Biosynthesis and Roles of Salicylic Acid in Balancing Stress Response and Growth in Plants
Abstract
:1. Introduction
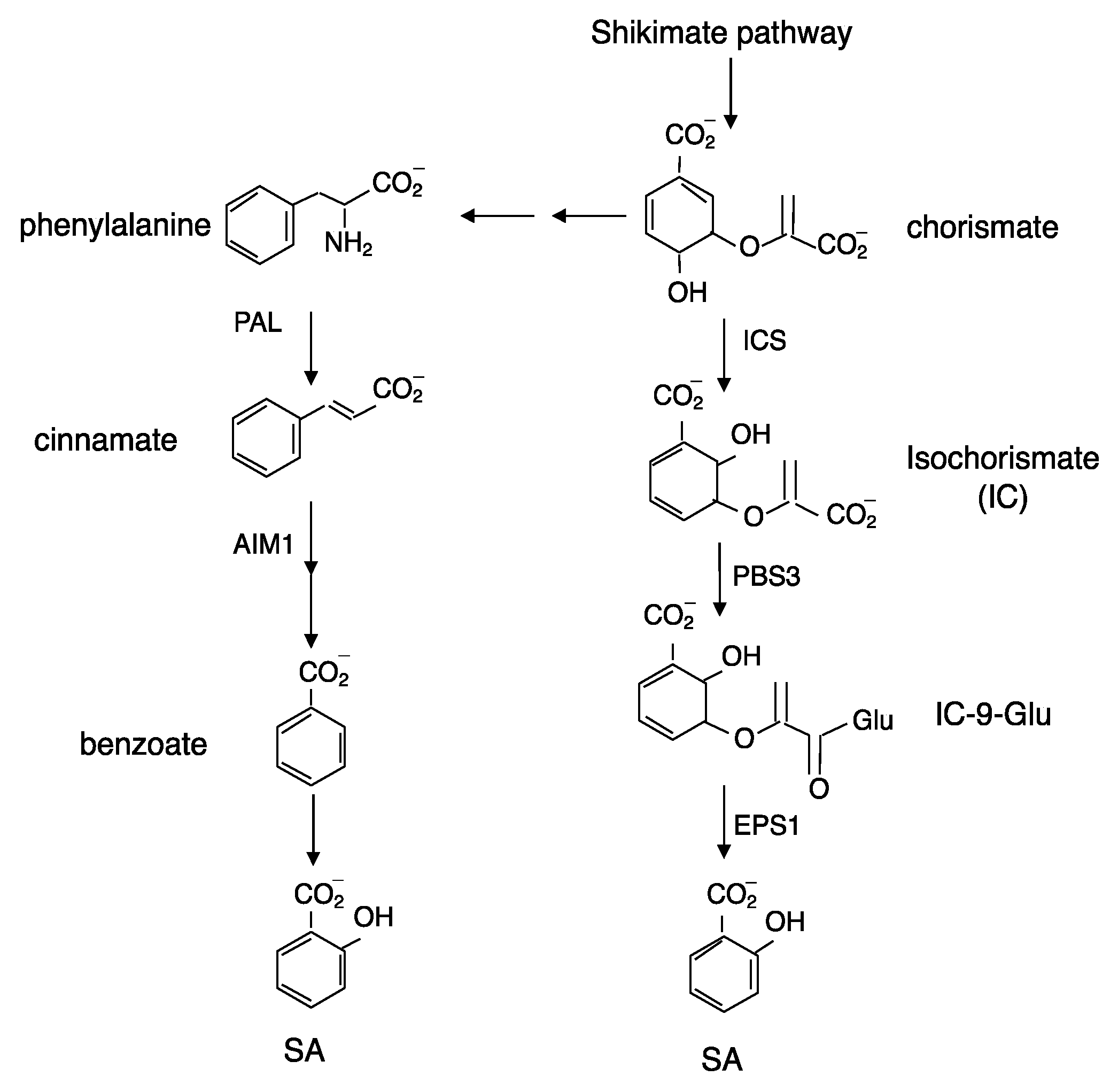
2. Biosynthesis of SA in Plants
3. Defense Crosstalk with Auxin in Plants
4. SA Regulation of Auxin Biosynthesis and Metabolism
5. SA Regulation of Auxin Transport
6. SA Regulation of Auxin Signaling and Response
7. Summary and Prospect
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raskin, I.; Ehmann, A.; Melander, W.R.; Meeuse, B.J.D. Salicylic acid: A natural inducer of heat production in Arum lilies. Science 1987, 237, 1601–1602. [Google Scholar] [CrossRef] [PubMed]
- White, R.F. Acetylasalicylic acid (Aspirin) inducesresistance to tobacco mosaic virus in tobacco. Virology 1979, 99, 410–412. [Google Scholar] [CrossRef]
- Yalpani, N.; Silverman, P.; Wilson, T.M.; Kleier, D.A.; Raskin, I. Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 1991, 3, 809–818. [Google Scholar] [PubMed] [Green Version]
- Metraux, J.-P.; Signer, H.; Ryals, J.A.; Ward, E.; Wyss-Benz, M.; Gaudin, J.; Raschorf, K.; Schmid, E.; Blum, W.; Inverardi, B. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 1990, 250, 1004–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malamy, J.; Carr, J.P.; Klessig, D.F.; Raskin, I. Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 1990, 250, 1002–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef]
- Chen, Z.; Zheng, Z.; Huang, J.; Lai, Z.; Fan, B. Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 2009, 4, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.K. PBS3 and EPS1 Complete Salicylic Acid Biosynthesis from Isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef] [Green Version]
- Rekhter, D.; Ludke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Klambt, H.D. Conversion in plants of benzoic acid to salicylic acid and its b-D-glucoside. Nature 1962, 196, 491. [Google Scholar] [CrossRef]
- Leon, J.; Shulaev, V.; Yalpani, N.; Lawton, M.A.; Raskin, I. Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc. Natl. Acad. Sci. USA 1995, 92, 10413–10417. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Zhao, H.; Ruan, W.; Deng, M.; Wang, F.; Peng, J.; Luo, J.; Chen, Z.; Yi, K. ABNORMAL INFLORESCENCE MERISTEM1 Functions in Salicylic Acid Biosynthesis to Maintain Proper Reactive Oxygen Species Levels for Root Meristem Activity in Rice. Plant Cell 2017, 29, 560–574. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [Green Version]
- Serino, L.; Reimmann, C.; Baur, H.; Beyeler, M.; Visca, P.; Haas, D. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol. Gen. Genet. 1995, 249, 217–228. [Google Scholar] [CrossRef]
- Garcion, C.; Lohmann, A.; Lamodiere, E.; Catinot, J.; Buchala, A.; Doermann, P.; Metraux, J.P. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 2008, 147, 1279–1287. [Google Scholar] [CrossRef] [Green Version]
- Jagadeeswaran, G.; Raina, S.; Acharya, B.R.; Maqbool, S.B.; Mosher, S.L.; Appel, H.M.; Schultz, J.C.; Klessig, D.F.; Raina, R. Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 2007, 51, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.W.; Lu, H.; Jung, H.W.; Greenberg, J.T. A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. Mol. Plant Microbe Interact. 2007, 20, 1192–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobuta, K.; Okrent, R.A.; Stoutemyer, M.; Rodibaugh, N.; Kempema, L.; Wildermuth, M.C.; Innes, R.W. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007, 144, 1144–1156. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Qualley, A.; Fan, B.; Dudareva, N.; Chen, Z. An Important Role of a BAHD Acyl Transferase-like Protein in Plant Innate Immunity. Plant J. 2009, 57, 1040–1053. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Wang, B.; Aryal, B.; Garcion, C.; Abou-Mansour, E.; Heck, S.; Geisler, M.; Mauch, F.; Nawrath, C.; Metraux, J.P. Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol. 2013, 162, 1815–1821. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, K.; Motomura, Y.; Yagi, Y.; Nomura, H.; Kikuchi, S.; Nakai, M.; Shiina, T. Chloroplast envelope localization of EDS5, an essential factor for salicylic acid biosynthesis in Arabidopsis thaliana. Plant Signal. Behav. 2013, 8, e23603. [Google Scholar] [CrossRef] [Green Version]
- Shine, M.B.; Yang, J.W.; El-Habbak, M.; Nagyabhyru, P.; Fu, D.Q.; Navarre, D.; Ghabrial, S.; Kachroo, P.; Kachroo, A. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 2016, 212, 627–636. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Iyer, S.; Caplan, A.; Klessig, D.F.; Fan, B. Differential accumulation of salicylic acid and salicylic acid-sensitive catalase in different rice tissues. Plant Physiol. 1997, 114, 193–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoo, S.; Inoue, S.; Suzuki, N.; Amakawa, N.; Matsui, H.; Nakagami, H.; Takahashi, A.; Arai, R.; Katou, S. Comparative analysis of plant isochorismate synthases reveals structural mechanisms underlying their distinct biochemical properties. Biosci. Rep. 2018, 38, BSR20171457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staswick, P.E.; Tiryaki, I.; Rowe, M.L. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 2002, 14, 1405–1415. [Google Scholar] [CrossRef] [Green Version]
- Terol, J.; Domingo, C.; Talon, M. The GH3 family in plants: Genome wide analysis in rice and evolutionary history based on EST analysis. Gene 2006, 371, 279–290. [Google Scholar] [CrossRef] [PubMed]
- van Butselaar, T.; Van den Ackerveken, G. Salicylic Acid Steers the Growth-Immunity Tradeoff. Trends Plant Sci. 2020, 25, 566–576. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [Green Version]
- Kunkel, B.N.; Johnson, J.M.B. Auxin Plays Multiple Roles during Plant-Pathogen Interactions. Cold Spring Harb. Perspect. Biol. 2021, 13, a040022. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Agnew, J.L.; Cohen, J.D.; He, P.; Shan, L.; Sheen, J.; Kunkel, B.N. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA 2007, 104, 20131–20136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, F.; Wu, S.; Sun, W.; Coaker, G.; Kunkel, B.; He, P.; Shan, L. The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 2013, 162, 1018–1029. [Google Scholar] [CrossRef] [Green Version]
- Glickmann, E.; Gardan, L.; Jacquet, S.; Hussain, S.; Elasri, M.; Petit, A.; Dessaux, Y. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol. Plant Microbe Interact. 1998, 11, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Kidd, B.N.; Kadoo, N.Y.; Dombrecht, B.; Tekeoglu, M.; Gardiner, D.M.; Thatcher, L.F.; Aitken, E.A.; Schenk, P.M.; Manners, J.M.; Kazan, K. Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Mol. Plant Microbe Interact. 2011, 24, 733–748. [Google Scholar] [CrossRef] [Green Version]
- Yamada, T. The role of auxin in plant-disease development. Annu. Rev. Phytopathol. 1993, 31, 253–273. [Google Scholar] [CrossRef] [PubMed]
- Mutka, A.M.; Fawley, S.; Tsao, T.; Kunkel, B.N. Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J. 2013, 74, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Zhu, B.; Liu, W.; Cheng, X.; Lin, D.; He, C.; Shi, H. Heat shock protein 90 co-chaperone modules fine-tune the antagonistic interaction between salicylic acid and auxin biosynthesis in cassava. Cell Rep. 2021, 34, 108717. [Google Scholar] [CrossRef] [PubMed]
- Armengot, L.; Caldarella, E.; Marques-Bueno, M.M.; Martinez, M.C. The Protein Kinase CK2 Mediates Cross-Talk between Auxin- and Salicylic Acid-Signaling Pathways in the Regulation of PINOID Transcription. PLoS ONE 2016, 11, e0157168. [Google Scholar] [CrossRef] [Green Version]
- Armengot, L.; Marques-Bueno, M.M.; Jaillais, Y. Regulation of polar auxin transport by protein and lipid kinases. J. Exp. Bot. 2016, 67, 4015–4037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armengot, L.; Marques-Bueno, M.M.; Soria-Garcia, A.; Muller, M.; Munne-Bosch, S.; Martinez, M.C. Functional interplay between protein kinase CK2 and salicylic acid sustains PIN transcriptional expression and root development. Plant J. 2014, 78, 411–423. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu. Rev. Plant. Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.M.; Liu, W.C.; Lu, Y.T. CATALASE2 Coordinates SA-Mediated Repression of Both Auxin Accumulation and JA Biosynthesis in Plant Defenses. Cell Host Microbe 2017, 21, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Silva, H.; Klessig, D.F. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 1993, 262, 1883–1886. [Google Scholar] [CrossRef]
- Durner, J.; Klessig, D.F. Salicylic acid is a modulator of tobacco and mammalian catalases. J. Biol. Chem. 1996, 271, 28492–28501. [Google Scholar] [CrossRef] [Green Version]
- Park, J.E.; Park, J.Y.; Kim, Y.S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.Y.; Kim, J.; Lee, Y.H.; Park, C.M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef] [Green Version]
- Westfall, C.S.; Sherp, A.M.; Zubieta, C.; Alvarez, S.; Schraft, E.; Marcellin, R.; Ramirez, L.; Jez, J.M. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 13917–13922. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, Q.; Li, Z.; Staswick, P.E.; Wang, M.; Zhu, Y.; He, Z. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007, 145, 450–464. [Google Scholar] [CrossRef] [Green Version]
- LeClere, S.; Tellez, R.; Rampey, R.A.; Matsuda, S.P.; Bartel, B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002, 277, 20446–20452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Shen, H.; Wang, M.; Li, Q.; He, Z. Salicyloyl-aspartate synthesized by the acetyl-amido synthetase GH3.5 is a potential activator of plant immunity in Arabidopsis. Acta Biochim. Biophys. Sin. 2013, 45, 827–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okrent, R.A.; Brooks, M.D.; Wildermuth, M.C. Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 2009, 284, 9742–9754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Adamowski, M.; Qi, L.; Alotaibi, S.S.; Friml, J. PIN-mediated polar auxin transport regulations in plant tropic responses. New Phytol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.R. A Fresh Look at the Role of Auxin in PIN Trafficking. Plant Physiol. 2016, 172, 821–822. [Google Scholar] [CrossRef]
- Zwiewka, M.; Bilanovicova, V.; Seifu, Y.W.; Nodzynski, T. The Nuts and Bolts of PIN Auxin Efflux Carriers. Front. Plant. Sci. 2019, 10, 985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhonukshe, P.; Tanaka, H.; Goh, T.; Ebine, K.; Mahonen, A.P.; Prasad, K.; Blilou, I.; Geldner, N.; Xu, J.; Uemura, T.; et al. Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 2008, 456, 962–966. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Tejos, R.; Beck, M.; Himschoot, E.; Li, H.; Robatzek, S.; Vanneste, S.; Friml, J. Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proc. Natl. Acad. Sci. USA 2013, 110, 7946–7951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Hu, T.; Yan, X.; Meng, T.; Wang, Y.; Wang, Q.; Zhang, X.; Gu, Y.; Sanchez-Rodriguez, C.; Gadeyne, A.; et al. Differential Regulation of Clathrin and Its Adaptor Proteins during Membrane Recruitment for Endocytosis. Plant Physiol. 2016, 171, 215–229. [Google Scholar] [CrossRef]
- Habets, M.E.J.; Offringa, R. PIN-driven polar auxin transport in plant developmental plasticity: A key target for environmental and endogenous signals. New Phytol. 2014, 203, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Marhava, P.; Bassukas, A.E.L.; Zourelidou, M.; Kolb, M.; Moret, B.; Fastner, A.; Schulze, W.X.; Cattaneo, P.; Hammes, U.Z.; Schwechheimer, C.; et al. A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 2018, 558, 297–300. [Google Scholar] [CrossRef]
- Zourelidou, M.; Absmanner, B.; Weller, B.; Barbosa, I.C.; Willige, B.C.; Fastner, A.; Streit, V.; Port, S.A.; Colcombet, J.; de la Fuente van Bentem, S.; et al. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. Elife 2014, 3, e02860. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Abas, M.; Verstraeten, I.; Glanc, M.; Molnar, G.; Hajny, J.; Lasak, P.; Petrik, I.; Russinova, E.; Petrasek, J.; et al. Salicylic Acid Targets Protein Phosphatase 2A to Attenuate Growth in Plants. Curr. Biol. 2020, 30, 381–395.e8. [Google Scholar] [CrossRef] [Green Version]
- Ke, M.; Ma, Z.; Wang, D.; Sun, Y.; Wen, C.; Huang, D.; Chen, Z.; Yang, L.; Tan, S.; Li, R.; et al. Salicylic acid regulates PIN2 auxin transporter hyperclustering and root gravitropic growth via Remorin-dependent lipid nanodomain organisation in Arabidopsis thaliana. New Phytol. 2021, 229, 963–978. [Google Scholar] [CrossRef] [PubMed]
- Kleine-Vehn, J.; Leitner, J.; Zwiewka, M.; Sauer, M.; Abas, L.; Luschnig, C.; Friml, J. Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 2008, 105, 17812–17817. [Google Scholar] [CrossRef] [Green Version]
- Duggan, J.; Jamal, G.; Tilley, M.; Davis, B.; McKenzie, G.; Vere, K.; Somekh, M.G.; O’Shea, P.; Harris, H. Functional imaging of microdomains in cell membranes. Eur. Biophys. J. 2008, 37, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Jing, Y.; Xiao, J.; Zhang, Y.; Zhu, Y.; Julian, R.; Lin, J. Membrane microdomains and the cytoskeleton constrain AtHIR1 dynamics and facilitate the formation of an AtHIR1-associated immune complex. Plant J. 2017, 90, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Sun, Y.; Ma, Z.; Ke, M.; Cui, Y.; Chen, Z.; Chen, C.; Ji, C.; Tran, T.M.; Yang, L.; et al. Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc. Natl. Acad. Sci. USA 2019, 116, 21274–21284. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454–1467.e15. [Google Scholar] [CrossRef]
- Manohar, M.; Tian, M.; Moreau, M.; Park, S.W.; Choi, H.W.; Fei, Z.; Friso, G.; Asif, M.; Manosalva, P.; von Dahl, C.C.; et al. Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 2014, 5, 777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Despres, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Cheng, Y.T.; Qu, N.; Zhao, Q.; Bi, D.; Li, X. Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 2006, 48, 647–656. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [Green Version]
- Quint, M.; Gray, W.M. Auxin signaling. Curr. Opin. Plant Biol. 2006, 9, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Paciorek, T.; Friml, J. Auxin signaling. J. Cell Sci. 2006, 119, 1199–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dharmasiri, N.; Estelle, M. Auxin signaling and regulated protein degradation. Trends Plant Sci. 2004, 9, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Caruana, J.C.; Dhar, N.; Raina, R. Overexpression of Arabidopsis microRNA167 induces salicylic acid-dependent defense against Pseudomonas syringae through the regulation of its targets ARF6 and ARF8. Plant Direct 2020, 4, e00270. [Google Scholar] [CrossRef]
- Wang, Y.; Li, K.; Chen, L.; Zou, Y.; Liu, H.; Tian, Y.; Li, D.; Wang, R.; Zhao, F.; Ferguson, B.J.; et al. MicroRNA167-Directed Regulation of the Auxin Response Factors GmARF8a and GmARF8b Is Required for Soybean Nodulation and Lateral Root Development. Plant Physiol. 2015, 168, 984–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.H.; Han, S.J.; Yoon, E.K.; Lee, W.S. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006, 34, 1892–1899. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ru, P.; Xu, L.; Ma, H.; Huang, H. Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res. 2006, 16, 457–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dempsey, D.A.; Klessig, D.F. How does the multifaceted plant hormone salicylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol. 2017, 15, 23. [Google Scholar] [CrossRef] [Green Version]
- Pokotylo, I.; Kravets, V.; Ruelland, E. Salicylic Acid Binding Proteins (SABPs): The Hidden Forefront of Salicylic Acid Signalling. Int. J. Mol. Sci. 2019, 20, 4377. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, Q.; Hu, H.; Fan, B.; Zhu, C.; Chen, Z. Biosynthesis and Roles of Salicylic Acid in Balancing Stress Response and Growth in Plants. Int. J. Mol. Sci. 2021, 22, 11672. https://doi.org/10.3390/ijms222111672
Zhong Q, Hu H, Fan B, Zhu C, Chen Z. Biosynthesis and Roles of Salicylic Acid in Balancing Stress Response and Growth in Plants. International Journal of Molecular Sciences. 2021; 22(21):11672. https://doi.org/10.3390/ijms222111672
Chicago/Turabian StyleZhong, Qinling, Hongliang Hu, Baofang Fan, Cheng Zhu, and Zhixiang Chen. 2021. "Biosynthesis and Roles of Salicylic Acid in Balancing Stress Response and Growth in Plants" International Journal of Molecular Sciences 22, no. 21: 11672. https://doi.org/10.3390/ijms222111672
APA StyleZhong, Q., Hu, H., Fan, B., Zhu, C., & Chen, Z. (2021). Biosynthesis and Roles of Salicylic Acid in Balancing Stress Response and Growth in Plants. International Journal of Molecular Sciences, 22(21), 11672. https://doi.org/10.3390/ijms222111672





