Auxin Metabolite Profiling in Isolated and Intact Plant Nuclei
Abstract
:1. Introduction
2. Results
2.1. Comparison of DC and FCM as Nucleus Isolation Methods
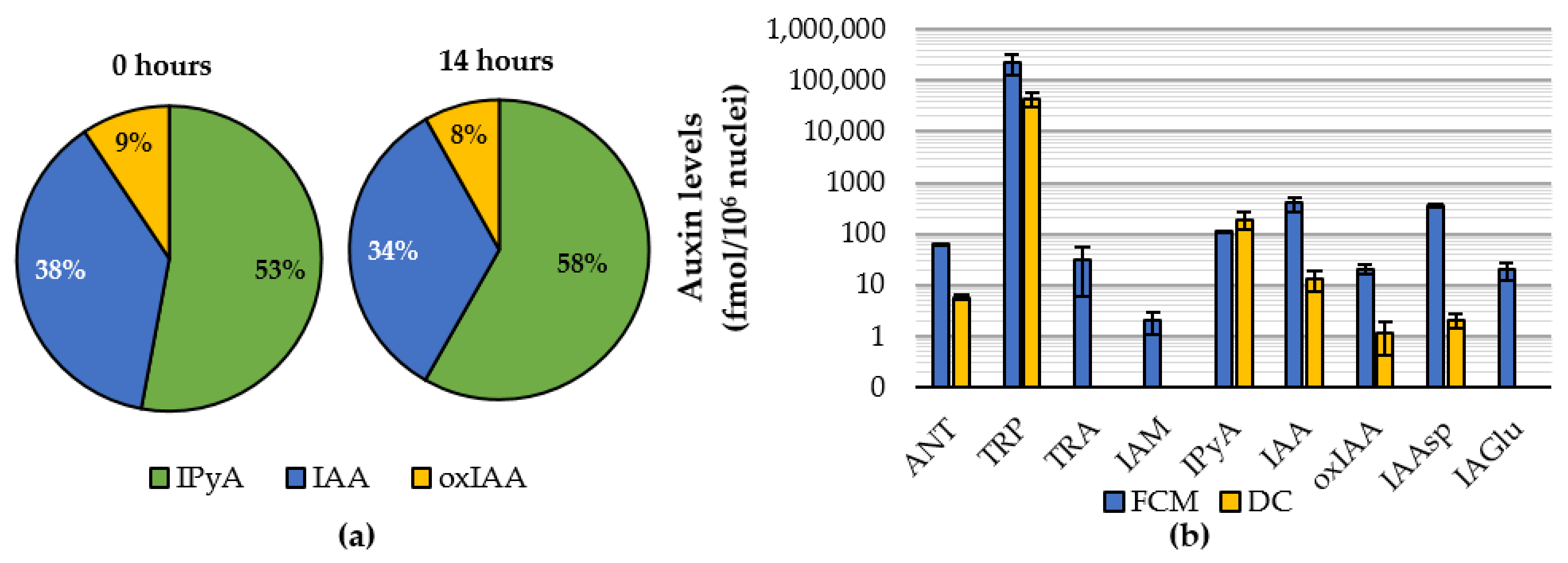
2.2. Auxinome of Isolated Nuclei
2.3. Feeding Experiments with Indole and IAA
3. Discussion
4. Materials and Methods
4.1. Cultivation of Cell Lines
4.2. Protoplast Preparation
4.3. Nuclei Isolation by Differential Centrifugation
4.4. Nuclei Isolation by FCM
4.5. Microscopy Analysis
4.6. Immunoblot Analysis
4.7. Auxin Purification and Determination
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brumos, J.; Robles, L.M.; Yun, J.; Vu, T.C.; Jackson, S.; Alonso, J.M.; Stepanova, A.N. Local Auxin Biosynthesis is a key regulator of plant development. Dev. Cell 2018, 47, 306–318. [Google Scholar] [CrossRef] [Green Version]
- Di Mambro, R.; De Ruvo, M.; Pacifici, E.; Salvi, E.; Sozzani, R.; Benfey, P.N.; Busch, W.; Novak, O.; Ljung, K.; Di Paola, L.; et al. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2017, 114, E7641–E7649. [Google Scholar] [CrossRef] [Green Version]
- Grones, P.; Majda, M.; Doyle, S.M.; Van Damme, D.; Robert, S. Fluctuating auxin response gradients determine pavement cell-shape acquisition. Proc. Natl. Acad. Sci. USA 2020, 117, 16027–16034. [Google Scholar] [CrossRef]
- Skalický, V.; Kubeš, M.; Napier, R.; Novák, O. Auxins and Cytokinins—The role of subcellular organization on homeostasis. Int. J. Mol. Sci. 2018, 19, 3115. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Dhonukshe, P.; Brewer, P.B.; Friml, J. Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell. Mol. Life Sci. 2006, 63, 2738–2754. [Google Scholar] [CrossRef]
- Barbez, E.; Kubeš, M.; Rolčík, J.; Béziat, C.; Pěnčík, A.; Wang, B.; Rosquete, M.R.; Zhu, J.; Dobrev, P.I.; Lee, Y.; et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 2012, 485, 119–122. [Google Scholar] [CrossRef] [Green Version]
- Dindas, J.; Scherzer, S.; Roelfsema, M.R.G.; Von Meyer, K.; Müller, H.M.; Al-Rasheid, K.A.S.; Palme, K.; Dietrich, P.; Becker, D.; Bennett, M.J.; et al. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Fendrych, M.; Akhmanova, M.; Merrin, J.; Glanc, M.; Hagihara, S.; Takahashi, K.; Uchida, N.; Torii, K.U.; Friml, J. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat. Plants 2018, 4, 453–459. [Google Scholar] [CrossRef]
- Retzer, K.; Singh, G.; Napier, R. It starts with TIRs. Nat. Plants 2018, 4, 410–411. [Google Scholar] [CrossRef]
- Simonini, S.; Mas, P.J.; Mas, C.M.V.S.; Østergaard, L.; Hart, D.J. Auxin sensing is a property of an unstructured domain in the Auxin Response Factor ETTIN of Arabidopsis thaliana. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Kubeš, M.; Napier, R. Non-canonical auxin signalling: Fast and curious. J. Exp. Bot. 2019, 70, 2609–2614. [Google Scholar] [CrossRef]
- Mravec, J.; Skůpa, P.; Bailly, A.; Hoyerová, K.; Křeček, P.; Bielach, A.; Petrášek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.-D.; et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef]
- Bosco, C.D.; Dovzhenko, A.; Liu, X.; Woerner, N.; Rensch, T.; Eismann, M.; Eimer, S.; Hegermann, J.; Paponov, I.A.; Ruperti, B.; et al. The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J. 2012, 71, 860–870. [Google Scholar] [CrossRef]
- Ding, Z.; Wang, B.; Moreno, I.; Dupláková, N.; Simon, S.; Carraro, N.; Reemmer, J.; Pěnčík, A.; Chen, X.; Tejos, R.; et al. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012, 3, 941. [Google Scholar] [CrossRef] [Green Version]
- Feraru, E.; Vosolsobě, S.; Feraru, M.I.; Petrášek, J.; Kleine-Vehn, J. Evolution and structural diversification of PILS Putative Auxin carriers in plants. Front. Plant Sci. 2012, 3, 227. [Google Scholar] [CrossRef] [Green Version]
- Ranocha, P.; Dima, O.; Nagy, R.; Felten, J.; Corratgé-Faillie, C.; Novák, O.; Morreel, K.; Lacombe, B.; Martinez, Y.; Pfrunder, S.; et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 2013, 4, 2625. [Google Scholar] [CrossRef]
- Middleton, A.M.; Dal Bosco, C.; Chlap, P.; Bensch, R.; Harz, H.; Ren, F.; Bergmann, S.; Wend, S.; Weber, W.; Hayashi, K.-I.; et al. Data-driven modeling of intracellular auxin fluxes indicates a dominant role of the er in controlling nuclear auxin uptake. Cell Rep. 2018, 22, 3044–3057. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef] [Green Version]
- Kriechbaumer, V.; Wang, P.; Hawes, C.; Abell, B.M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 2012, 70, 292–302. [Google Scholar] [CrossRef]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [Green Version]
- Casanova-Sáez, R.; Mateo-Bonmatí, E.; Ljung, K. Auxin metabolism in plants. Cold Spring Harb. Perspect. Biol. 2021, 13, a039867. [Google Scholar] [CrossRef]
- Jin, S.-H.; Ma, X.-M.; Kojima, M.; Sakakibara, H.; Wang, Y.W.; Hou, B.-K. Overexpression of glucosyltransferase UGT85A1 influences trans-zeatin homeostasis and trans-zeatin responses likely through O-glucosylation. Planta 2013, 237, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell Online 2005, 17, 616–627. [Google Scholar] [CrossRef] [Green Version]
- Cano, A.; Sánchez-García, A.B.; Albacete, A.; González-Bayón, R.; Justamante, M.S.; Ibáñez, S.; Acosta, M.; Pérez-Pérez, J.M. Enhanced conjugation of auxin by gh3 enzymes leads to poor adventitious rooting in carnation stem cuttings. Front. Plant Sci. 2018, 9, 566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Mambro, R.; Svolacchia, N.; Dello Ioio, R.; Pierdonati, E.; Salvi, E.; Pedrazzini, E.; Vitale, A.; Perilli, S.; Sozzani, R.; Benfey, P.N.; et al. The lateral root cap acts as an auxin sink that controls meristem size. Curr. Biol. 2019, 29, 1199–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez Carranza, A.P.; Singh, A.; Steinberger, K.; Panigrahi, K.; Palme, K.; Dovzhenko, A.; Dal Bosco, C. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Yang, Y.; Chen, S.; Ning, N.; Hu, H. Arabidopsis IAR4 modulates primary root growth under salt stress through ros-mediated modulation of auxin distribution. Front. Plant Sci. 2019, 10, 522. [Google Scholar] [CrossRef]
- Porco, S.; Pěnčík, A.; Rashed, A.; Voß, U.; Casanova-Sáez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R.; Swarup, R.; Swarup, K.; et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Lin, J.E.; Harris, C.; Campos Mastrotti Pereira, F.; Wu, F.; Blakeslee, J.J.; Peer, W.A. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, 11010–11015. [Google Scholar] [CrossRef] [Green Version]
- Novák, O.; Napier, R.; Ljung, K. Zooming in on plant hormone analysis: Tissue- and cell-specific approaches. Annu. Rev. Plant Biol. 2017, 68, 323–348. [Google Scholar] [CrossRef]
- Pertoft, H. Fractionation of cells and subcellular particles with Percoll. J. Biochem. Biophys. Methods 2000, 44, 1–30. [Google Scholar] [CrossRef]
- Folta, K.M.; Kaufman, L.S. Isolation of Arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat. Protoc. 2007, 1, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Tan, H.T.; Chung, M.C.M. Subcellular fractionation methods and strategies for proteomics. Proteomics 2010, 10, 3935–3956. [Google Scholar] [CrossRef]
- Saxena, P.K.; Fowke, L.C.; King, J. An efficient procedure for isolation of nuclei from plant protoplasts. Protoplasma 1985, 128, 184–189. [Google Scholar] [CrossRef]
- Zhang, H.-B.; Zhao, X.; Ding, X.; Paterson, A.H.; Wing, R.A. Preparation of megabase-size DNA from plant nuclei. Plant J. 1995, 7, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Vertommen, A.; Panis, B.; Swennen, R.; Carpentier, S.C. Challenges and solutions for the identification of membrane proteins in non-model plants. J. Proteomics 2011, 74, 1165–1181. [Google Scholar] [CrossRef]
- Zini, N.; Matteucci, A.; Squarzoni, S.; Galanzi, A.; Rizzoli, R.; Papa, S. Electron microscopy microsampling of isolated nuclei sorted by flow cytometry. Cytometry 1986, 7, 605–608. [Google Scholar] [CrossRef]
- Šimková, H.; Číhalíková, J.; Vrána, J.; Lysák, M.A.; Doležel, J. Preparation of HMW DNA from plant nuclei and chromosomes isolated from root tips. Biol. Plant. 2003, 46, 369–373. [Google Scholar] [CrossRef]
- Zhang, C.; Barthelson, R.A.; Lambert, G.M.; Galbraith, D.W. Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 2008, 147, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Deal, R.B.; Henikoff, S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 2010, 18, 1030–1040. [Google Scholar] [CrossRef] [Green Version]
- Deal, R.B.; Henikoff, S. The INTACT method for cell type–specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 2011, 6, 56–68. [Google Scholar] [CrossRef]
- Seifertová, D.; Klíma, P.; Pařezová, M.; Petrášek, J.; Zažímalová, E.; Opatrný, Z. Plant cell lines in cell morphogenesis research. Methods Mol. Biol. 2014, 1080, 215–229. [Google Scholar] [CrossRef]
- Gigli-Bisceglia, N.; Engelsdorf, T.; Hamann, T. Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell Mol. Life Sci. 2020, 77, 2049–2077. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Copeland, C. Nuclear extraction from Arabidopsis thaliana. Bio Protoc. 2012, 2, e306. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koláčková, V.; Perničková, K.; Vrána, J.; Duchoslav, M.; Jenkins, G.; Phillips, D.; Turkosi, E.; Šamajová, O.; Sedlářová, M.; Šamaj, J.; et al. Nuclear disposition of alien chromosome introgressions into wheat and rye using 3D-FISH. Int. J. Mol. Sci. 2019, 20, 4143. [Google Scholar] [CrossRef] [Green Version]
- Perničková, K.; Koláčková, V.; Lukaszewski, A.J.; Fan, C.; Vrána, J.; Duchoslav, M.; Jenkins, G.; Phillips, D.; Šamajová, O.; Sedlářová, M.; et al. Instability of alien chromosome introgressions in wheat associated with improper positioning in the nucleus. Int. J. Mol. Sci. 2019, 20, 1448. [Google Scholar] [CrossRef] [Green Version]
- Antoniadi, I.; Plačková, L.; Simonovik, B.; Doležal, K.; Turnbull, C.; Ljung, K.; Novák, O. Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. Plant Cell 2015, 27, 1955–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrovská, B.; Jeřábková, H.; Chamrád, I.; Vrána, J.; Lenobel, R.; Uřinovská, J.; Šebela, M.; Doležel, J. Proteomic analysis of barley cell nuclei purified by flow sorting. Cytogenet. Genome Res. 2014, 143, 78–86. [Google Scholar] [CrossRef]
- Novák, O.; Hényková, E.; Sairanen, I.; Kowalczyk, M.; Pospíšil, T.; Ljung, K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012, 72, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Polanská, L.; Vičánková, A.; Nováková, M.; Malbeck, J.; Dobrev, P.I.; Brzobohatý, B.; Vaňková, R.; Macháčková, I. Altered cytokinin metabolism affects cytokinin, auxin, and abscisic acid contents in leaves and chloroplasts, and chloroplast ultrastructure in transgenic tobacco. J. Exp. Bot. 2007, 58, 637–649. [Google Scholar] [CrossRef] [Green Version]
- Včelařová, L.; Skalický, V.; Chamrád, I.; Lenobel, R.; Kubeš, M.; Pěnčík, A.; Novák, O. Auxin metabolome profiling in the Arabidopsis endoplasmic reticulum using an optimised organelle isolation protocol. Int. J. Mol. Sci. 2021, 22, 9370. [Google Scholar] [CrossRef] [PubMed]
- Šafář, J.; Noa-Carrazana, J.C.; Vrána, J.; Bartoš, J.; Alkhimova, O.; Sabau, X.; Šimková, H.; Lheureux, F.; Caruana, M.-L.; Doležel, J.; et al. Creation of a BAC resource to study the structure and evolution of the banana (Musa balbisiana) genome. Genome 2004, 47, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C.R.; Somerville, S.C.; Ogren, W.L. Isolation of photosynthetically active protoplasts and chloroplastids from Arabidopsis thaliana. Plant Sci. Lett. 1981, 21, 89–96. [Google Scholar] [CrossRef]
- Robert, S.; Zouhar, J.; Carter, C.J.; Raikhel, N. Isolation of intact vacuoles from Arabidopsis rosette leaf–derived protoplasts. Nat. Protoc. 2007, 2, 259–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersson, S.V.; Johansson, A.I.; Kowalczyk, M.; Makoveychuk, A.; Wang, J.Y.; Moritz, T.; Grebe, M.; Benfey, P.N.; Sandberg, G.; Ljung, K. An Auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 2009, 21, 1659–1668. [Google Scholar] [CrossRef]
- McKeown, P.; Pendle, A.F.; Shaw, P.J. Preparation of arabidopsis nuclei and nucleoli. Methods Mol. Biol. 2008, 463, 67–75. [Google Scholar] [CrossRef]
- Tarkowská, D.; Novák, O.; Floková, K.; Tarkowski, P.; Turečková, V.; Grúz, J.; Rolčík, J.; Strnad, M. Quo vadis plant hormone analysis? Planta 2014, 240, 55–76. [Google Scholar] [CrossRef]
- Pěnčík, A.; Casanova-Sáez, R.; Pilařová, V.; Žukauskaitė, A.; Pinto, R.; Luis Micol, J.; Ljung, K.; Novák, O. Ultra-rapid auxin metabolite profiling for high-throughput mutant screening in Arabidopsis. J. Exp. Bot. 2018, 69, 2569–2579. [Google Scholar] [CrossRef] [Green Version]
- Béziat, C.; Barbez, E.; Feraru, M.I.; Lucyshyn, D.; Kleine-Vehn, J. Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition. Nat. Plants 2017, 3, 17105. [Google Scholar] [CrossRef] [Green Version]
- May, M.J.; Leaver, C.J. Oxidative Stimulation of Glutathione Synthesis in Arabidopsis thaliana Suspension Cultures. Plant Physiol. 1993, 103, 621–627. [Google Scholar] [CrossRef] [Green Version]
- Nagata, T.; Nemoto, Y.; Hasezawa, S. Tobacco BY-2 Cell Line as the “HeLa” Cell in the Cell Biology of Higher Plants. Int. Rev. Cytol. 1992, 132, 1–30. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P.G.; Karol, K.G.; Mandoli, D.F.; Kuehl, J.; Arumuganathan, K.; Ellis, M.W.; Mishler, B.D.; Kelch, D.G.; Olmstead, R.G.; Boore, J.L. The first complete chloroplast genome sequence of a lycophyte, Huperzia lucidula (Lycopodiaceae). Gene 2005, 350, 117–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thibivilliers, S.; Anderson, D.; Libault, M. Isolation of plant root nuclei for single cell RNA sequencing. Curr. Protoc. Plant Biol. 2020, 5, e20120. [Google Scholar] [CrossRef]

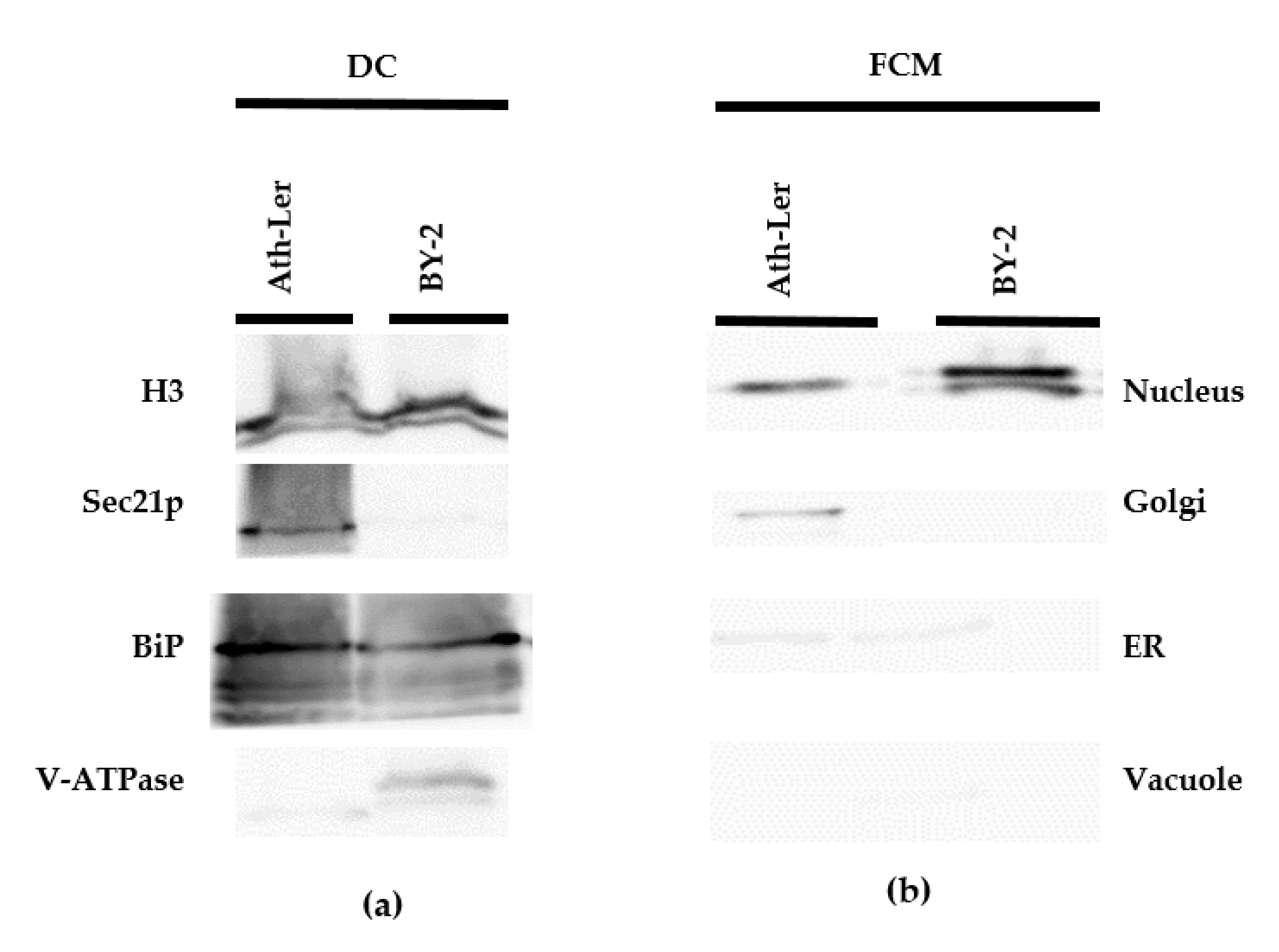

| Parameters | DC | GC | FCM 1 | AP |
|---|---|---|---|---|
| Yield | +++ | ++ | + | − |
| Purity | − | + | ++ | +++ |
| Instrumentation | +++ | +++ | − | + |
| Duration | + | − | ++ | +++ |
| Cell-type specificity | − | − | +++1 | +++ |
| Simultaneous multi-organelle isolation | − | − | +++ | − |
| Downstream application | G, P, (M) | G, T, P | G, P, T, M | G, T |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skalický, V.; Vojtková, T.; Pěnčík, A.; Vrána, J.; Juzoń, K.; Koláčková, V.; Sedlářová, M.; Kubeš, M.F.; Novák, O. Auxin Metabolite Profiling in Isolated and Intact Plant Nuclei. Int. J. Mol. Sci. 2021, 22, 12369. https://doi.org/10.3390/ijms222212369
Skalický V, Vojtková T, Pěnčík A, Vrána J, Juzoń K, Koláčková V, Sedlářová M, Kubeš MF, Novák O. Auxin Metabolite Profiling in Isolated and Intact Plant Nuclei. International Journal of Molecular Sciences. 2021; 22(22):12369. https://doi.org/10.3390/ijms222212369
Chicago/Turabian StyleSkalický, Vladimír, Tereza Vojtková, Aleš Pěnčík, Jan Vrána, Katarzyna Juzoń, Veronika Koláčková, Michaela Sedlářová, Martin F. Kubeš, and Ondřej Novák. 2021. "Auxin Metabolite Profiling in Isolated and Intact Plant Nuclei" International Journal of Molecular Sciences 22, no. 22: 12369. https://doi.org/10.3390/ijms222212369
APA StyleSkalický, V., Vojtková, T., Pěnčík, A., Vrána, J., Juzoń, K., Koláčková, V., Sedlářová, M., Kubeš, M. F., & Novák, O. (2021). Auxin Metabolite Profiling in Isolated and Intact Plant Nuclei. International Journal of Molecular Sciences, 22(22), 12369. https://doi.org/10.3390/ijms222212369







