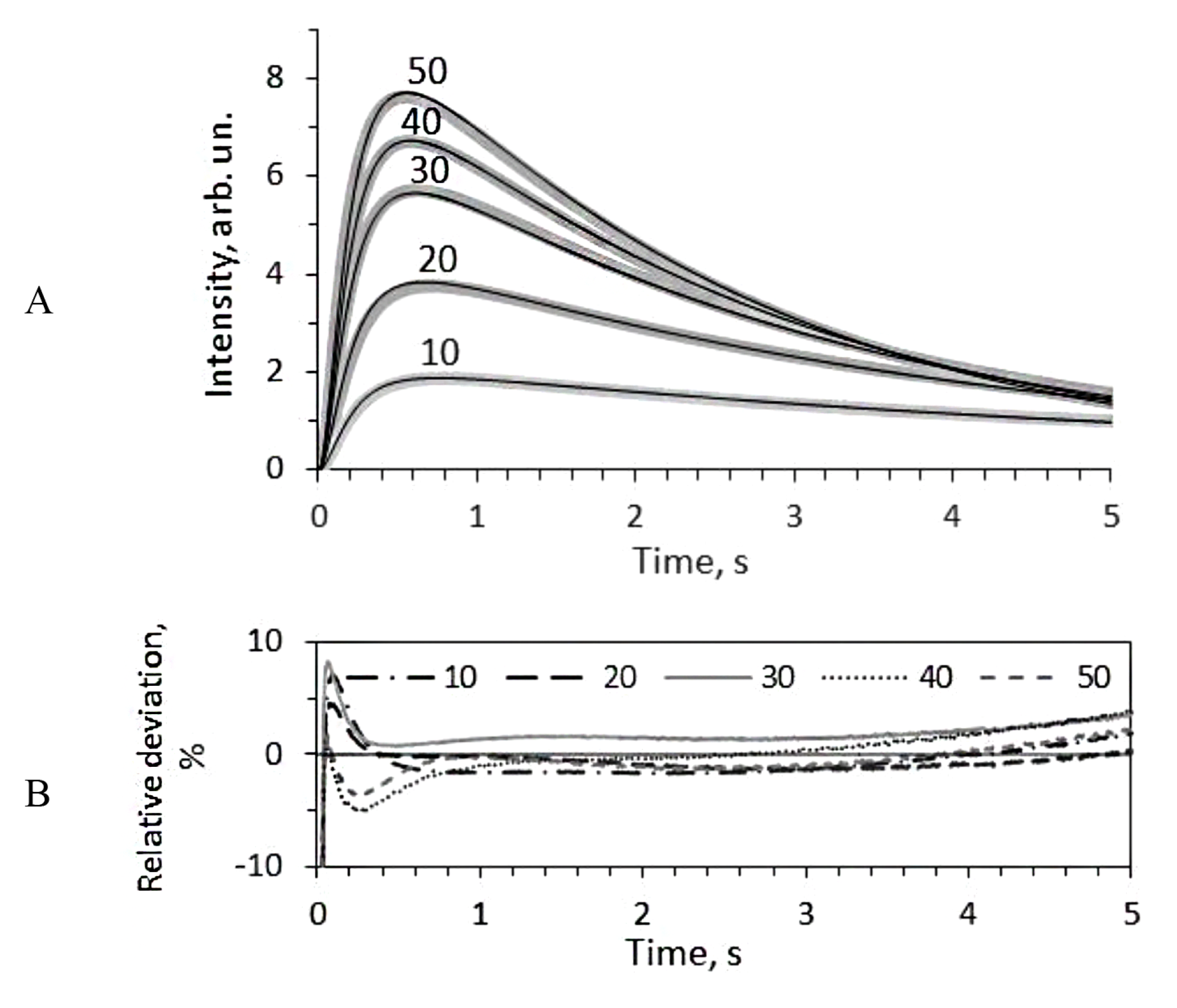
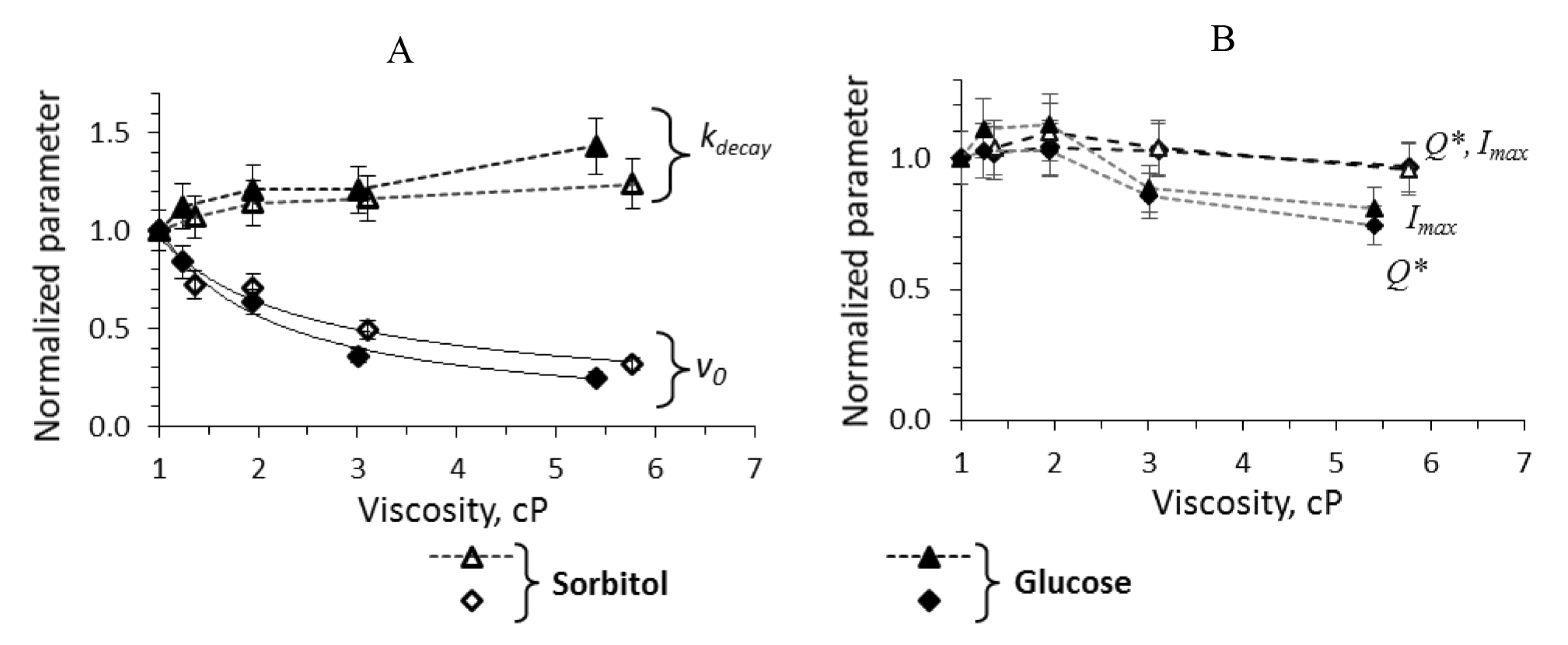
3.1. Kinetic Effects of Media with Glycerol and Sucrose on the Bioluminescent Reaction Catalyzed by Bacterial Luciferase
3.2. Mathematical Modeling of the Kinetics of the Bacterial Bioluminescent Reaction
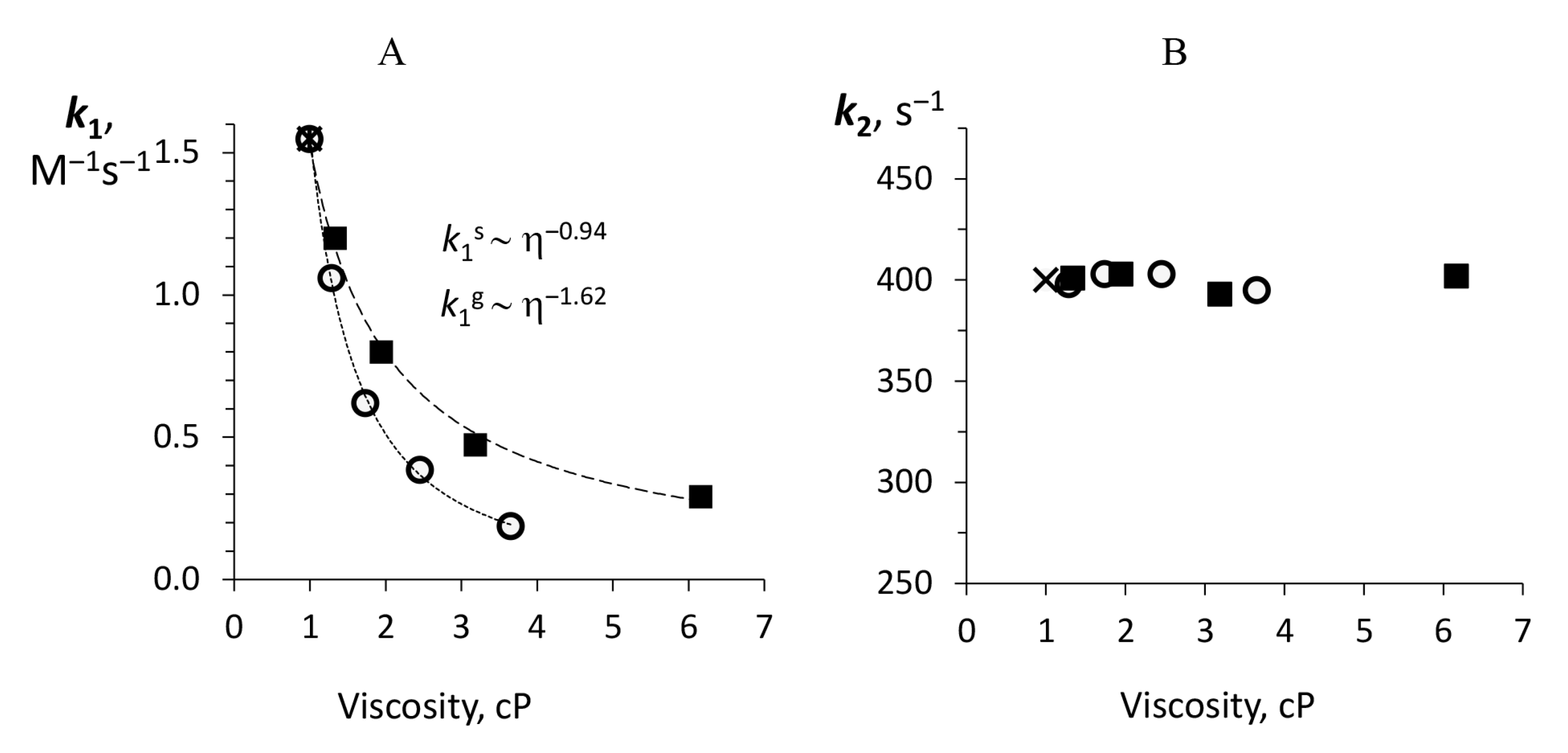
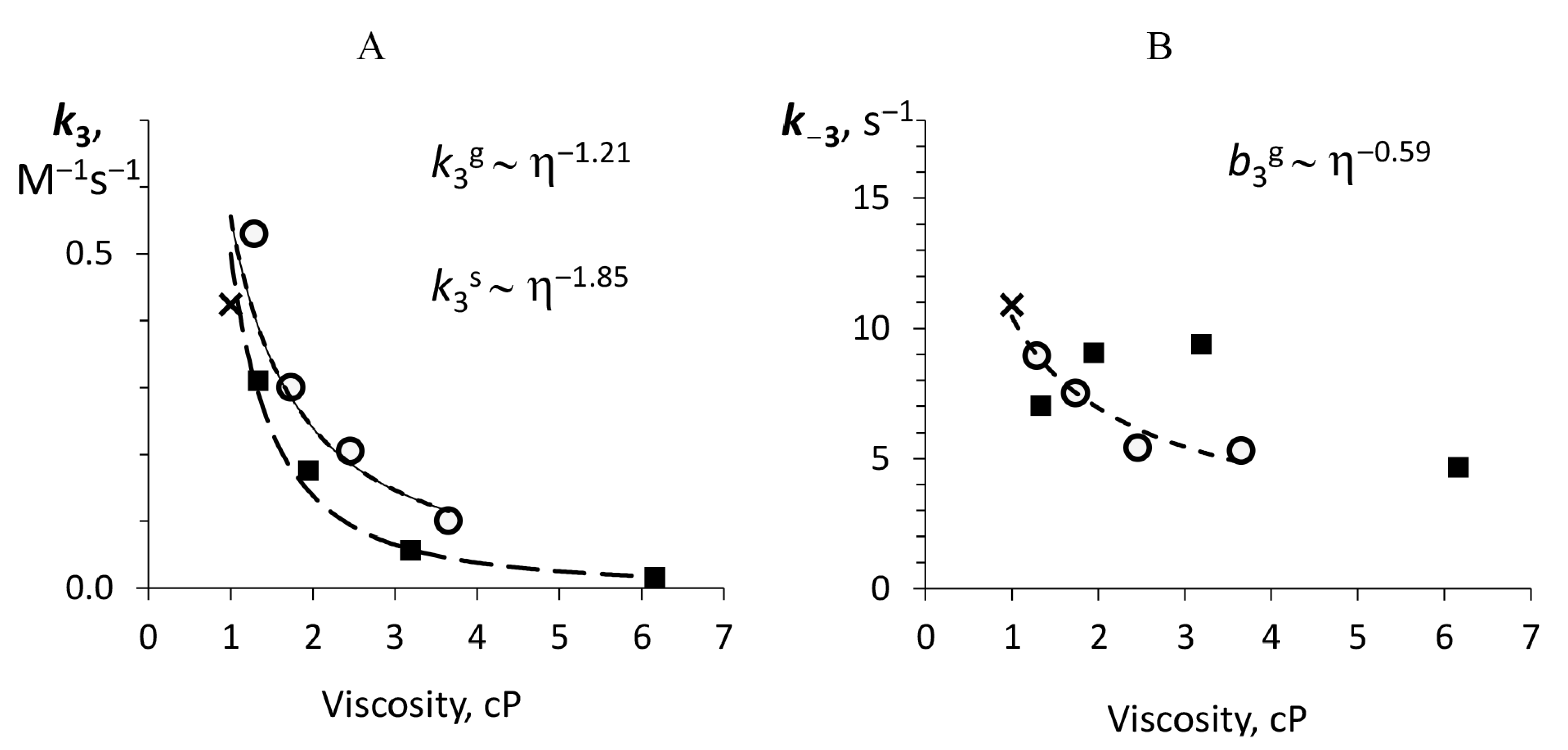
3.3. Dependence of the Individual Rate Constants on Media Viscosity
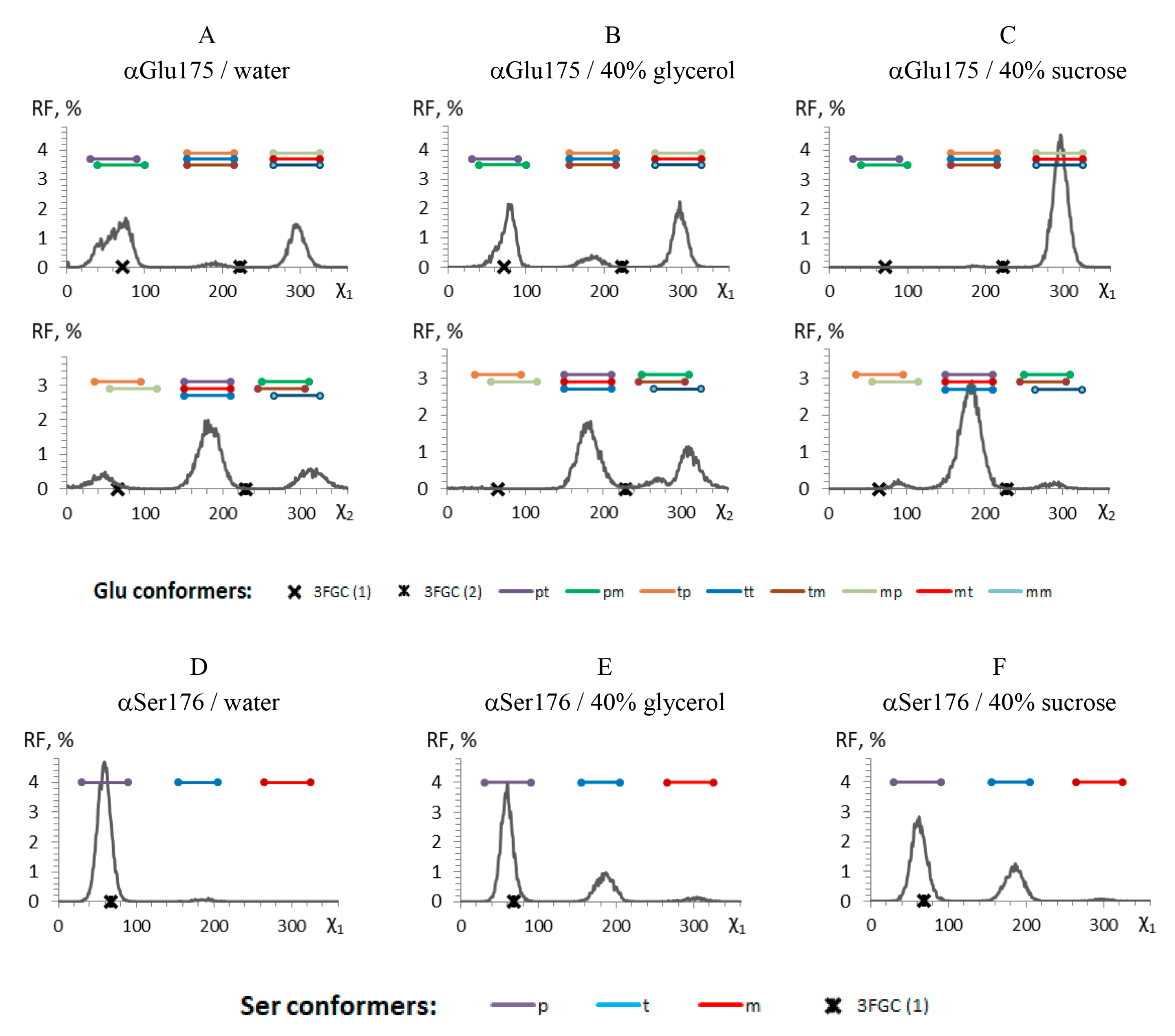
3.4. Effects of Glycerol and Sucrose Molecules on Bacterial Luciferase Structure
3.5. The Dependence of Luciferase Kinetics on the Molecular Size of Viscogenic Cosolvent
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Balcells, C.; Pastor, I.; Pitulice, L.; Hernández, C.; Via, M.; Garcés, J.L.; Madurga, S.; Vilaseca, E.; Isvoran, A.; Cascante, M.; et al. Macromolecular crowding upon in-vivo-like enzyme-kinetics: Effect of enzyme-obstacle size ratio. New Front. Chem. 2015, 24, 3. [Google Scholar]
- Schavemaker, P.E.; Boersma, A.J.; Poolman, B. How important is protein diffusion in prokaryotes? Front. Mol. Biosci. 2018, 5, 93. [Google Scholar] [CrossRef] [PubMed]
- Rivas, G.; Minton, A.P. Biochemical Reactions in Cytomimetic Media. Front. Mol. Biosci. 2020, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Shimolina, L.E.; Izquierdo, M.A.; López-Duarte, I.; Bull, J.A.; Shirmanova, M.V.; Klapshina, L.G.; Zagaynova, E.V.; Kuimova, M.K. Imaging tumor microscopic viscosity in vivo using molecular rotors. Sci. Rep. 2017, 7, 41097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Persson, L.B.; Ambati, V.S.; Brandman, O. Cellular control of viscosity counters changes in temperature and energy availability. Cell 2020, 183, 1572–1585. [Google Scholar] [CrossRef]
- Lim, W.A.; Raines, R.T.; Knowles, J.R. Triosephosphate isomerase catalysis is diffusion controlled. Appendix: Analysis of triose phosphate equilibria in aqueous solution by phosphorus-31 NMR. Biochemistry 1988, 27, 1165–1167. [Google Scholar] [CrossRef]
- Vagenende, V.; Yap, M.G.; Trout, B.L. Molecular anatomy of preferential interaction coefficients by elucidating protein solvation in mixed solvents: Methodology and application for lysozyme in aqueous glycerol. J. Phys. Chem. B 2009, 113, 11743–11753. [Google Scholar] [CrossRef]
- Saunders, A.J.; Davis-Searles, P.R.; Allen, D.L.; Pielak, G.J.; Erie, D.A. Osmolyte-induced changes in protein conformational equilibria. Biopolym. Orig. Res. Biomol. 2000, 53, 293–307. [Google Scholar] [CrossRef]
- Gadda, G.; Sobrado, P. Kinetic solvent viscosity effects as probes for studying the mechanisms of enzyme action. Biochemistry 2018, 57, 3445–3453. [Google Scholar] [CrossRef]
- Nakatani, H.; Dunford, H. Meaning of diffusion-controlled association rate constants in enzymology. J. Phys. Chem. 1979, 83, 2662–2665. [Google Scholar] [CrossRef]
- Verma, R.; Mitchell-Koch, K. In Silico studies of small molecule interactions with enzymes reveal aspects of catalytic function. Catalysts 2017, 7, 212. [Google Scholar] [CrossRef] [Green Version]
- Masson, P.; Lushchekina, S.; Schopfer, L.M.; Lockridge, O. Effects of viscosity and osmotic stress on the reaction of human butyrylcholinesterase with cresyl saligenin phosphate, a toxicant related to aerotoxic syndrome: Kinetic and molecular dynamics studies. Biochem. J. 2013, 454, 387–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loose, M.; Kruse, K.; Schwille, P. Protein self-organization: Lessons from the min system. Annu. Rev. Biophys. 2011, 40, 315–336. [Google Scholar] [CrossRef] [PubMed]
- Lipkow, K.; Odde, D.J. Model for protein concentration gradients in the cytoplasm. Cell. Mol. Bioeng. 2008, 1, 84–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sashi, P.; Bhuyan, A.K. Viscosity dependence of some protein and enzyme reaction rates: Seventy-five years after Kramers. Biochemistry 2015, 54, 4453–4461. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.; Hastings, J.W. Bioluminescence; Harvard University Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Haddock, S.H.; Moline, M.A.; Case, J.F. Bioluminescence in the sea. Annu. Rev. Mar. Sci. 2010, 2, 443–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J. Perspectives on bioluminescence mechanisms. Photochem. Photobiol. 2017, 93, 389–404. [Google Scholar] [CrossRef] [PubMed]
- Brodl, E.; Winkler, A.; Macheroux, P. Molecular mechanisms of bacterial bioluminescence. Comput. Struct. Biotechnol. J. 2018, 16, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, P. Biochemistry and genetics of bacterial bioluminescence. In Bioluminescence: Fundamentals and Applications in Biotechnology-Volume 1; Springer: Berlin/Heidelberg, Germany, 2014; pp. 37–64. [Google Scholar]
- Vannier, T.; Hingamp, P.; Turrel, F.; Tanet, L.; Lescot, M.; Timsit, Y. Diversity and evolution of bacterial bioluminescence genes in the global ocean. NAR Genom. Bioinform. 2020, 2, lqaa018. [Google Scholar] [CrossRef] [Green Version]
- Abu-Soud, H.; Mullins, L.S.; Baldwin, T.O.; Raushel, F.M. Stopped-flow kinetic analysis of the bacterial luciferase reaction. Biochemistry 1992, 31, 3807–3813. [Google Scholar] [CrossRef]
- Lei, B.; Ding, Q.; Tu, S.C. Identity of the emitter in the bacterial luciferase luminescence reaction: Binding and fluorescence quantum yield studies of 5-decyl-4a-hydroxy-4a, 5-dihydroriboflavin-5′-phosphate as a model. Biochemistry 2004, 43, 15975–15982. [Google Scholar] [CrossRef] [PubMed]
- A Kratasyuk, V.; N Esimbekova, E. Applications of luminous bacteria enzymes in toxicology. Comb. Chem. High Throughput Screen. 2015, 18, 952–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esimbekova, E.; Kondik, A.; Kratasyuk, V. Bioluminescent enzymatic rapid assay of water integral toxicity. Environ. Monit. Assess. 2013, 185, 5909–5916. [Google Scholar] [CrossRef] [PubMed]
- Gregor, C.; Gwosch, K.C.; Sahl, S.J.; Hell, S.W. Strongly enhanced bacterial bioluminescence with the ilux operon for single-cell imaging. Proc. Natl. Acad. Sci. USA 2018, 115, 962–967. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Müller, F.; Visser, A.J. The sensitized bioluminescence mechanism of bacterial luciferase. Photochem. Photobiol. 2019, 95, 679–704. [Google Scholar] [CrossRef] [Green Version]
- Lisitsa, A.; Sukovatyi, L.; Kratasyuk, V.; Nemtseva, E. Viscous media slow down the decay of the key intermediate in bacterial bioluminescent reaction. Dokl. Biochem. Biophys. 2020, 492, 162–165. [Google Scholar] [CrossRef]
- Campbell, Z.T.; Weichsel, A.; Montfort, W.R.; Baldwin, T.O. Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the β subunit. Biochemistry 2009, 48, 6085–6094. [Google Scholar] [CrossRef]
- Sukovatyi, L.; Lisitsa, A.; Kratasyuk, V.; Nemtseva, E. The Effect of Osmolytes on the Bioluminescent Reaction of Bacteria: Structural and Dynamic Properties. Biophysics 2020, 65, 966–971. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- Grant, B.J.; Rodrigues, A.P.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef] [Green Version]
- RC Team. R: A Language and Environment for Statistical Computing; RC Team: Vienna, Austria, 2013. [Google Scholar]
- Haddad, Y.; Adam, V.; Heger, Z. Rotamer dynamics: Analysis of rotamers in molecular dynamics simulations of proteins. Biophys. J. 2019, 116, 2062–2072. [Google Scholar] [CrossRef] [PubMed]
- Abu-Soud, H.M.; Clark, A.C.; Francisco, W.A.; Baldwin, T.O.; Raushel, F.M. Kinetic destabilization of the hydroperoxy flavin intermediate by site-directed modification of the reactive thiol in bacterial luciferase. J. Biol. Chem. 1993, 268, 7699–7706. [Google Scholar] [CrossRef]
- Tinikul, R.; Pitsawong, W.; Sucharitakul, J.; Nijvipakul, S.; Ballou, D.P.; Chaiyen, P. The transfer of reduced flavin mononucleotide from LuxG oxidoreductase to luciferase occurs via free diffusion. Biochemistry 2013, 52, 6834–6843. [Google Scholar] [CrossRef]
- Suadee, C.; Nijvipakul, S.; Svasti, J.; Entsch, B.; Ballou, D.P.; Chaiyen, P. Luciferase from Vibrio campbellii is more thermostable and binds reduced FMN better than its homologues. J. Biochem. 2007, 142, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Tinikul, R.; Chaiyen, P. Structure, mechanism, and mutation of bacterial luciferase. In Bioluminescence: Fundamentals and Applications in Biotechnology; Springer: Berlin/Heidelberg, Germany, 2014; Volume 3, pp. 47–74. [Google Scholar]
- Lovell, S.C.; Word, J.M.; Richardson, J.S.; Richardson, D.C. The penultimate rotamer library. Proteins Struct. Funct. Bioinform. 2000, 40, 389–408. [Google Scholar] [CrossRef]
- Moore, C.; Lei, B.; Tu, S.C. Relationship between the conserved α subunit arginine 107 and effects of phosphate on the activity and stability of Vibrio harveyi luciferase. Arch. Biochem. Biophys. 1999, 370, 45–50. [Google Scholar] [CrossRef]
- Tang, Y.Q.; Luo, Y.; Liu, Y.J. Theoretical Study on Role of Aliphatic Aldehyde in Bacterial Bioluminescence. J. Photochem. Photobiol. A Chem. 2021, 419, 113446. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Telis, V.R.N.; Telis-Romero, J.; Mazzotti, H.; Gabas, A.L. Viscosity of aqueous carbohydrate solutions at different temperatures and concentrations. Int. J. Food Prop. 2007, 10, 185–195. [Google Scholar] [CrossRef]
- Zhu, C.; Ma, Y.; Zhou, C. Densities and viscosities of sugar alcohol aqueous solutions. J. Chem. Eng. Data 2010, 55, 3882–3885. [Google Scholar] [CrossRef]
- Chavez, A.L.; Birch, G.G. The hydrostatic and hydrodynamic volumes of polyols in aqueous solutions and their sweet taste. Chem. Senses 1997, 22, 149–161. [Google Scholar] [CrossRef] [Green Version]
- Bárcena-Uribarri, I.; Thein, M.; Maier, E.; Bonde, M.; Bergström, S.; Benz, R. Use of nonelectrolytes reveals the channel size and oligomeric constitution of the Borrelia burgdorferi P66 porin. PLoS ONE 2013, 8, e78272. [Google Scholar] [CrossRef]
- Hastings, J.; Balny, C. The oxygenated bacterial luciferase-flavin intermediate. Reaction products via the light and dark pathways. J. Biol. Chem. 1975, 250, 7288–7293. [Google Scholar] [CrossRef]
- Cline, T.W.; Hastings, J. Mutationally altered bacterial luciferase. Implications for subunit functions. Biochemistry 1972, 11, 3359–3370. [Google Scholar] [CrossRef]
- Deeva, A.A.; Temlyakova, E.A.; Sorokin, A.A.; Nemtseva, E.V.; Kratasyuk, V.A. Structural distinctions of fast and slow bacterial luciferases revealed by phylogenetic analysis. Bioinformatics 2016, 32, 3053–3057. [Google Scholar] [CrossRef] [PubMed]
- Grote, R.F.; Hynes, J.T. The stable states picture of chemical reactions. II. Rate constants for condensed and gas phase reaction models. J. Chem. Phys. 1980, 73, 2715–2732. [Google Scholar] [CrossRef]
- Adams, J.A.; Taylor, S.S. Energetic limits of phosphotransfer in the catalytic subunit of cAMP-dependent protein kinase as measured by viscosity experiments. Biochemistry 1992, 31, 8516–8522. [Google Scholar] [CrossRef]
- Loveridge, E.J.; Evans, R.M.; Allemann, R.K. Solvent effects on environmentally coupled hydrogen tunnelling during catalysis by dihydrofolate reductase from Thermotoga maritima. Chem. Eur. J. 2008, 14, 10782–10788. [Google Scholar] [CrossRef]
- Coll, R.J.; Fink, A.L. Cryoenzymology of human plasmin catalysis: Comparison of cryosolvents and reactions with nitrophenyl ester and anilide substrates. Cryobiology 1987, 24, 332–344. [Google Scholar] [CrossRef]
- Hou, C.; Liu, Y.J.; Ferré, N.; Fang, W.H. Understanding bacterial bioluminescence: A theoretical study of the entire process, from reduced flavin to light emission. Chem. Eur. J. 2014, 20, 7979–7986. [Google Scholar] [CrossRef]
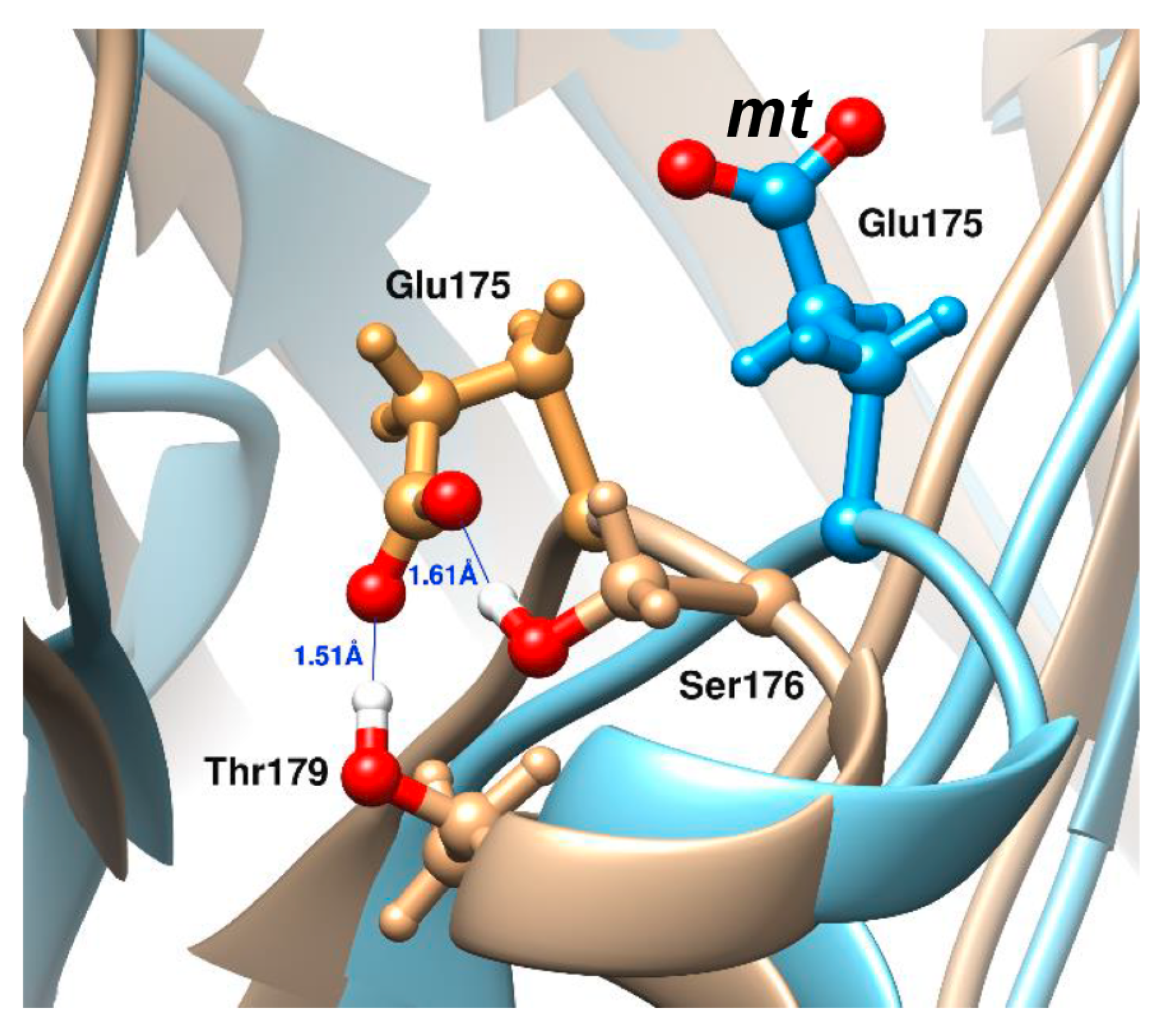
- Hosseinkhani, S.; Szittner, R.; Meighen, E.A. Random mutagenesis of bacterial luciferase: Critical role of Glu175 in the control of luminescence decay. Biochem. J. 2005, 385, 575–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, M.H.; Romsos, D.R.; Leveille, G.A. Effect of glycerol on lipogenic enzyme activities and on fatty acid synthesis in the rat and chicken. J. Nutr. 1976, 106, 1668–1677. [Google Scholar] [PubMed] [Green Version]
- Goguadze, N.G.; Hammerstad-pedersen, J.M.; Khoshtariya, D.E.; Ulstrup, J. Conformational dynamics and solvent viscosity effects in carboxypeptidase-A-catalyzed benzoylglycylphenyllactate hydrolysis. Eur. J. Biochem. 1991, 200, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.G.; Munoz-Clares, R.A.; Uribe, S. Trehalose-mediated inhibition of the plasma membrane H+-ATPase from Kluyveromyces lactis: Dependence on viscosity and temperature. J. Bacteriol. 2002, 184, 4384–4391. [Google Scholar] [PubMed] [Green Version]
- Lushchekina, S.V.; Inidjel, G.; Martinez, N.; Masson, P.; Trovaslet-Leroy, M.; Nachon, F.; Koza, M.M.; Seydel, T.; Peters, J. Impact of sucrose as osmolyte on molecular dynamics of mouse acetylcholinesterase. Biomolecules 2020, 10, 1664. [Google Scholar] [CrossRef] [PubMed]


| Rate Constant * | Buffer | Cosolvent | 10% | 20% | 30% | 40% | |
|---|---|---|---|---|---|---|---|
| , s | 7.85 | Glycerol | 7.50 | 5.05 | 3.66 | 2.60 | 0.81 ± 0.05 |
| Sucrose | 5.69 | 4.25 | 3.29 | 2.64 | |||
| , s | 0.15 | Glycerol | 0.15 | 0.15 | 0.12 | 0.10 | 0.84 ± 0.11 |
| Sucrose | 0.15 | 0.12 | 0.09 | 0.05 | 0.82 ± 0.06 | ||
| , Ms | 1.55 | Glycerol | 1.06 | 0.62 | 0.38 | 0.19 | 1.62 ± 0.04 |
| Sucrose | 1.21 | 0.82 | 0.47 | 0.29 | 0.94 ± 0.03 | ||
| , s | 400 | Glycerol | 398 | 403 | 403 | 395 | N/A |
| Sucrose | 401 | 403 | 393 | 402 | N/A | ||
| , Ms | 0.42 | Glycerol | 0.53 | 0.30 | 0.21 | 0.10 | 1.21 ± 0.35 |
| Sucrose | 0.31 | 0.18 | 0.06 | 0.02 | 1.85 ± 0.15 | ||
| , s | 10.90 | Glycerol | 8.95 | 7.50 | 5.42 | 5.30 | 0.59 ± 0.07 |
| Sucrose | 7.02 | 9.07 | 9.40 | 4.67 | N/A | ||
| , s | 0.46 | Glycerol | 0.45 | 0.47 | 0.43 | 0.42 | N/A |
| Sucrose | 0.38 | 0.64 | 1.69 | 5.13 | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lisitsa, A.E.; Sukovatyi, L.A.; Bartsev, S.I.; Deeva, A.A.; Kratasyuk, V.A.; Nemtseva, E.V. Mechanisms of Viscous Media Effects on Elementary Steps of Bacterial Bioluminescent Reaction. Int. J. Mol. Sci. 2021, 22, 8827. https://doi.org/10.3390/ijms22168827
Lisitsa AE, Sukovatyi LA, Bartsev SI, Deeva AA, Kratasyuk VA, Nemtseva EV. Mechanisms of Viscous Media Effects on Elementary Steps of Bacterial Bioluminescent Reaction. International Journal of Molecular Sciences. 2021; 22(16):8827. https://doi.org/10.3390/ijms22168827
Chicago/Turabian StyleLisitsa, Albert E., Lev A. Sukovatyi, Sergey I. Bartsev, Anna A. Deeva, Valentina A. Kratasyuk, and Elena V. Nemtseva. 2021. "Mechanisms of Viscous Media Effects on Elementary Steps of Bacterial Bioluminescent Reaction" International Journal of Molecular Sciences 22, no. 16: 8827. https://doi.org/10.3390/ijms22168827







