Structure and Formation Mechanism of Antimicrobial Peptides Temporin B- and L-Induced Tubular Membrane Protrusion
Abstract
1. Introduction
2. Materials and Methods
2.1. All-Atom Simulation Parameters and Protocol
2.2. Coarse-Gained Simulation
2.3. Simulation Analyses
2.3.1. Potential of Mean Force
2.3.2. Lipid Orientation Order
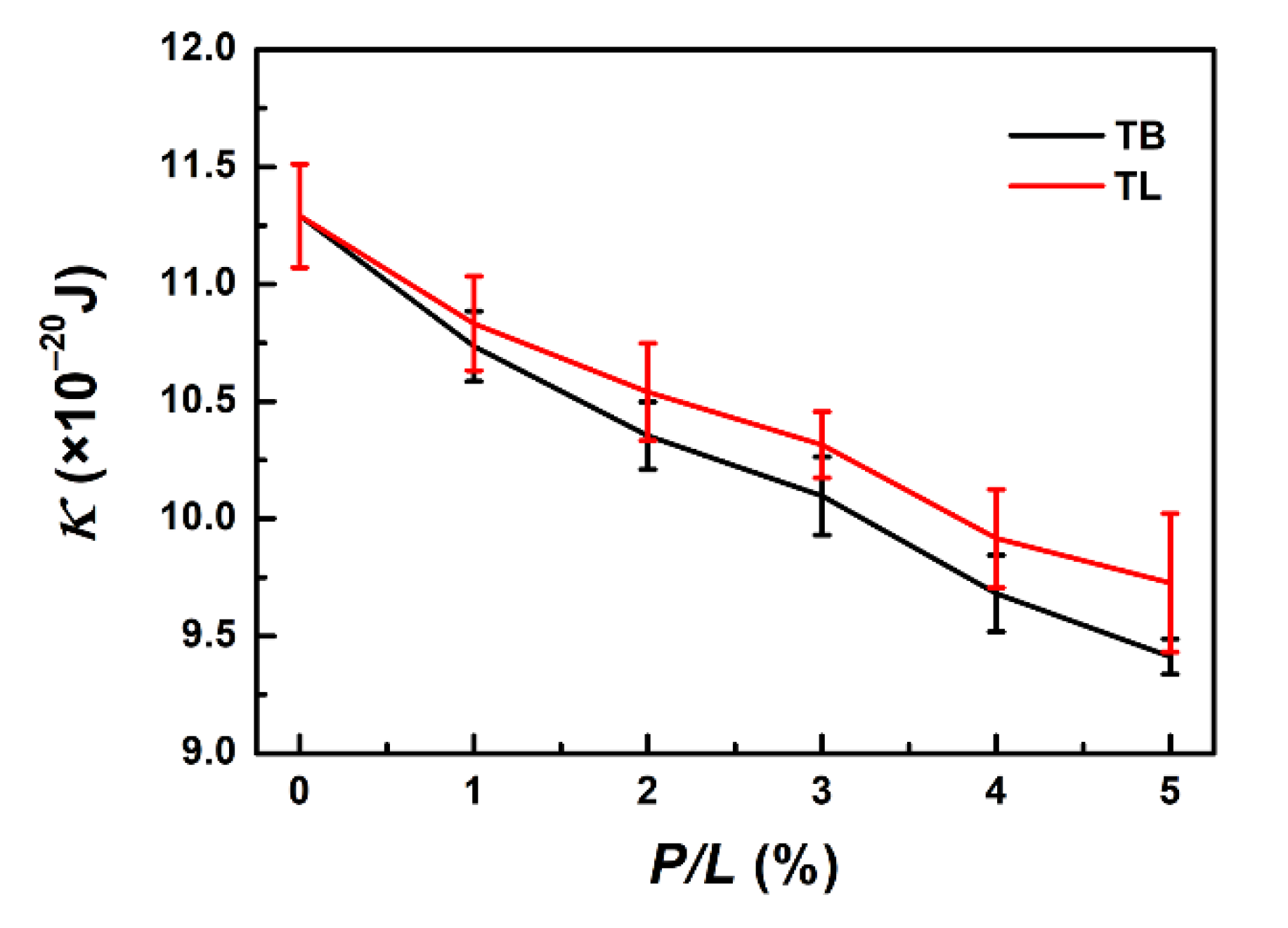
2.3.3. Membrane Bending Rigidity
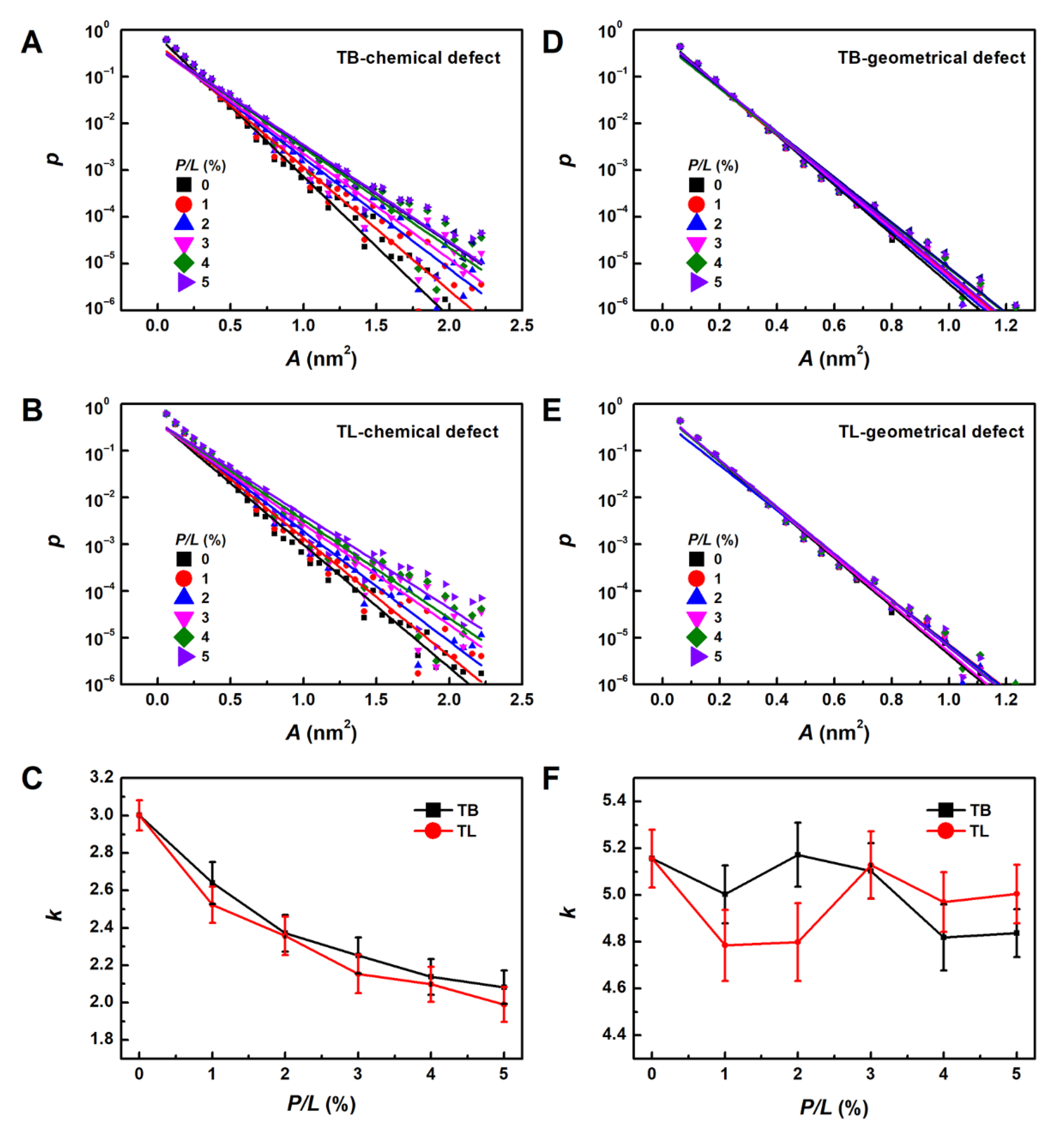
2.3.4. Lipid Packing Defect
3. Results
3.1. Temporin B and L Form Stable Helical Dimers at Water–Gas Interface
3.2. Temporin B and L Maintain α-Helix Conformations at Lipid Membrane Surface
3.3. Temporin B and L Squeeze out Lipids from Membrane
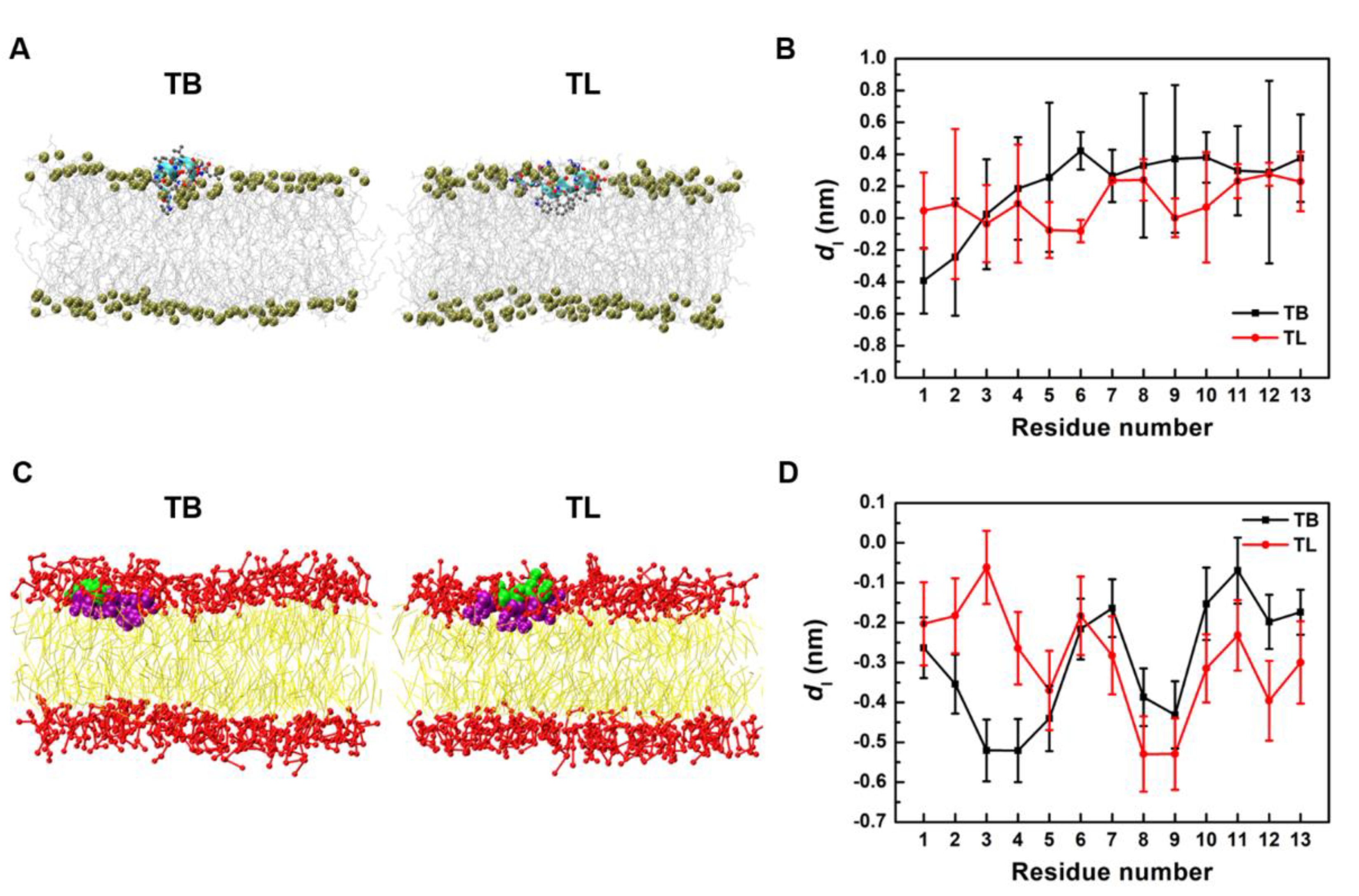
3.4. Temporin B and L Penetrate Shallowly into Membrane
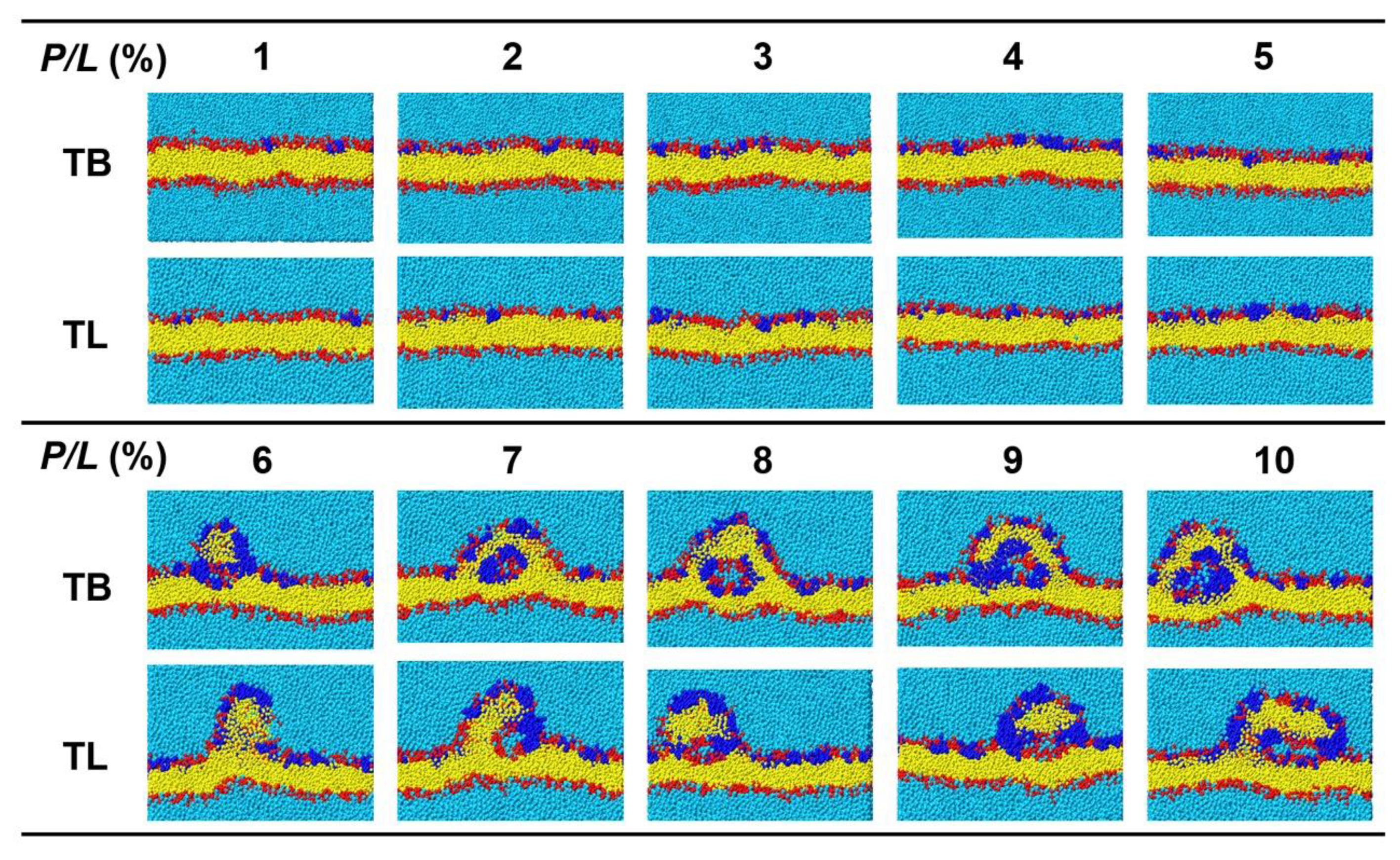
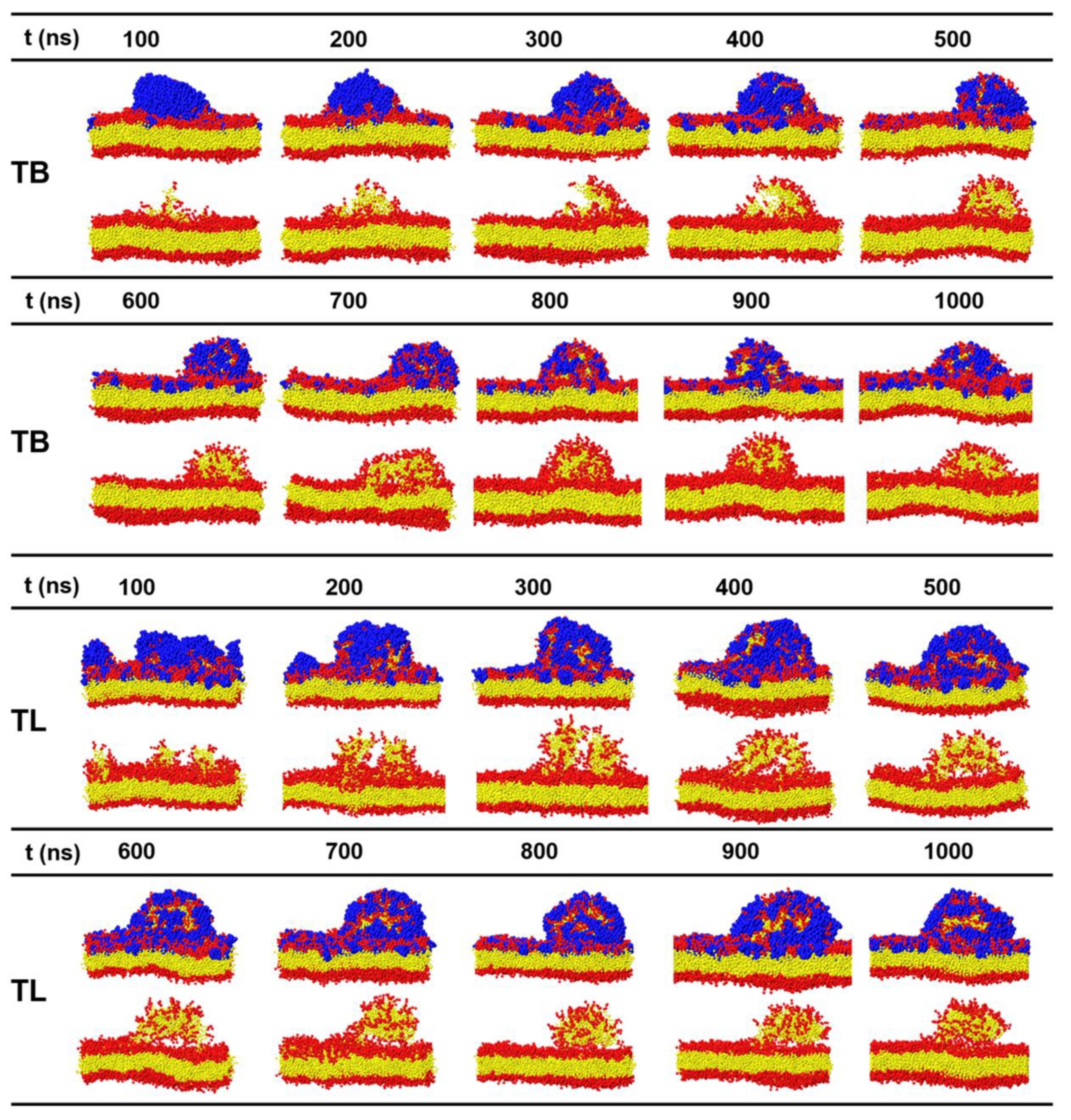
3.5. Temporin B and L Induce Tubule-like Membrane Protrusions
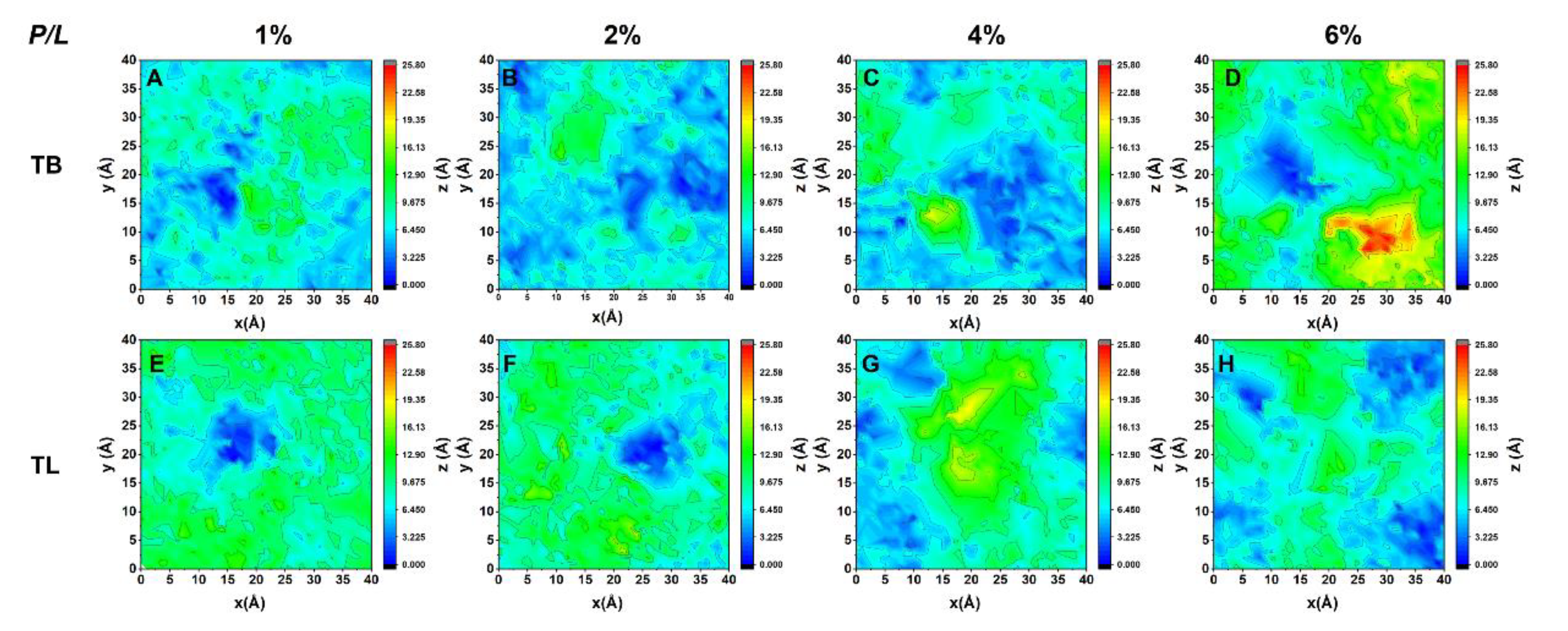
3.6. Temporin B and L Create Lipid Packing Defects
3.7. Binding of TB and TL Enhances the Flexibility of a Membrane
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.N.; Ferre, R.; Castanho, M.A. Antimicrobial peptides: Linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Lehrer, R. Cationic peptides: A new source of antibiotics. Trends Biotechnol. 1998, 16, 82–88. [Google Scholar] [CrossRef]
- Mookherjee, N.; Hancock, R.E. Cationic host defence peptides: Innate immune regulatory peptides as a novel approach for treating infections. Cell. Mol. Life Sci. 2007, 64, 922–933. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Powers, J.P.; Hancock, R.E. The relationship between peptide structure and antibacterial activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Murase, O.; Fujii, N.; Miyajima, K. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 1996, 35, 11361–11368. [Google Scholar] [CrossRef]
- Matsuzaki, K. Why and how are peptide–lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta 1999, 1462, 1–10. [Google Scholar] [CrossRef]
- He, K.; Ludtke, S.J.; Heller, W.T.; Huang, H.W. Mechanism of alamethicin insertion into lipid bilayers. Biophys. J. 1996, 71, 2669–2679. [Google Scholar] [CrossRef]
- Yang, L.; Harroun, T.A.; Weiss, T.M.; Ding, L.; Huang, H.W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001, 81, 1475–1485. [Google Scholar] [CrossRef]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Pouny, Y.; Rapaport, D.; Mor, A.; Nicolas, P.; Shai, Y. Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membranes. Biochemistry 1992, 31, 12416–12423. [Google Scholar] [CrossRef] [PubMed]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Simmaco, M.; Mignogna, G.; Canofeni, S.; Miele, R.; Mangoni, M.L.; Barra, D. Temporins, antimicrobial peptides from the european red frog rana temporaria. Eur. J. Biochem. 1996, 242, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Mangoni, M.L.; Rinaldi, A.C.; di Giulio, A.; Mignogna, G.; Bozzi, A.; Barra, D.; Simmaco, M. Structure-function relationships of temporins, small antimicrobialpeptides from amphibian skin. Eur. J. Biochem. 2000, 267, 1447–1454. [Google Scholar] [CrossRef]
- Rinaldi, A.C.; di Giulio, A.; Liberi, M.; Gualtieri, G.; Oratore, A.; Schinina, M.E.; Simmaco, M.; Bozzi, A. Effects of temporins on molecular dynamics and membrane permeabilization in lipid vesicles. J. Pept. Res. 2001, 58, 213–220. [Google Scholar] [CrossRef]
- Zhao, H.X.; Rinaldi, A.C.; di Giulio, A.; Simmaco, M.; Kinnunen, P.K.J. Interactions of the antimicrobial peptides temporins with model biomembranes. Comparison of temporins B and L. Biochemistry 2002, 41, 4425–4436. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Saugar, J.M.; Dellisanti, M.; Barra, D.; Simmaco, M.; Rivas, L. Temporins, small antimicrobial peptides with leishmanicidal activity. J. Biol. Chem. 2005, 280, 984–990. [Google Scholar] [CrossRef]
- Marcocci, M.E.; Amatore, D.; Villa, S.; Casciaro, B.; Aimola, P.; Franci, G.; Grieco, P.; Galdiero, M.; Palamara, A.T.; Mangoni, M.L.; et al. The amphibian antimicrobial peptide Temporin B inhibits in vitro herpes simplex virus 1 infection. Antimicrob. Agents Chemother. 2018, 62, e02367-17. [Google Scholar] [CrossRef]
- Rinaldi, A.C.; Mangoni, M.L.; Rufo, A.; Luzi, C.; Barra, D.; Zhao, H.X.; Kinnunen, P.K.J.; Bozzi, A.; di Giulio, A.; Simmaco, M. Temporin L: Antimicrobial, haemolytic and cytotoxic activities, and effects on membrane permeabilization in lipid vesicles. Biochem. J. 2002, 368, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Mangoni, M.L.; Papo, N.; Barra, D.; Simmaco, M.; Bozzi, A.; di Giulio, A.; Rinaldi, A. Effects of the antimicrobial peptide temporin L on cell morphology, membrane and viability of escherichia coli. Biochem. J. 2004, 380, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Carotenuto, A.; Malfi, S.; Saviello, M.R.; Campiglia, P.; Gomez-Monterrey, I.; Mangoni, M.L.; Gaddi, L.M.H.; Novellino, E.; Grieco, P. A different molecular mechanism underlying antimicrobial and hemolytic actions of temporins A and L. J. Med. Chem. 2008, 51, 2354–2362. [Google Scholar] [CrossRef] [PubMed]
- Saviello, M.R.; Malfi, S.; Campiglia, P.; Cavalli, A.; Grieco, P.; Novellino, E.; Carotenuto, A. New insight into the mechanism of action of the temporin antimicrobial peptides. Biochemistry 2010, 49, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.X.; Jutila, A.; Nurminen, T.; Wickstrom, S.A.; Keski-Oja, J.; Kinnunen, P.K.J. Binding of endostatin to phosphatidylserine-containing membranes and formation of amyloid-like fibers. Biochemistry 2005, 44, 2857–2863. [Google Scholar] [CrossRef] [PubMed]
- Kinnunen, P.K.; Domanov, Y.A.; Mattila, J.P.; Varis, T. Formation of lipid/peptide tubules by IAPP and temporin B on supported lipid membranes. Soft Matter 2015, 11, 9188–9200. [Google Scholar] [CrossRef] [PubMed]
- Domanov, Y.A.; Kinnunen, P.K. Antimicrobial peptides temporins B and L induce formation of tubular lipid protrusions from supported phospholipid bilayers. Biophys. J. 2006, 91, 4427–4439. [Google Scholar] [CrossRef]
- Lin, J.H.; Baumgaertner, A. Stability of a melittin pore in a lipid bilayer: A molecular dynamics study. Biophys. J. 2000, 78, 1714–1724. [Google Scholar] [CrossRef][Green Version]
- Leontiadou, H.; Mark, A.E.; Marrink, S.J. Antimicrobial peptides in action. J. Am. Chem. Soc. 2006, 128, 12156–12161. [Google Scholar] [CrossRef]
- Herce, H.D.; Garcia, A.E. Molecular dynamics simulations suggest a mechanism for translocation of the HIV-1 TAT peptide across lipid membranes. Proc. Natl. Acad. Sci. USA 2007, 104, 20805–20810. [Google Scholar] [CrossRef]
- Cirac, A.D.; Moiset, G.; Mika, J.T.; Kocer, A.; Salvador, P.; Poolman, B.; Marrink, S.J.; Sengupta, D. The molecular basis for antimicrobial activity of pore-forming cyclic peptides. Biophys. J. 2011, 100, 2422–2431. [Google Scholar] [CrossRef]
- Gao, L.; Fang, W. Effects of induced tension and electrostatic interactions on the mechanisms of antimicrobial peptide translocation across lipid bilayer. Soft Matter 2009, 5, 3312. [Google Scholar] [CrossRef]
- Chen, L.; Jia, N.; Gao, L.; Fang, W.; Golubovic, L. Effects of antimicrobial peptide revealed by simulations: Translocation, pore formation, membrane corrugation and euler buckling. Int. J. Mol. Sci. 2013, 14, 7932–7958. [Google Scholar] [CrossRef]
- Sun, H.; Chen, L.; Gao, L.; Fang, W. Nanodomain formation of ganglioside gm1 in lipid membrane: Effects of cholera toxin-mediated cross-linking. Langmuir 2015, 31, 9105–9114. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Gao, L.; Fang, W.; Golubovic, L. How the antimicrobial peptides kill bacteria: Computational physics insights. Commun. Comput. Phys. 2015, 11, 709–725. [Google Scholar] [CrossRef]
- Chen, L.; Li, X.; Gao, L.; Fang, W. Theoretical insight into the relationship between the structures of antimicrobial peptides and their actions on bacterial membranes. J. Phys. Chem. B 2015, 119, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wan, M.; Fu, L.; Zhang, S.; Wang, S.; Gao, L.; Fang, W. Peptide-lipid interaction sites affect vesicles’ responses to antimicrobial peptides. Biophys. J. 2018, 115, 1518–1529. [Google Scholar] [CrossRef]
- Zhang, S.; Fu, L.; Wan, M.; Song, J.; Gao, L.; Fang, W. Peripheral antimicrobial peptide gomesin induces membrane protrusion, folding, and laceration. Langmuir 2019, 35, 13233–13242. [Google Scholar] [CrossRef]
- Fu, L.; Wan, M.; Zhang, S.; Gao, L.; Fang, W. Polymyxin B loosens lipopolysaccharide bilayer but stiffens phospholipid bilayer. Biophys. J. 2020, 118, 138–150. [Google Scholar] [CrossRef]
- Van der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Pronk, S.; Pall, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; van Gunsteren, W.F. Definition and testing of the gromos force-field versions 54a7 and 54b7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Kukol, A. Lipid models for united-atom molecular dynamics simulations of proteins. J. Chem. Theory Comput. 2009, 5, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, A.; Saravanan, R.; Mohanram, H.; Mangoni, M.L.; Bhattacharjya, S. NMR structures and interactions of temporin-1tl and temporin-1tb with lipopolysaccharide micelles: Mechanistic insights into outer membrane permeabilization and synergistic activity. J. Biol. Chem. 2011, 286, 24394–24406. [Google Scholar] [CrossRef]
- Manzo, G.; Ferguson, P.M.; Hind, C.K.; Clifford, M.; Gustilo, V.B.; Ali, H.; Bansal, S.S.; Bui, T.T.; Drake, A.F.; Atkinson, R.A.; et al. Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities. Sci. Rep. 2019, 9, 10934. [Google Scholar] [CrossRef] [PubMed]
- Van der Spoel, D.; van Maaren, P.J.; Berendsen, H.J.C. A systematic study of water models for molecular simulation: Derivation of water models optimized for use with a reaction field. J. Chem. Phys. 1998, 108, 10220–10230. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh ewald: An Nlog(N) method for ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular-dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef]
- Monticelli, L.; Kandasamy, S.K.; Periole, X.; Larson, R.G.; Tieleman, D.P.; Marrink, S.-J. The MARTINI coarse-grained force field: Extension to proteins. J. Chem. Theory Comput. 2008, 4, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Kästner, J. Umbrella sampling. Wires Comput. Mol. Sci. 2011, 1, 932–942. [Google Scholar] [CrossRef]
- Kumar, S.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A.; Rosenberg, J.M. The weighted histogram analysis method for free-energy calculations on biomolecules.1. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Watson, M.C.; Brandt, E.G.; Welch, P.M.; Brown, F.L. Determining biomembrane bending rigidities from simulations of modest size. Phys. Rev. Lett. 2012, 109, 028102. [Google Scholar] [CrossRef]
- Levine, Z.A.; Venable, R.M.; Watson, M.C.; Lerner, M.G.; Shea, J.E.; Pastor, R.W.; Brown, F.L. Determination of biomembrane bending moduli in fully atomistic simulations. J. Am. Chem. Soc. 2014, 136, 13582–13585. [Google Scholar] [CrossRef]
- Vanni, S.; Vamparys, L.; Gautier, R.; Drin, G.; Etchebest, C.; Fuchs, P.F.; Antonny, B. Amphipathic lipid packing sensor motifs: Probing bilayer defects with hydrophobic residues. Biophys. J. 2013, 104, 575–584. [Google Scholar] [CrossRef]
- Vamparys, L.; Gautier, R.; Vanni, S.; Bennett, W.F.; Tieleman, D.P.; Antonny, B.; Etchebest, C.; Fuchs, P.F. Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature. Biophys. J. 2013, 104, 585–593. [Google Scholar] [CrossRef]
- Arouri, A.; Kiessling, V.; Tamm, L.; Dathe, M.; Blume, A. Morphological changes induced by the action of antimicrobial peptides on supported lipid bilayers. J. Phys. Chem. B 2011, 115, 158–167. [Google Scholar] [CrossRef]
- Oren, Z.; Lerman, J.C.; Gudmundsson, G.H.; Agerberth, B.; Shai, Y. Structure and organization of the human antimicrobial peptide ll-37 in phospholipid membranes: Relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 1999, 341, 501–513. [Google Scholar] [CrossRef]
- Feder, R.; Dagan, A.; Mor, A. Structure-activity relationship study of antimicrobial dermaseptin s4 showing the consequences of peptide oligomerization on selective cytotoxicity. J. Biol. Chem. 2000, 275, 4230–4238. [Google Scholar] [CrossRef] [PubMed]
- Zai, Y.; Xi, X.; Ye, Z.; Ma, C.; Zhou, M.; Chen, X.; Siu, S.W.I.; Chen, T.; Wang, L.; Kwok, H.F. Aggregation and its influence on the bioactivities of a novel antimicrobial peptide, temporin-pf, and its analogues. Int. J. Mol. Sci. 2021, 22, 4509. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Ma, M.; Shao, Z.; Zhang, J.; Fu, L.; Li, X.; Fang, W.; Gao, L. Structure and Formation Mechanism of Antimicrobial Peptides Temporin B- and L-Induced Tubular Membrane Protrusion. Int. J. Mol. Sci. 2021, 22, 11015. https://doi.org/10.3390/ijms222011015
Zhang S, Ma M, Shao Z, Zhang J, Fu L, Li X, Fang W, Gao L. Structure and Formation Mechanism of Antimicrobial Peptides Temporin B- and L-Induced Tubular Membrane Protrusion. International Journal of Molecular Sciences. 2021; 22(20):11015. https://doi.org/10.3390/ijms222011015
Chicago/Turabian StyleZhang, Shan, Ming Ma, Zhuang Shao, Jincheng Zhang, Lei Fu, Xiangyuan Li, Weihai Fang, and Lianghui Gao. 2021. "Structure and Formation Mechanism of Antimicrobial Peptides Temporin B- and L-Induced Tubular Membrane Protrusion" International Journal of Molecular Sciences 22, no. 20: 11015. https://doi.org/10.3390/ijms222011015
APA StyleZhang, S., Ma, M., Shao, Z., Zhang, J., Fu, L., Li, X., Fang, W., & Gao, L. (2021). Structure and Formation Mechanism of Antimicrobial Peptides Temporin B- and L-Induced Tubular Membrane Protrusion. International Journal of Molecular Sciences, 22(20), 11015. https://doi.org/10.3390/ijms222011015




