Metabolomic Approaches to Investigate the Effect of Metformin: An Overview
Abstract
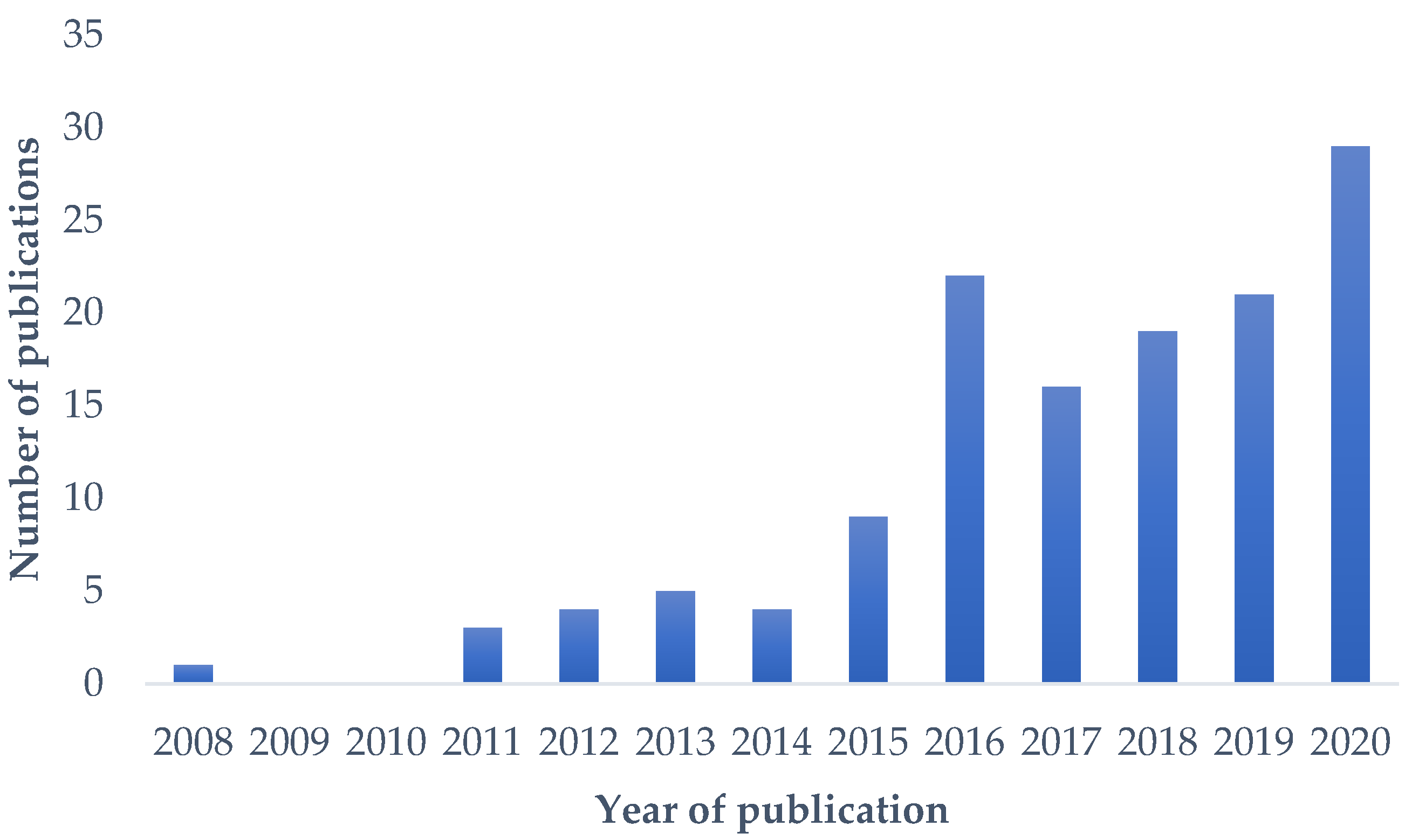
:1. Introduction
2. Metabolomics
3. Plasma Metabolome and Metformin Treatment
3.1. Effect of Metformin Treatment in Non-Diabetic Condition
3.2. Effect of Metformin Treatment in Insuline-Resistant Condition
4. Urinary Metabolome and Metformin Treatment
4.1. Effect of Metformin Treatment in Insuline-Resistant In Vivo Models
4.2. Effect of Metformin Treatment on Human Urine Metabolome
5. Cell and Tissue Metabolome and Metformin Treatment
6. Conclusions
Funding
Conflicts of Interest
References
- Rojas, L.B.; Gomes, M.B. Metformin: An old but still the best treatment for type 2 diabetes. Diabetol Metab Syndr 2013, 5, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UK Prospective Diabetes Study (UKPDS) Group. Effect of intensive blood-glucose control with metformin on complications in overweight patients with type 2 diabetes (UKPDS 34). Lancet 1998, 352, 854–865. [Google Scholar] [CrossRef]
- Tanret, G. An alkaloid extracted from Galega officinalis. Compt. Rend. 1914, 158, 1182–1184. [Google Scholar]
- Mooney, M.H.; Fogarty, S.; Stevenson, C.; Gallagher, A.M.; Palit, P.; Hawley, S.A.; Hardie, D.G.; Coxon, G.D.; Waigh, R.D.; Tate, R.J.; et al. Mechanisms underlying the metabolic actions of galegine that contribute to weight loss in mice. Br. J. Pharmacol. 2008, 153, 1669–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, T.A.; Campero, C.M.; Chayer, R.; Cosentino, B.; Caracino, M. Experimental toxicity of verbesina encelioides in sheep and isolation of galegine. Vet. Hum. Toxicol. 1996, 38, 417–419. [Google Scholar] [PubMed]
- Perla, V.; Jayanty, S.S. Biguanide related compounds in traditional antidiabetic functional foods. Food Chem. 2013, 138, 1574–1580. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Woo, S.L.; Hu, X.; Botchlett, R.; Chen, L.; Huo, Y.; Wu, C. Metformin and metabolic diseases: A focus on hepatic aspects. Front. Med. 2015, 9, 173–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diabetes Prevention Program Research Group. Long-term safety, tolerability, and weight loss associated with metformin in the Diabetes Prevention Program Outcomes Study. Diabetes Care 2012, 35, 731–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masarwa, R.; Brunetti, V.C.; Aloe, S.; Henderson, M.; Platt, R.W.; Filion, K.B. Efficacy and Safety of Metformin for Obesity: A Systematic Review. Pediatrics 2021, 147, e20201610. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.F.; Wang, H.J.; Liu, Z.; Geng, S.K.; Wang, H.T.; Wang, X.D.; Li, T.; Morel, L.; Wan, W.G.; Lu, L.J.; et al. Safety and efficacy of metformin in systemic lupus erythematosus: A multicentre, randomised, double-blind, placebo-controlled trial. Lancet Rheumatol. 2020, 2, E210–E216. [Google Scholar] [CrossRef]
- Jonker, J.W.; Wagenaar, E.; Van Eijl, S.; Schinkel, A.H. Deficiency in the organic cation transporters 1 and 2 (Oct1/Oct2 [Slc22a1/Slc22a2]) in mice abolishes renal secretion of organic cations. Mol. Cell Biol. 2003, 23, 7902–7908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.S.; Jonker, J.W.; Kato, Y.; Kusuhara, H.; Schinkel, A.H.; Sugiyama, Y. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J. Pharmacol. Exp. Ther. 2002, 302, 510–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, Y.; Sheardown, S.A.; Brown, C.; Owen, R.P.; Zhang, S.; Castro, R.A.; Ianculescu, A.G.; Yue, L.; Lo, J.C.; Burchard, E.G.; et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Investig. 2007, 117, 1422–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, Y.; Brown, C.; Castro, R.A.; Shi, R.J.; Lin, E.T.; Owen, R.P.; Sheardown, S.A.; Yue, L.; Burchard, E.G.; Brett, C.M.; et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin. Pharmacol. Ther. 2008, 83, 273–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rena, G.; Hardie, D.G.; Pearson, E.R. The mechanisms of action of metformin. Diabetologia 2017, 60, 1577–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foretz, M.; Guigas, B.; Bertrand, L.; Pollak, M.; Viollet, B. Metformin: From mechanisms of action to therapies. Cell Metab. 2014, 20, 953–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaMoia, T.E.; Shulman, G.I. Cellular and Molecular Mechanisms of Metformin Action. Endocr Rev. 2021, 42, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.A.; Birnbaum, M.J. An energetic tale of AMPK-independent effects of metformin. J. Clin. Investig. 2010, 120, 2267–2270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridges, H.R.; Jones, A.J.; Pollak, M.N.; Hirst, J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. Biochem. J. 2014, 462, 475–487. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; An, H.; Liu, T.; Qin, C.; Sesaki, H.; Guo, S.; Radovick, S.; Hussain, M.; Maheshwari, A.; Wondisford, F.E.; et al. Metformin Improves Mitochondrial Respiratory Activity through Activation of AMPK. Cell Rep. 2019, 29, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Musi, N.; Hirshman, M.F.; Nygren, J.; Svanfeldt, M.; Bavenholm, P.; Rooyackers, O.; Zhou, G.; Williamson, J.M.; Ljunqvist, O.; Efendic, S.; et al. Metformin increases AMP-activated protein kinase activity in skeletal muscle of subjects with type 2 diabetes. Diabetes 2002, 51, 2074–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, J.M.; Treebak, J.T.; Schjerling, P.; Goodyear, L.; Wojtaszewski, J.F. Two weeks of metformin treatment induces AMPK-dependent enhancement of insulin-stimulated glucose uptake in mouse soleus muscle. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1099–E1109. [Google Scholar] [CrossRef] [PubMed]
- McCreight, L.J.; Mari, A.; Coppin, L.; Jackson, N.; Umpleby, A.M.; Pearson, E.R. Metformin increases fasting glucose clearance and endogenous glucose production in non-diabetic individuals. Diabetologia 2020, 63, 444–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gormsen, L.C.; Sondergaard, E.; Christensen, N.L.; Brosen, K.; Jessen, N.; Nielsen, S. Metformin increases endogenous glucose production in non-diabetic individuals and individuals with recent-onset type 2 diabetes. Diabetologia 2019, 62, 1251–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCreight, L.J.; Bailey, C.J.; Pearson, E.R. Metformin and the gastrointestinal tract. Diabetologia 2016, 59, 426–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Hu, N. Effects of Metformin on the Gut Microbiota in Obesity and Type 2 Diabetes Mellitus. Diabetes Metab. Syndr. Obes. 2020, 13, 5003–5014. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.Y.; Lee, S.J.; Lee, S.; Kim, S.J.; Han, S.J.; Kim, H.J.; Kim, D.J.; Kim, Y.S.; Woo, J.T.; Ahn, K.J.; et al. Failure of monotherapy in clinical practice in patients with type 2 diabetes: The Korean National Diabetes Program. J. Diabetes Investig. 2018, 9, 1144–1152. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.B.; Conner, C.; Nichols, G.A. Secondary failure of metformin monotherapy in clinical practice. Diabetes Care 2010, 33, 501–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryor, R.; Martinez-Martinez, D.; Quintaneiro, L.; Cabreiro, F. The Role of the Microbiome in Drug Response. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 417–435. [Google Scholar] [CrossRef] [PubMed]
- Tannenbaum, C.; Day, D.; Matera, A. Age and sex in drug development and testing for adults. Pharmacol. Res. 2017, 121, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Boullata, J.I. Drug and nutrition interactions: Not just food for thought. J. Clin. Pharm. Ther. 2013, 38, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Krzyszczyk, P.; Acevedo, A.; Davidoff, E.J.; Timmins, L.M.; Marrero-Berrios, I.; Patel, M.; White, C.; Lowe, C.; Sherba, J.J.; Hartmanshenn, C.; et al. The growing role of precision and personalized medicine for cancer treatment. Technology Singap World Sci. 2018, 6, 79–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steuer, A.E.; Brockbals, L.; Kraemer, T. Metabolomic Strategies in Biomarker Research-New Approach for Indirect Identification of Drug Consumption and Sample Manipulation in Clinical and Forensic Toxicology? Front. Chem. 2019, 7, 319. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Li, H.; Peng, X.X. Functional metabolomics: From biomarker discovery to metabolome reprogramming. Protein Cell 2015, 6, 628–637. [Google Scholar] [CrossRef] [Green Version]
- Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007, 387, 525–527. [Google Scholar] [CrossRef]
- Bjerrum, J.T. Metabonomics Methods and Protocols; Humana Press: Totowa, NJ, USA, 2015. [Google Scholar]
- Connor, S.C.; Wu, W.; Sweatman, B.C.; Manini, J.; Haselden, J.N.; Crowther, D.J.; Waterfield, C.J. Effects of feeding and body weight loss on the 1H-NMR-based urine metabolic profiles of male Wistar Han rats: Implications for biomarker discovery. Biomarkers 2004, 9, 156–179. [Google Scholar] [CrossRef]
- Bingol, K. Recent Advances in Targeted and Untargeted Metabolomics by NMR and MS/NMR Methods. High. Throughput 2018, 7, 9. [Google Scholar] [CrossRef] [Green Version]
- Beger, R.D.; Schmidt, M.A.; Kaddurah-Daouk, R. Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites 2020, 10, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaddurah-Daouk, R.; Weinshilboum, R.; Network, P.R. Metabolomic Signatures for Drug Response Phenotypes: Pharmacometabolomics Enables Precision Medicine. Clin. Pharmacol. Ther. 2015, 98, 71–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, G.M.; Waikar, S.S. Biomarkers in nephrology: Core Curriculum 2013. Am. J. Kidney Dis. 2013, 62, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Fiehn, O.; Garvey, W.T.; Newman, J.W.; Lok, K.H.; Hoppel, C.L.; Adams, S.H. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS ONE 2010, 5, e15234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, V.O.; Townsend, R.R.; Feldman, H.I.; Pappan, K.L.; Kensicki, E.; Vander Jagt, D.L. Plasma metabolomic profiles in different stages of CKD. Clin. J. Am. Soc. Nephrol. 2013, 8, 363–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allman, E.L.; Painter, H.J.; Samra, J.; Carrasquilla, M.; Llinas, M. Metabolomic Profiling of the Malaria Box Reveals Antimalarial Target Pathways. Antimicrob Agents Chemother 2016, 60, 6635–6649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perng, W.; Rifas-Shiman, S.L.; Sordillo, J.; Hivert, M.F.; Oken, E. Metabolomic Profiles of Overweight/Obesity Phenotypes during Adolescence: A Cross-Sectional Study in Project Viva. Obesity 2020, 28, 379–387. [Google Scholar] [CrossRef]
- Paige, L.A.; Mitchell, M.W.; Krishnan, K.R.; Kaddurah-Daouk, R.; Steffens, D.C. A preliminary metabolomic analysis of older adults with and without depression. Int. J. Geriatr. Psychiatry 2007, 22, 418–423. [Google Scholar] [CrossRef]
- Rhodes, C.J.; Ghataorhe, P.; Wharton, J.; Rue-Albrecht, K.C.; Hadinnapola, C.; Watson, G.; Bleda, M.; Haimel, M.; Coghlan, G.; Corris, P.A.; et al. Plasma Metabolomics Implicates Modified Transfer RNAs and Altered Bioenergetics in the Outcomes of Pulmonary Arterial Hypertension. Circulation 2017, 135, 460–475. [Google Scholar] [CrossRef] [PubMed]
- Oresic, M.; Hyotylainen, T.; Herukka, S.K.; Sysi-Aho, M.; Mattila, I.; Seppanan-Laakso, T.; Julkunen, V.; Gopalacharyulu, P.V.; Hallikainen, M.; Koikkalainen, J.; et al. Metabolome in progression to Alzheimer’s disease. Transl. Psychiatry 2011, 1, e57. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Huo, T.; Li, N.; Xiong, Z.; Li, F. Lysophosphatidylcholine—Biomarker of Metformin action: Studied using UPLC/MS/MS. Biomed. Chromatogr. 2009, 23, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Dahabiyeh, L.A.; Mujammami, M.; Arafat, T.; Benabdelkamel, H.; Alfadda, A.A.; Abdel Rahman, A.M. A Metabolic Pattern in Healthy Subjects Given a Single Dose of Metformin: A Metabolomics Approach. Front. Pharmacol. 2021, 12, 705932. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A Branched-Chain Amino Acid-Related Metabolic Signature that Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef] [Green Version]
- Krebs, M.; Krssak, M.; Bernroider, E.; Anderwald, C.; Brehm, A.; Meyerspeer, M.; Nowotny, P.; Roth, E.; Waldhausl, W.; Roden, M. Mechanism of amino acid-induced skeletal muscle insulin resistance in humans. Diabetes 2002, 51, 599–605. [Google Scholar] [CrossRef] [Green Version]
- Preiss, D.; Rankin, N.; Welsh, P.; Holman, R.R.; Kangas, A.J.; Soininen, P.; Wurtz, P.; Ala-Korpela, M.; Sattar, N. Effect of metformin therapy on circulating amino acids in a randomized trial: The CAMERA study. Diabet. Med. 2016, 33, 1569–1574. [Google Scholar] [CrossRef] [PubMed]
- Walford, G.A.; Davis, J.; Warner, A.S.; Ackerman, R.J.; Billings, L.K.; Chamarthi, B.; Fanelli, R.R.; Hernandez, A.M.; Huang, C.; Khan, S.Q.; et al. Branched chain and aromatic amino acids change acutely following two medical therapies for type 2 diabetes mellitus. Metabolism 2013, 62, 1772–1778. [Google Scholar] [CrossRef] [Green Version]
- Rotroff, D.M.; Oki, N.O.; Liang, X.; Yee, S.W.; Stocker, S.L.; Corum, D.G.; Meisner, M.; Fiehn, O.; Motsinger-Reif, A.A.; Giacomini, K.M.; et al. Pharmacometabolomic Assessment of Metformin in Non-diabetic, African Americans. Front. Pharmacol. 2016, 7, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, T.; Cai, S.; Lu, X.; Sha, Y.; Yu, M.; Li, F. Metabonomic study of biochemical changes in the serum of type 2 diabetes mellitus patients after the treatment of metformin hydrochloride. J. Pharm Biomed. Anal. 2009, 49, 976–982. [Google Scholar] [CrossRef]
- Xu, T.; Brandmaier, S.; Messias, A.C.; Herder, C.; Draisma, H.H.M.; Demirkan, A.; Yu, Z.H.; Ried, J.S.; Haller, T.; Heier, M.; et al. Effects of Metformin on Metabolite Profiles and LDL Cholesterol in Patients With Type 2 Diabetes. Diabetes Care 2015, 38, 1858–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, J.; Brandmaier, S.; Leonhardt, J.; Scheerer, M.F.; Mohney, R.P.; Xu, T.; Bi, J.; Rotter, M.; Troll, M.; Chi, S.; et al. Metformin Effect on Nontargeted Metabolite Profiles in Patients With Type 2 Diabetes and in Multiple Murine Tissues. Diabetes 2016, 65, 3776–3785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breier, M.; Wahl, S.; Prehn, C.; Ferrari, U.; Sacco, V.; Weise, M.; Grallert, H.; Adamski, J.; Lechner, A. Immediate reduction of serum citrulline but no change of steroid profile after initiation of metformin in individuals with type 2 diabetes. J. Steroid Biochem. 2017, 174, 114–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irving, B.A.; Carter, R.E.; Soop, M.; Weymiller, A.; Syed, H.; Karakelides, H.; Bhagra, S.; Short, K.R.; Tatpati, L.; Barazzoni, R.; et al. Effect of insulin sensitizer therapy on amino acids and their metabolites. Metabolism 2015, 64, 720–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huhtala, M.S.; Tertti, K.; Pellonpera, O.; Ronnemaa, T. Amino acid profile in women with gestational diabetes mellitus treated with metformin or insulin. Diabetes Res. Clin. Pract 2018, 146, 8–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safai, N.; Suvitaival, T.; Ali, A.; Spegel, P.; Al-Majdoub, M.; Carstensen, B.; Vestergaard, H.; Ridderstrale, M.; Grp, C.T. Effect of metformin on plasma metabolite profile in the Copenhagen Insulin and Metformin Therapy (CIMT) trial. Diabetic Med. 2018, 35, 944–953. [Google Scholar] [CrossRef]
- Tomasova, P.; Buganova, M.; Pelantova, H.; Holubova, M.; Sediva, B.; Zelezna, B.; Haluzik, M.; Maletinska, L.; Kunes, J.; Kuzma, M. Metabolomics Based on MS in Mice with Diet-Induced Obesity and Type 2 Diabetes Mellitus: The Effect of Vildagliptin, Metformin, and Their Combination. Appl. Biochem. Biotechnol. 2019, 188, 165–184. [Google Scholar] [CrossRef]
- Sonnet, D.S.; O’Leary, M.N.; Gutierrez, M.A.; Nguyen, S.M.; Mateen, S.; Hsu, Y.; Mitchell, K.P.; Lopez, A.J.; Vockley, J.; Kennedy, B.K.; et al. Metformin inhibits Branched Chain Amino Acid (BCAA) derived ketoacidosis and promotes metabolic homeostasis in MSUD. Sci. Rep. 2016, 6, 28775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zemdegs, J.; Martin, H.; Pintana, H.; Bullich, S.; Manta, S.; Marques, M.A.; Moro, C.; Laye, S.; Ducrocq, F.; Chattipakorn, N.; et al. Metformin Promotes Anxiolytic and Antidepressant-Like Responses in Insulin-Resistant Mice by Decreasing Circulating Branched-Chain Amino Acids. J. Neurosci. 2019, 39, 5935–5948. [Google Scholar] [CrossRef] [Green Version]
- Sundelin, E.; Gormsen, L.C.; Jensen, J.B.; Vendelbo, M.H.; Jakobsen, S.; Munk, O.L.; Christensen, M.; Brosen, K.; Frokiaer, J.; Jessen, N. Genetic Polymorphisms in Organic Cation Transporter 1 Attenuates Hepatic Metformin Exposure in Humans. Clin. Pharmacol. Ther. 2017, 102, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, Y.; Sun, T.Q.; Li, Y.; Markovtsov, V.; Uy, G.; Gross, L.; Goff, D.A.; Shaw, S.J.; Boralsky, L.; Singh, R.; et al. Global metabolite profiling of mice with high-fat diet-induced obesity chronically treated with AMPK activators R118 or metformin reveals tissue-selective alterations in metabolic pathways. BMC Res. Notes 2014, 7, 674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunn, W.B.; Bailey, N.J.; Johnson, H.E. Measuring the metabolome: Current analytical technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wu, X.; Wang, X. Urine metabolomics. Clin. Chim Acta 2012, 414, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yin, P.; Shao, Y.; Wang, Z.; Wang, B.; Lehmann, R.; Xu, G. Which is the urine sample material of choice for metabolomics-driven biomarker studies? Anal. Chim. Acta 2020, 1105, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Pelantova, H.; Buganova, M.; Holubova, M.; Sediva, B.; Zemenova, J.; Sykora, D.; Kavalkova, P.; Haluzik, M.; Zelezna, B.; Maletinska, L.; et al. Urinary metabolomic profiling in mice with diet-induced obesity and type 2 diabetes mellitus after treatment with metformin, vildagliptin and their combination. Mol. Cell Endocrinol. 2016, 431, 88–100. [Google Scholar] [CrossRef]
- Dong, Y.; Chen, Y.T.; Yang, Y.X.; Shou, D.; Li, C.Y. Urinary Metabolomic Profiling in Zucker Diabetic Fatty Rats with Type 2 Diabetes Mellitus Treated with Glimepiride, Metformin, and Their Combination. Molecules 2016, 21, 1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, L.L.; Sun, Q.H.; Liu, G.R.; Guo, J.Y. Urinary Metabolomics Study of the Intervention Effect of Hypoglycemic Decoction on Type 2 Diabetes Mellitus Rats Model. Evid. Based Complement. Altern. Med. 2019, 2019, 1394641. [Google Scholar] [CrossRef] [PubMed]
- Maulidiani, M.; Abas, F.; Rudiyanto, R.; Abd Kadir, N.H.; Zolkeflee, N.K.Z.; Lajise, N.H. Analysis of urinary metabolic alteration in type 2 diabetic rats treated with metformin using the metabolomics of quantitative spectral deconvolution H-1 NMR spectroscopy. Microchem. J. 2020, 153, 104513. [Google Scholar] [CrossRef]
- Zhu, Y.; Feng, Y.; Shen, L.; Xu, D.; Wang, B.; Ruan, K.; Cong, W. Effect of metformin on the urinary metabolites of diet-induced-obese mice studied by ultra performance liquid chromatography coupled to time-of-flight mass spectrometry (UPLC-TOF/MS). J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2013, 925, 110–116. [Google Scholar] [CrossRef]
- Mediani, A.; Abas, F.; Maulidiani, M.; Abu Bakar Sajak, A.; Khatib, A.; Tan, C.P.; Ismail, I.S.; Shaari, K.; Ismail, A.; Lajis, N.H. Metabolomic analysis and biochemical changes in the urine and serum of streptozotocin-induced normal- and obese-diabetic rats. J. Physiol. Biochem. 2018, 74, 403–416. [Google Scholar] [CrossRef]
- Lee, Y.F.; Sim, X.Y.; Teh, Y.H.; Ismail, M.N.; Greimel, P.; Murugaiyah, V.; Ibrahim, B.; Gam, L.H. The effects of high-fat diet and metformin on urinary metabolites in diabetes and prediabetes rat models. Biotechnol. Appl. Biochem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Chung, J.Y.; Cho, S.K.; Shin, H.W.; Jang, I.J.; Park, J.W.; Yu, K.S.; Cho, J.Y. Antihyperglycemic mechanism of metformin occurs via the AMPK/LXRalpha/POMC pathway. Sci. Rep. 2015, 5, 8145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Jeong, G.H.; Lee, I.K.; Yoon, Y.R.; Liu, K.H.; Gu, N.; Shin, K.H. A Pharmacometabolomic Approach to Predict Response to Metformin in Early-Phase Type 2 Diabetes Mellitus Patients. Molecules 2018, 23, 1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zukunft, S.; Prehn, C.; Rohring, C.; Moller, G.; Hrabe de Angelis, M.; Adamski, J.; Tokarz, J. High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metabolomics 2018, 14, 18. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Sun, H.; Xu, H.; Qiu, S.; Wang, X. Cell metabolomics. OMICS 2013, 17, 495–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Riano, C.; Garcia, A.; Barbas, C. Metabolomics studies in brain tissue: A review. J. Pharm. Biomed. Anal. 2016, 130, 141–168. [Google Scholar] [CrossRef] [PubMed]
- Aljofan, M.; Riethmacher, D. Anticancer activity of metformin: A systematic review of the literature. Future Sci. OA 2019, 5, FSO410. [Google Scholar] [CrossRef] [Green Version]
- Janzer, A.; German, N.J.; Gonzalez-Herrera, K.N.; Asara, J.M.; Haigis, M.C.; Struhl, K. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 10574–10579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soliman, G.A.; Steenson, S.M.; Etekpo, A.H. Effects of Metformin and a Mammalian Target of Rapamycin (mTOR) ATP-Competitive Inhibitor on Targeted Metabolomics in Pancreatic Cancer Cell Line. Metabolomics 2016, 6, 183. [Google Scholar]
- Zhang, J.; Hang, C.; Jiang, T.; Yi, S.; Shao, W.; Li, W.; Lin, D. Nuclear Magnetic Resonance-Based Metabolomic Analysis of the Anticancer Effect of Metformin Treatment on Cholangiocarcinoma Cells. Front. Oncol. 2020, 10, 570516. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Romero, I.L.; Litchfield, L.M.; Lengyel, E.; Locasale, J.W. Metformin Targets Central Carbon Metabolism and Reveals Mitochondrial Requirements in Human Cancers. Cell Metab. 2016, 24, 728–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, M.; Qi, H.; Xia, T.; Zhao, X.; Wang, W.; Wang, Z.; Lu, C.; Ning, Z.; Chen, H.; Li, T.; et al. Metabolomics profiling of metformin-mediated metabolic reprogramming bypassing AMPKalpha. Metabolism 2019, 91, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Tian, N.; Wang, J.; Yang, M.; Kong, L. Metabolic switching in the hypoglycemic and antitumor effects of metformin on high glucose induced HepG2 cells. J. Pharm. Biomed. Anal. 2018, 156, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Riera-Borrull, M.; Garcia-Heredia, A.; Fernandez-Arroyo, S.; Hernandez-Aguilera, A.; Cabre, N.; Cuyas, E.; Luciano-Mateo, F.; Camps, J.; Menendez, J.A.; Joven, J. Metformin Potentiates the Benefits of Dietary Restraint: A Metabolomic Study. Int. J. Mol. Sci. 2017, 18, 2263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Y.; Tong, Y.; Guo, Y.; Lang, X.; Huang, X.; Xie, X.; Guan, Y.; Li, Z. Metformin Attenuates the Metabolic Disturbance and Depression-like Behaviors Induced by Corticosterone and Mediates the Glucose Metabolism Pathway. Pharmacopsychiatry 2021, 54, 131–141. [Google Scholar] [PubMed]
- Li, W.; Chaudhari, K.; Shetty, R.; Winters, A.; Gao, X.; Hu, Z.; Ge, W.P.; Sumien, N.; Forster, M.; Liu, R.; et al. Metformin Alters Locomotor and Cognitive Function and Brain Metabolism in Normoglycemic Mice. Aging Dis. 2019, 10, 949–963. [Google Scholar] [CrossRef] [Green Version]


| Name | Change | Method | Metabolic Pathway | Refs. |
|---|---|---|---|---|
| Alanine | up | LC-MS, NMR | Alanine and aspartate metabolism | [24,55,59] |
| Glucuronic acid | down | LC-MS | Amino sugar metabolism | [24,55,61] |
| 3-Hydroxymethylglutaric acid | up | LC-MS | BCAA metabolism | [24,55] |
| Hippuric acid | up or down b | LC-MS | Benzoate metabolism Gut microbiota metabolism | [24,61] |
| Linoleyl Carnitine | down | LC-MS | Fatty acid metabolism (Acyl carnitine) | [24,55] |
| gamma-Glutamylleucine | up | LC-MS | Gamma-glutamyl amino acid metabolism | [24,55] |
| Glucose | up | LC-MS | Glycolysis, gluconeogenesis and pyruvate metabolism | [24,55] |
| Aminoadipic acid | up | LC-MS | Lysine metabolism | [24,55] |
| 2-Ketobutyric acid | up | LC-MS | Methionine, cysteine, SAM and taurine metabolism | [24,55] |
| Phenylacetate | up | LC-MS | Phenylalanine metabolism | [24,55] |
| 16:0 LPC | down | LC-MS | Phosphatidylcholine metabolism | [24,54,55] |
| 18:0 LPC | down | LC-MS | Phosphatidylcholine metabolism | [24,54,55] |
| Arachidonic acid | up | LC-MS | Polyunsaturated fatty acid metabolism | [24,61] |
| Cholic acid | down | LC-MS | Primary bile acid metabolism | [24,55] |
| Hypoxanthine | up or down b | LC-MS | Purine metabolism | [24,61] |
| Inosine | down | LC-MS | Purine metabolism | [24,61] |
| Sphinganine 1-phosphate | down | LC-MS | Sphingolipid synthesis | [24,55] |
| Dopamine3-O-sulfate | up | LC-MS | Tyrosine metabolism | [24,55,61] |
| Tyrosine | down | LC-MS, NMR | Tyrosine metabolism | [59,60,61] |
| Arginine | up or down b | LC-MS | Urea cycle | [24,55] |
| Citrulline | down | LC-MS | Urea cycle | [24,55] |
| Ornithine | down | LC-MS | Urea cycle | [24,61] |
| Proline | up | LC-MS | Urea cycle | [24,55] |
| Name | Change | Method | Metabolic Pathway | Refs. |
|---|---|---|---|---|
| Alanine | up | LC-MS, NMR | Alanine and aspartate metabolism | [24,65,67] |
| Isoleucine | up | LC-MS, NMR | BCAA metabolism | [60,67,68] |
| Leucine | up | LC-MS | BCAA metabolism | [24,60,68] |
| gamma-glutamyltyrosine | down | LC-MS | Gamma-glutamyl amino acid | [24,64] |
| Glycine | up | LC-MS, NMR | Glycine, serine and threonine metabolism | [24,67] |
| Glutaroyl carnitine | down | LC-MS | Lysine metabolism | [24,64] |
| Phenylalanine | up | LC-MS | Phenylalanine metabolism | [24,62] |
| PC ae C36:4 c | down | LC-MS | Phosphatidylcholine metabolism | [63,65] |
| PC ae C38:5 c | down | LC-MS | Phosphatidylcholine metabolism | [63,65] |
| PC ae C38:6 c | down | LC-MS | Phosphatidylcholine metabolism | [63,65] |
| Choline | down | LC-MS | Phospholipid metabolism | [24,64] |
| Trimethylamine-N-oxide | up | LC-MS, NMR | Phospholipid metabolism Gut microbiota metabolism | [24,62] |
| Inosine | up or down b | LC-MS | Purine metabolism | [24,64] |
| Cholesterol | down | LC-MS | Sterol metabolism | [24,64] |
| Malate | up | LC-MS | TCA Cycle | [24,64] |
| 3-Indoxyl sulfate | up | LC-MS | Tryptophan metabolism Gut microbiota metabolism | [24,64] |
| Tryptophan | up or down b | LC-MS | Tryptophan metabolism | [24,62] |
| 3-(4-hydroxyphenyl)lactate | down | LC-MS | Tyrosine metabolism | [24,64] |
| Arginine | down | LC-MS | Urea cycle | [24,64] |
| Citrulline | down | LC-MS | Urea cycle | [24,64,65] |
| Ornithine | down | LC-MS | Urea cycle | [24,64] |
| Proline | up | LC-MS | Urea cycle | [24,65] |
| No. | Name | Changes | Biospecimen | Animal | Method | Metabolic Pathway | Refs. |
|---|---|---|---|---|---|---|---|
| 1 | 1-Methylhistamine | up | Urine | Rat | LC-MS | Histidine metabolism | [79] |
| 2 | alpha-ketoglutaric acid | down | Urine | Rat | NMR | TCA cycle | [80] |
| 3 | Carnitine | up | Blood | Mice | LC-MS | Lipid metabolism | [69] |
| 4 | Citric acid | down | Urine | Rat | LC-MS, NMR | TCA cycle | [78,79,80] |
| 5 | Cortisol | down | Urine | Rat | LC-MS | Lipid metabolism | [79] |
| 6 | Fumarate | down | Urine | Rat | NMR | TCA cycle | [80] |
| 7 | Isocitric acid | down | Urine | Rat | LC-MS | TCA cycle | [79] |
| 8 | Isoleucine | down | Blood | Mice | LC-MS | BCAA metabolism | [69,70,71] |
| 9 | Leucine | down | Blood | Mice | LC-MS | BCAA metabolism | [70,71] |
| 10 | Malic acid | down | Urine | Mice | LC-MS | TCA cycle | [81] |
| 11 | N-carbamoyl-β-alanine | down | Urine | Rat | NMR | Uracil metabolism | [77,78] |
| 12 | Pantothenic acid | down | Urine | Mice | LC-MS | CoA biosynthesis | [81] |
| 13 | Phenylalanine | up | Urine | Rat | LC-MS | Phenylalanine, tyrosine metabolism | [79] |
| 14 | Sphingosine | down | Urine | Rat | LC-MS | Sphinganine metabolsim | [78] |
| 15 | Succinate | down | Urine | Rat | NMR | TCA cycle | [80] |
| 16 | Succinoadenosine | up | Urine | Rat | LC-MS | Purine metabolism | [78] |
| 17 | Valine | down | Blood | Mice | LC-MS | BCAA metabolism | [70,71] |
| 18 | Trimethylamine | up | Urine | Mice | LC-MS | Gut microbiota metabolism | [81] |
| 19 | Phenylacetylglycine | up | Urine | Mice | LC-MS | Gut microbiota metabolism | [81] |
| 20 | Indoxyl sulfate | up | Urine | Mice | LC-MS | Gut microbiota metabolism | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.W. Metabolomic Approaches to Investigate the Effect of Metformin: An Overview. Int. J. Mol. Sci. 2021, 22, 10275. https://doi.org/10.3390/ijms221910275
Kim HW. Metabolomic Approaches to Investigate the Effect of Metformin: An Overview. International Journal of Molecular Sciences. 2021; 22(19):10275. https://doi.org/10.3390/ijms221910275
Chicago/Turabian StyleKim, Hyun Woo. 2021. "Metabolomic Approaches to Investigate the Effect of Metformin: An Overview" International Journal of Molecular Sciences 22, no. 19: 10275. https://doi.org/10.3390/ijms221910275
APA StyleKim, H. W. (2021). Metabolomic Approaches to Investigate the Effect of Metformin: An Overview. International Journal of Molecular Sciences, 22(19), 10275. https://doi.org/10.3390/ijms221910275






