Discovering the DNA-Binding Consensus of the Thermus thermophilus HB8 Transcriptional Regulator TTHA1359
Abstract
1. Introduction
2. Results
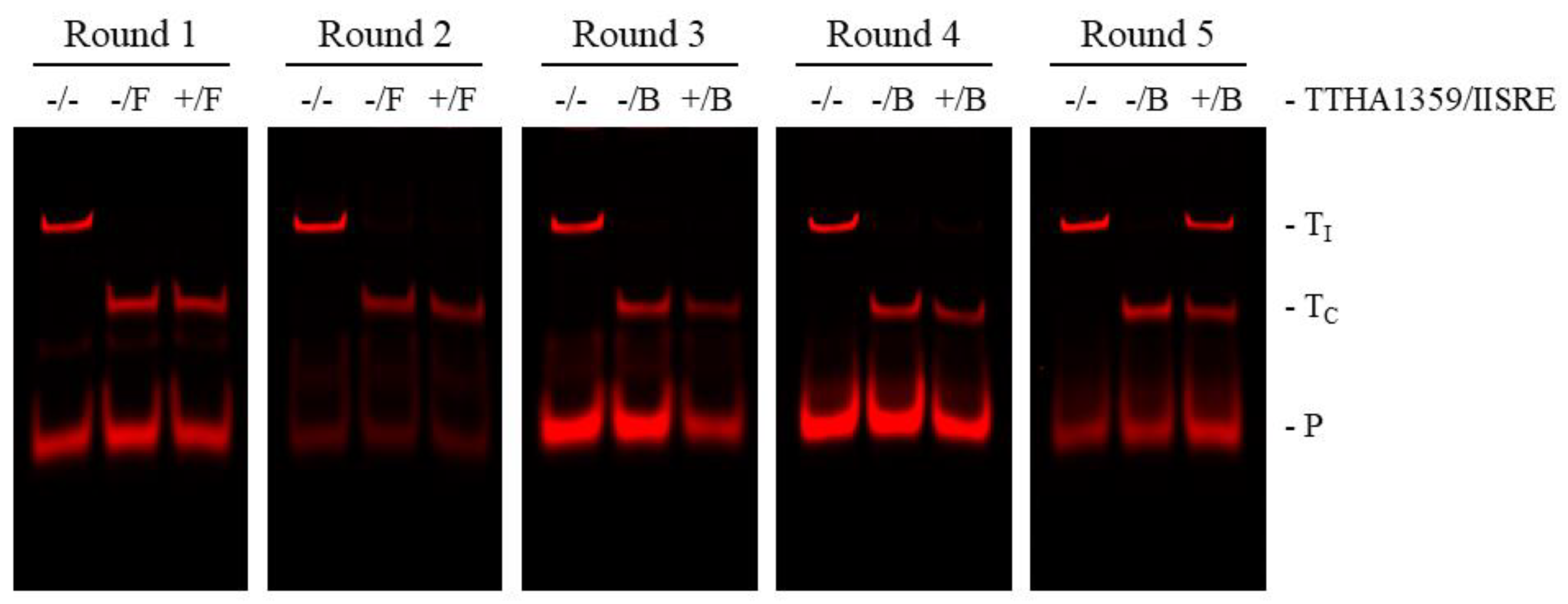
2.1. REPSA Selection of TTHA1359-Binding Sequences
2.2. Identification of Consensus TTHA1359-Binding Sequences
2.3. Biophysical Characterization of TTHA1359-DNA Binding
2.4. Exploration of Potential Regulatory TTHA1359-DNA Binding Sites in the T. thermophilus HB8 Genome
2.5. In Vitro Validation of TTHA1359-DNA Binding Sites in the T. thermophilus HB8 Genome
2.6. Bioinformatic Analysis of Potential TTHA1359-Regulated Genes
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides
4.2. TTHA1359 Protein
4.3. TTHA1359 Consensus Sequence Determination
4.4. Protein-DNA Binding Assays
4.5. Bioinformatic Determination of Candidate Regulated Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Browning, D.F.; Busby, S.J. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004, 2, 57–65. [Google Scholar] [CrossRef]
- Mejía-Almonte, C.; Busby, S.J.; Wade, J.T.; van Helden, J.; Arkin, A.P.; Stormo, G.D.; Eilbeck, K.; Palsson, B.O.; Galagan, J.E.; Collado-Vides, J. Redefining fundamental concepts of transcription initiation in bacteria. Nat. Rev. Genet. 2020, 21, 1–16. [Google Scholar] [CrossRef]
- Körner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the Bacterial Superfamily of Crp-Fnr Transcription Regulators: Exploiting the Metabolic Spectrum by Controlling Alternative Gene Programs. FEMS Microbiol. Rev. 2003, 27, 559–592. [Google Scholar] [CrossRef]
- Lawson, C.L.; Swigon, D.; Murakami, K.S.; Darst, S.A.; Berman, H.M.; Ebright, R.H. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 2004, 14, 10–20. [Google Scholar] [CrossRef]
- Spiro, S.; Guest, J.R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 1990, 6, 399–428. [Google Scholar] [PubMed]
- Ebright, R.H.; Ebright, Y.W.; Gunasekera, A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989, 17, 10295–10305. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Constantinidou, C.; Hobman, J.L.; Minchin, S.D. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004, 32, 5874–5893. [Google Scholar] [CrossRef] [PubMed]
- Grainger, D.C.; Hurd, D.; Harrison, M.; Holdstock, J.; Busby, S.J. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. USA 2005, 102, 17693–17698. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.J.; Kiley, P.J. Characterization of the dimerization domain in the FNR transcription factor. J. Biol. Chem. 2001, 276, 45744–45750. [Google Scholar] [CrossRef] [PubMed]
- Eiglmeier, K.; Honore, N.; Iuchi, S.; Lin, E.C.; Cole, S.T. Molecular genetic analysis of FNR-dependent promoters. Mol. Microbiol. 1989, 3, 869–878. [Google Scholar] [CrossRef]
- Salmon, K.; Hung, S.P.; Mekjian, K.; Baldi, P.; Hatfield, G.W.; Gunsalus, R.P. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 2003, 278, 29837–29855. [Google Scholar] [PubMed]
- Kang, Y.; Weber, K.D.; Qiu, Y.; Kiley, P.J.; Blattner, F.R. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 2005, 187, 1135–1160. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Imahori, K. Description of Thermus thermophilus (Yoshida and Oshima) comb. Nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Evol. Microbiol. 1974, 24, 102–112. [Google Scholar] [CrossRef]
- Yokoyama, S.; Hirota, H.; Kigawa, T.; Yabuki, T.; Shirouzu, M.; Terada, T.; Ito, Y.; Matsuno, Y.; Kuroda, Y.; Nishimura, Y.; et al. Structural genomics projects in Japan. Nat. Struct. Biol. 2000, 7, 943–945. [Google Scholar] [CrossRef]
- Ebihara, A.; Shinkai, A.; Nakagawa, N.; Masui, R.; Miki, K.; Yokoyama, S.; Kuramitsu, S. Structural-Biological Whole Cell Project of Thermus thermophilus HB8: From Structural Genomics to Systems Biology. Nihon Kessho Gakkaishi 2006, 48, 403–410. [Google Scholar] [CrossRef][Green Version]
- Thermus Thermophilus HB8, Genome Assembly. Available online: https://www.ncbi.nlm.nih.gov/genome/461?genome_assembly_id=166855 (accessed on 7 August 2021).
- Shinkai, A.; Kira, S.; Nakagawa, N.; Kashihara, A.; Kuramitsu, S.; Yokoyama, S. Transcription activation mediated by a cyclic AMP receptor protein from Thermus thermophilus HB8. J. Bacteriol. 2007, 189, 3891–3901. [Google Scholar] [CrossRef] [PubMed]
- Agari, Y.; Kashihara, A.; Yokoyama, S.; Kuramitsu, S.; Shinkai, A. Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator. Mol. Microbiol. 2008, 70, 60–75. [Google Scholar] [CrossRef]
- Agari, Y.; Kuramitsu, S.; Shinkai, A. X-ray crystal structure of TTHB099, a CRP/FNR superfamily transcriptional regulator from Thermus thermophilus HB8, reveals a DNA-binding protein with no required allosteric effector molecule. Proteins 2012, 80, 1490–1494. [Google Scholar] [CrossRef]
- Agari, Y.; Kuramitsu, S.; Shinkai, A. Identification of novel genes regulated by the oxidative stress-responsive transcriptional activator SdrP in Thermus thermophilus HB8. FEMS Microbiol. Lett. 2010, 313, 127–134. [Google Scholar] [CrossRef]
- Van Dyke, M.W.; Beyer, M.D.; Clay, E.; Hiam, K.J.; McMurry, J.L.; Xie, Y. Identification of preferred DNA-binding sites for the Thermus thermophilus transcriptional regulator SbtR by the combinatorial approach REPSA. PLoS ONE 2016, 11, e0159408. [Google Scholar] [CrossRef]
- Lee, M.; Um, H.; Van Dyke, M.W. Identification and characterization of preferred DNA-binding sites for the Thermus thermophilus transcriptional regulator FadR. PLoS ONE 2017, 12, e0184796. [Google Scholar] [CrossRef]
- Cox, J.S.; Moncja, K.; Mckinnes, M.; Van Dyke, M.W. Identification and characterization of preferred DNA-binding sites for the Thermus thermophilus HB8 transcriptional regulator TTHA0973. Int. J. Mol. Sci. 2019, 20, 3336. [Google Scholar] [CrossRef]
- Cox, J.S.; Van Dyke, M.W. General and Genomic DNA-binding specificity for the Thermus thermophilus HB8 transcription factor TTHB023. Biomolecules 2020, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Moncja, K.; Van Dyke, M.W. Determination and dissection of DNA-binding specificity for the Thermus thermophilus HB8 transcriptional regulator TTHB099. Int. J. Mol. Sci. 2020, 21, 7929. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Soberón-Chávez, G.; Alcaraz, L.D.; Morales, E.; Ponce-Soto, G.Y.; Servín-González, L. The transcriptional regulators of the CRP family regulate different essential bacterial functions and can be inherited vertically and horizontally. Front. Microbiol. 2017, 8, 959. [Google Scholar] [CrossRef] [PubMed]
- Concepcion, J.; Witte, K.; Wartchow, C.; Choo, S.; Yao, D.; Persson, H.; Wei, J.; Li, P.; Heidecker, B.; Ma, W.; et al. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb. Chem. High Throughput Screen. 2009, 12, 791–800. [Google Scholar] [CrossRef]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic annotation of microbial genomes and metagenomic sequences. In Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies; Li, R.W., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2011; pp. 61–78. [Google Scholar]
- Van Dyke, M.; Gracien, I. Restriction endonuclease protection assays using infrared-fluorescent probes. Protocolsio 2020. Available online: doi.org/10.17504/protocols.io.bi5ikg4e (accessed on 7 August 2021).
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Berg, O.G.; von Hippel, P.H. Selection of DNA binding sites by regulatory proteins II. The binding of cyclic AMP receptor protein to recognition sites. J. Mol. Biol. 1988, 200, 709–723. [Google Scholar]
- Van Dyke, M.W.; Van Dyke, N.; Sunavala-Dossabhoy, G. REPSA: General combinatorial approach for identifying preferred ligand-DNA binding sequences. Methods 2007, 42, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Van Dyke, M. Direct double-stranded DNA quantitation from PCR reactions. Protocolsio 2017. Available online: doi.org/10.17504/protocols.io.k5pcy5n (accessed on 7 August 2021).
- Van Dyke, M.; Cox, J. DNA analysis by native polyacrylamide gel electrophoresis and infrared fluorescence imaging. Protocols.io. 2017. Available online: doi.org/10.17504/protocols.io.mcyc2xw (accessed on 7 August 2021).





| Name | Sequence | R2 | |||
|---|---|---|---|---|---|
| wt | ATTGTGACACACATCACAAT | 3.447 | |||
| wt_p1 | cTTGTGACACACATCACAAT | ⸺⸺⸺⸺⸺⸺ Ambiguous ⸺⸺⸺⸺⸺⸺ | |||
| wt_p2 | AgTGTGACACACATCACAAT | 36.78 | |||
| wt_p3 | ATgGTGACACACATCACAAT | 86.53 | |||
| wt_p4 | ATTtTGACACACATCACAAT | ⸺⸺⸺⸺⸺⸺ Ambiguous ⸺⸺⸺⸺⸺⸺ | |||
| wt_p5 | ATTGgGACACACATCACAAT | ⸺⸺⸺⸺⸺⸺ Ambiguous ⸺⸺⸺⸺⸺⸺ | |||
| wt_p6 | ATTGTtACACACATCACAAT | 74.67 | |||
| wt_p7 | ATTGTGgCACACATCACAAT | 23.35 | |||
| wt_s5 | ATTGTGAcacacTCACAAT | ⸺⸺⸺⸺⸺⸺ Ambiguous ⸺⸺⸺⸺⸺⸺ | |||
| wt_s7 | ATTGTGAcacacacTCACAAT | ⸺⸺⸺⸺⸺⸺ Ambiguous ⸺⸺⸺⸺⸺⸺ | |||
| CRP_Ec | AAATGTGATCTAGATCACATTT | 1.011 | |||
| ctrl | ATACGAAAAACACACAC | ⸺⸺⸺⸺⸺⸺ Ambiguous ⸺⸺⸺⸺⸺⸺ | |||
| Start | End | p-Value | Q-Value | Sequence | Loc | Gene | Op |
|---|---|---|---|---|---|---|---|
| 917,761 | 917,782 | 1.01 × 10−8 | 0.0125 | AAATGTGAACATATTCACTTTC | −376 | TTHA0973 | 1/6 |
| 898,965 | 898,986 | 1.48 × 10−8 | 0.0125 | AATCGTGAAGTTTATCACATAT | −64 | TTHA0953 | S |
| 1503 | 1524 | 1.78 × 10−8 | 0.0125 | GAAAGTGAGATAACTCACATAT | +624 | TTHC002 | S |
| 402,440 | 402,461 | 1.01 × 10−8 | 0.0532 | TAAAGTGCTTTATTTCACAAAA | −34 | TTHA0425 | S |
| 809,120 | 809,141 | 2.64 × 10−7 | 0.104 | AATTGTGCTGGGCCACACAAAT | +975 | TTHA0843 | 1/3 |
| 931,166 | 931,187 | 2.95 × 10−7 | 0.104 | ATTAGTGAAACTTTTCACGATT | −95 | TTHA0987 | S |
| 752,613 | 752,634 | 3.71 × 10−7 | 0.105 | GAAGGTAACTTCAAACACTTTC | −44 | TTHA0784 | S |
| 418,352 | 418,373 | 3.97 × 10−7 | 0.105 | AATTGTCAACGGGATTACGTAT ATACGTAATCCCGTTGACAATT | −90 −54 | TTHA0446 TTHA0447 | S 1/5 |
| 81,405 | 81,426 | 5.35 × 10−7 | 0.113 | CCATGTGTTTTAGTTTACTTTA TAAAGTAAACTAAAACACATGG | −46 −18/+4 | TTHA0080 TTHA0081 | 1/2 1/3 |
| 899,588 | 899,609 | 5.82 × 10−7 | 0.113 | AATCGTGAATAAAATCACTAGG | −22 | TTHA0954 | 1/2 |
| 932,531 | 932,552 | 5.88 × 10−7 | 0.113 | TCTTGTACTTTTATTCACGATT | +1250 | TTHA0987 | S |
| 871,755 | 871,776 | 1.69 × 10−6 | 0.298 | ACTTGTCAGCAAAATTACGATG | +620 | TTHA0911 | S |
| 613,187 | 613,208 | 2.35 × 10−6 | 0.383 | CAATGTCCTTTTAAGCTCAATT | +306 | TTHA0644 | 2/3 |
| 496,704 | 496,725 | 3.11 × 10−6 | 0.471 | GAAAGAGAATGTTAGCACATTT AAATGTGCTAACATTCTCTTTC | −36 −34 | TTHA0533 TTHA0534 | 1/2 1/2 |
| Operon | Gene | Product | Role | LogFC | Adj. p-Value |
|---|---|---|---|---|---|
| S | TTHA0953 | UP (HTH MarR family) | Transcription | 1.29 | 0.111 |
| S | TTHA0425 | NADH dehydrogenase (ubiquinone) | Energy metabolism | −1.82 | 0.00416 |
| S | TTHA0446 | 50S ribosomal protein L34 (rpmH) | Translation | nd | nd |
| 1 | TTHA0447 | Branched-chain amino acid transporter ATP-binding protein | Quorum sensing | −0.0356 | 0.972 |
| 2 | TTHA0448 | Long-chain-fatty-acid--CoA ligase | Quorum sensing | −0.461 | 0.684 |
| 3 | TTHA0449 | Branched-chain amino acid ABC transporter, permease protein | Quorum sensing | −1.18 | 0.412 |
| 4 | TTHA0450 | Branched-chain amino acid ABC transporter, permease protein | Quorum sensing | −0.417 | 0.780 |
| 5 | TTHA0451 | Branched-chain amino acid ABC transporter, amino acid-binding protein | Quorum sensing | −1.35 | 0.384 |
| 1 | TTHA0080 | UP | ? | 1.03 | 0.219 |
| 2 | tRNA-Ala-3 | tRNAala | Translation | nd | nd |
| 1 | TTHA0081 | UP (Rad52_Rad22-like) | ? | 0.998 | 0.100 |
| 2 | TTHA0082 | Metallophosphoesterase | ? | 1.00 | 0.107 |
| 3 | TTHA0083 | 16S rRNA dimethyladenosine transferase | Translation | 0.410 | 0.558 |
| 1 | TTHA0954 | Mannosyl-3-phosphoglycerate synthase | Sugar metabolism | 0.530 | 0.306 |
| 2 | TTHA0955 | Mannosyl-3-phosphoglycerate phosphatase | Sugar metabolism | 0.0925 | 0.865 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teague, J.L.; Barrows, J.K.; Baafi, C.A.; Van Dyke, M.W. Discovering the DNA-Binding Consensus of the Thermus thermophilus HB8 Transcriptional Regulator TTHA1359. Int. J. Mol. Sci. 2021, 22, 10042. https://doi.org/10.3390/ijms221810042
Teague JL, Barrows JK, Baafi CA, Van Dyke MW. Discovering the DNA-Binding Consensus of the Thermus thermophilus HB8 Transcriptional Regulator TTHA1359. International Journal of Molecular Sciences. 2021; 22(18):10042. https://doi.org/10.3390/ijms221810042
Chicago/Turabian StyleTeague, Josiah L., John K. Barrows, Cynthia A. Baafi, and Michael W. Van Dyke. 2021. "Discovering the DNA-Binding Consensus of the Thermus thermophilus HB8 Transcriptional Regulator TTHA1359" International Journal of Molecular Sciences 22, no. 18: 10042. https://doi.org/10.3390/ijms221810042
APA StyleTeague, J. L., Barrows, J. K., Baafi, C. A., & Van Dyke, M. W. (2021). Discovering the DNA-Binding Consensus of the Thermus thermophilus HB8 Transcriptional Regulator TTHA1359. International Journal of Molecular Sciences, 22(18), 10042. https://doi.org/10.3390/ijms221810042








