Regulation of Cytosolic pH: The Contributions of Plant Plasma Membrane H+-ATPases and Multiple Transporters
Abstract
:1. Introduction
2. Roles of Plasma Membrane H+-ATPases and Multiple Transporters in Cytosolic pH Homeostasis
2.1. H+-ATPase Family
2.1.1. Plasma-Membrane-Located Family Members, Function and the Effect of Their Expression Level Changes on the Cytosolic pH
2.1.2. Mechanism of H+ Transport
2.1.3. Regulation by Extracellular/Cytosolic pH
2.2. NHX Family
2.2.1. Plasma-Membrane-Located Family Members, Function and the Effect of Their Expression Level Changes on the Cytosolic pH
2.2.2. Mechanism of H+ Transport
2.2.3. Regulation by Extracellular/Cytosolic pH
2.3. CHX Family
2.4. AMT Family and NRT Family
2.4.1. Plasma-Membrane-Located Family Members, Function and the Effect of Their Expression Level Changes on the Cytosolic pH
2.4.2. Mechanism of H+ Transport
2.4.3. Regulation by Extracellular/Cytosolic pH
2.5. PHT Family
2.5.1. Plasma-Membrane-Located Family Members, Function, and the Effect of Their Expression Level Changes on the Cytosolic pH
2.5.2. Mechanism of H+ Transport
2.5.3. Regulation by Extracellular/Cytosolic pH
2.6. KT/KUP/HAK Family
2.6.1. Plasma-Membrane-Located Family Members, Function and the Effect of Their Expression Level Changes on the Cytosolic pH
2.6.2. Mechanism of H+ Transport
2.6.3. Regulation by Extracellular/Cytosolic pH
3. Notable Issues in This Field
3.1. Not All Plasma Membrane Transporters Possess H+-Coupled Substrate Transport Mechanisms, and Using Transport Mechanisms of a Protein to Represent the Case of the Entire Family Is Not Suitable
3.2. Special Caution Is Needed When Drawing Conclusion to the H+ Transfer Mechanism of Transporters
4. Roles of H+ Transport in Genetic Plant Improvements and Stress Resistance
4.1. Increasing Yield
4.2. Acid Stress Resistance
5. Conclusions and Prospects
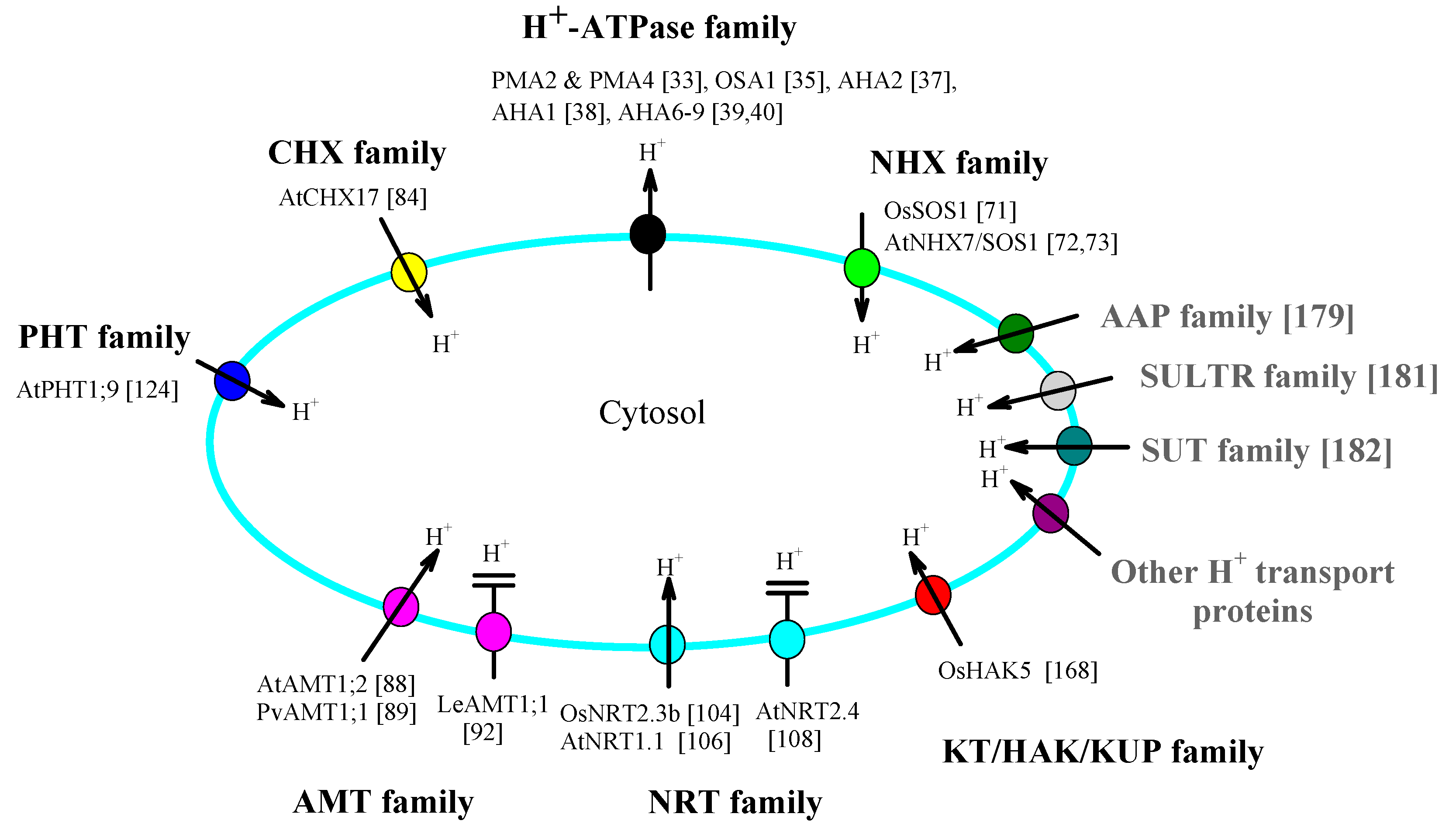
Author Contributions
Funding
Conflicts of Interest
References
- Schumacher, K. pH in the plant endomembrane system-an import and export business. Curr. Opin. Plant Biol. 2014, 22, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Blumwald, E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 2014, 22, 1–6. [Google Scholar] [CrossRef]
- Reguera, M.; Bassil, E.; Tajima, H.; Wimmer, M.; Chanoca, A.; Otegui, M.; Paris, N.; Blumwald, E. pH Regulation by NHX-Type Antiporters Is Required for Receptor-Mediated Protein Trafficking to the Vacuole in Arabidopsis. Plant Cell 2015, 27, 1200–1217. [Google Scholar] [CrossRef] [Green Version]
- Casey, J.; Grinstein, S.; Orlowski, J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010, 11, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Felle, H.H. pH: Signal and messenger in plant cells. Plant Biol. 2001, 3, 577–591. [Google Scholar] [CrossRef]
- Pittman, J. Multiple transport pathways for mediating intracellular pH homeostasis: The contribution of H+/ion exchangers. Front. Plant Sci. 2012, 3, 11. [Google Scholar] [CrossRef] [Green Version]
- Sze, H.; Chanroj, S. Plant endomembrane dynamics: Studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol. 2018, 177, 875–895. [Google Scholar] [CrossRef] [Green Version]
- Wegner, L.H.; Shabala, S. Biochemical pH clamp: The forgotten resource in membrane bioenergetics. New Phytol. 2020, 225, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Kurkdjian, A.; Guern, J. Intracellular pH: Measurement and importance in cell activity. Annu. Rev. Plant Biol. 1989, 40, 271–303. [Google Scholar] [CrossRef]
- Hochachka, P.W.; Somero, G.N. Biochemical Adaptation. Mechanism and Process. In Physiological Evolution; Oxford University Press: Oxford, UK, 2002; pp. 345–351. [Google Scholar]
- Niñoles, R.; Rubio, L.; García-Sánchez, M.; Fernández, J.; Bueso, E.; Alejandro, S.; Serrano, R. A dominant-negative form of Arabidopsis AP-3 β-adaptin improves intracellular pH homeostasis. Plant J. 2013, 74, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Fan, X.; Miller, A.J.; Xu, G. Plant nitrogen uptake and assimilation: Regulation of cellular pH homeostasis. J. Exp. Bot. 2020, 71, 4380–4392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, D. The fine control of cytosolic pH. Physiol. Plant. 1986, 67, 702–706. [Google Scholar] [CrossRef]
- Cosse, M.; Seidel, T. Plant proton pumps and cytosolic pH-homeostasis. Front. Plant Sci. 2021, 12, 846. [Google Scholar] [CrossRef] [PubMed]
- Wegner, L.H.; Li, X.; Zhang, J.; Yu, M.; Shabala, S.; Hao, Z. Biochemical and biophysical pH clamp controlling net H+ efflux across the plasma membrane of plant cells. New Phytol. 2021, 230, 408–415. [Google Scholar] [CrossRef]
- Isayenkov, S.; Dabravolski, S.A.; Pan, T.; Shabala, S. Phylogenetic diversity and physiological roles of plant monovalent cation/H+ antiporters. Front. Plant Sci. 2020, 11, 1451. [Google Scholar] [CrossRef]
- Falhof, J.; Pedersen, J.T.; Fuglsang, A.T.; Palmgren, M. Plasma Membrane H+-ATPase regulation in the center of plant physiology. Mol. Plant 2016, 9, 323–337. [Google Scholar] [CrossRef] [Green Version]
- Appelhagen, I.; Nordholt, N.; Seidel, T.; Spelt, K.; Koes, R.; Quattrochio, F.; Sagasser, M.; Weisshaar, B. Transparent testa 13 is a tonoplast P3A -ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J. 2015, 82, 840–849. [Google Scholar] [CrossRef]
- Li, Y.; Provenzano, S.; Bliek, M.; Spelt, C.; Appelhagen, I.; Faria, L.M.; Verweij, W.; Schubert, A.; Sagasser, M.; Seidel, T.; et al. Evolution of tonoplast P-ATPase transporters involved in vacuolar acidification. New Phytol. 2016, 211, 1092–1107. [Google Scholar] [CrossRef] [Green Version]
- Baxter, I.; Tchieu, J.; Sussman, M.; Boutry, M.; Palmgren, M.; Gribskov, M.; Harper, J.; Axelsen, K. Genomic comparison of P-Type ATPase ion pumps in Arabidopsis and Rice. Plant Physiol. 2003, 132, 618–628. [Google Scholar] [CrossRef] [Green Version]
- Kalampanayil, B.; Wimmers, L. Identification and characterization of a salt-stress-induced plasma membrane H+-ATPase in tomato. Plant Cell Environ. 2001, 24, 999–1000. [Google Scholar] [CrossRef]
- Santi, S.; Locci, G.; Monte, R.; Pinton, R.; Varanini, Z. Induction of nitrate uptake in maize roots: Expression of a putative high-affinity nitrate transporter and plasma membrane H+-ATPase isoforms. J. Exp. Bot. 2003, 54, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Okumura, M.; Inoue, S.; Takahashi, K.; Ishizaki, K.; Kohchi, T.; Kinoshita, T. Characterization of the Plasma Membrane H+-ATPase in the Liverwort Marchantia polymorpha. Plant Physiol. 2012, 159, 826–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wdowikowska, A.; Kłobus, G. The plasma membrane proton pump gene family in cucumber. Acta Physiol. Plant. 2016, 38, 1–14. [Google Scholar] [CrossRef]
- Stritzler, M.; García, M.N.; Schlesinger, M.; Cortelezzi, J.I.; Capiati, D. The plasma membrane H+-ATPase gene family in Solanum tuberosum L. Role of PHA1 in tuberization. J. Exp. Bot. 2017, 68, 4821–4837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arango, M.; Gévaudant, F.; Oufattole, M.; Boutry, M. The plasma membrane proton pump ATPase: The significance of gene subfamilies. Planta 2002, 216, 355–365. [Google Scholar] [CrossRef]
- Xu, Z.; Marowa, P.; Liu, H.; Du, H.; Zhang, C.; Li, Y. Genome-wide identification and analysis of P-Type plasma membrane H+-ATPase sub-gene family in sunflower and the role of HHA4 and HHA11 in the development of salt stress resistance. Genes 2020, 11, 361. [Google Scholar] [CrossRef] [Green Version]
- Palmgren, M. Plant plasam membrane H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Biol. 2001, 52, 817–845. [Google Scholar] [CrossRef] [Green Version]
- Morth, J.P.; Pedersen, B.P.; Buch-Pedersen, M.; Andersen, J.; Vilsen, B.; Palmgren, M.; Nissen, P. A structural overview of the plasma membrane Na+, K+-ATPase and H+-ATPase ion pumps. Nat. Rev. Mol. Cell Biol. 2010, 12, 60–70. [Google Scholar] [CrossRef]
- Elmore, J.; Coaker, G. The role of the plasma membrane H+-ATPase in plant-microbe interactions. Mol. Plant 2011, 4, 416–427. [Google Scholar] [CrossRef] [Green Version]
- Schaller, A.; Oecking, C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 1999, 11, 263–272. [Google Scholar]
- Frick, U.B.; Schaller, A. cDNA microarray analysis of fusicoccin-induced changes in gene expression in tomato plants. Planta 2002, 216, 83–94. [Google Scholar] [CrossRef]
- Luo, H.; Morsomme, P.; Boutry, M. The two major types of plant plasma membrane H+-ATPases show different enzymatic properties and confer differential pH sensitivity of yeast growth. Plant Physiol. 1999, 119, 627–634. [Google Scholar] [CrossRef] [Green Version]
- Robertson, W.; Clark, K.; Young, J.C.; Sussman, M. An Arabidopsis thaliana plasma membrane proton pump is essential for pollen development. Genetics 2004, 168, 1677–1687. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Wang, Y.; Chen, X.; Xu, F.; Ding, M.; Ye, W.; Kawai, Y.; Toda, Y.; Hayashi, Y.; Suzuki, T.; et al. Plasma membrane H+-ATPase overexpression increases rice yield via simultaneous enhancement of nutrient uptake and photosynthesis. Nat. Commun. 2021, 12, 735. [Google Scholar] [CrossRef]
- Harper, J.; Surowy, T.; Sussman, M. Molecular cloning and sequence of cDNA encoding the plasma membrane proton pump (H+-ATPase) of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1989, 86, 1234–1238. [Google Scholar] [CrossRef] [Green Version]
- Haruta, M.; Burch, H.L.; Nelson, R.B.; Barrett-Wilt, G.; Kline, K.G.; Mohsin, S.B.; Young, J.C.; Otegui, M.; Sussman, M. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 2010, 285, 17918–17929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, S.; Takemiya, A.; Sakamoto, T.; Kurata, T.; Tsutsumi, T.; Kinoshita, T.; Shimazaki, K. The plasma membrane H+-ATPase AHA1 plays a major role in stomatal opening in response to blue light. Plant Physiol. 2016, 171, 2731–2743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, W.; Zhang, D.; Song, T.; Xu, F.; Lin, S.; Xu, W.; Li, Q.; Zhu, Y.; Liang, J.; Zhang, J. Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. J. Exp. Bot. 2017, 68, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, R.D.; Portes, M.; Olsen, L.I.; Damineli, D.S.; Hayashi, M.; Nunes, C.O.; Pedersen, J.T.; Lima, P.T.; Campos, C.; Feijó, J.; et al. Plasma membrane H+-ATPases sustain pollen tube growth and fertilization. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Buch-Pedersen, M.J.; Pedersen, B.P.; Veierskov, B.; Nissen, P.; Palmgren, M.G. Protons and how they are transported by proton pumps. Pflug. Arch. 2009, 457, 573–579. [Google Scholar] [CrossRef]
- Pedersen, B.P.; Buch-Pedersen, M.J.; Morth, J.P.; Palmgren, M.G.; Nissen, P. Crystal structure of the plasma membrane proton pump. Nature 2007, 450, 1111–1114. [Google Scholar] [CrossRef] [PubMed]
- Buch-Pedersen, M.J.; Palmgren, M.G. Conserved Asp684 in transmembrane segment M6 of the plant plasma membrane P-type proton pump AHA2 is a molecular determinant of proton translocation. J. Biol. Chem. 2003, 278, 17845–17851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, R.D.; Olsen, L.I.; Ezike, C.V.; Pedersen, J.T.; Manstretta, R.; López-Marqués, R.L.; Palmgren, M. Roles of plasma membrane proton ATPases AHA2 and AHA7 in normal growth of roots and root hairs in Arabidopsis thaliana. Physiol. Plant. 2019, 166, 848–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmgren, M.; Christensen, G. Functional comparisons between plant plasma membrane H+-ATPase isoforms expressed in yeast. J. Biol. Chem. 1994, 269, 3027–3033. [Google Scholar] [CrossRef]
- Regenberg, B.; Villalba, J.M.; Lanfermeijer, F.; Palmgren, M. C-terminal deletion analysis of plant plasma membrane H+-ATPase: Yeast as a model system for solute transport across the plant plasma membrane. Plant Cell 1995, 7, 1655–1666. [Google Scholar]
- Olivari, C.; Pugliarello, M.; Rasi-Caldogno, F.; Michelis, M.I. Characteristics and regulatory properties of the H+-ATPase in a plasma membrane fraction purified from Arabidopsis thaliana. Bot. Acta 1993, 106, 13–19. [Google Scholar] [CrossRef]
- Zhu, Y.; Di, T.; Xu, G.; Chen, X.; Zeng, H.; Yan, F.; Shen, Q. Adaptation of plasma membrane H+-ATPase of rice roots to low pH as related to ammonium nutrition. Plant Cell Environ. 2009, 32, 1428–1440. [Google Scholar] [CrossRef]
- Liang, C.J.; Ge, Y.Q.; Su, L.; Bu, J.J. Response of plasma membrane H+-ATPase in rice (Oryza sativa) seedlings to simulated acid rain. Environ. Sci. Pollut. Res. Int. 2015, 22, 535–545. [Google Scholar] [CrossRef]
- Liang, C.; May, L.L. Comparison of plasma membrane H+-ATPase response to acid rain stress between rice and soybean. Environ. Sci. Pollut. Res. 2019, 27, 6389–6400. [Google Scholar] [CrossRef]
- Reid, R.; Field, L.; Pitman, M. Effects of external pH, fusicoccin and butyrate on the cytoplasmic pH in barley root tips measured by 31P-nuclear magnetic resonance spectroscopy. Planta 1985, 166, 341–347. [Google Scholar] [CrossRef]
- Raven, J.A. Sensing pH? Plant Cell Environ. 1990, 13, 721–729. [Google Scholar] [CrossRef]
- Bobik, K.; Boutry, M.; Duby, G. Activation of the plasma membrane H+-ATPase by acid stress. Plant Signal. Behav. 2010, 5, 681–683. [Google Scholar] [CrossRef] [Green Version]
- An, R.; Chen, Q.; Chai, M.; Lu, P.; Su, Z.; Qin, Z.; Chen, J.; Wang, X. AtNHX8, a member of the monovalent cation: Proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter. Plant J. 2007, 49, 718–728. [Google Scholar] [CrossRef]
- Fu, X.; Lu, Z.; Wei, H.; Zhang, J.; Yang, X.; Wu, A.; Ma, L.; Kang, M.; Lu, J.; Wang, H.; et al. Genome-wide identification and expression analysis of the NHX (Sodium/Hydrogen Antiporter) gene family in cotton. Front. Genet. 2020, 11, 964. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [Green Version]
- Aharon, G.S.; Apse, M.P.; Duan, S.; Hua, X.; Blumwald, E. Characterization of a family of vacuolar Na+/H+ antiporters in Arabidopsis thaliana. Plant Soil 2003, 253, 245–256. [Google Scholar] [CrossRef]
- Brett, C.L.; Donowitz, M.; Rao, R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 2005, 288, 223–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassil, E.; Tajima, H.; Liang, Y.; Ohto, M.; Ushijima, K.; Nakano, R.; Esumi, T.; Coku, A.; Belmonte, M.; Blumwald, E. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 2011, 23, 3482–3497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Jiang, X.; Zhan, K.; Cheng, X.; Chen, X.; Pardo, J.M.; Cui, D.Q. Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch Biochem. Biophys. 2008, 473, 8–15. [Google Scholar] [CrossRef]
- Olías, R.; Eljakaoui, Z.; Li, J.; MarínManzano, M.C.; Pardo, J.M.; Belver, A. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 2009, 32, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Feki, K.; Quintero, F.J.; Khoudi, H.; Leidi, E.; Masmoudi, K.; Pardo, J.M.; Brini, F. A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis. Plant Cell Rep. 2013, 33, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Q.S.; Barkla, B.J.; Vera-Estrella, R.; Zhu, J.K.; Schumaker, K.S. Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 2003, 132, 1041–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239. [Google Scholar] [CrossRef]
- Zorb, C.; Noll, A.; Karl, S.; Leib, K.; Yan, F.; Schubert, S. Molecular characterization of Na+/H+ antiporters (ZmNHX) of maize (Zea mays L.) and their expression under salt stress. J. Plant Physiol. 2005, 162, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Esmaili, M.; Overduin, M.; Fliegel, L. Expression and detergent free purification and reconstitution of the plant plasma membrane Na+/H+ antiporter SOS1 overexpressed in Pichia pastoris. Biochim. Biophys. Acta Biomembr. 2019, 1862, 183111. [Google Scholar] [CrossRef]
- Dragwidge, J.; Scholl, S.; Schumacher, K.; Gendall, A. NHX-type Na+(K+)/H+ antiporters are required for TGN/EE trafficking and endosomal ion homeostasis in Arabidopsis thaliana. J. Cell. Sci. 2019, 132, jcs226472. [Google Scholar] [CrossRef] [Green Version]
- Jegadeeson, V.; Kumari, K.; Pulipati, S.; Parida, A.; Venkataraman, G. Expression of wild rice Porteresia coarctata PcNHX1 antiporter gene (PcNHX1) in tobacco controlled by PcNHX1 promoter (PcNHX1p) confers Na+-specific hypocotyl elongation and stem-specific Na+ accumulation in transgenic tobacco. Plant Physiol. Biochem. 2019, 139, 161–170. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [Green Version]
- Mahi, H.E.; Pérez-Hormaeche, J.; Luca, A.D.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.M.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.; et al. A critical role of sodium flux via the plasma membrane Na+/H+ Exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef] [Green Version]
- Shabala, L.; Cuin, T.A.; Newman, I.A.; Shabala, S. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 2005, 222, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Babourina, O.; Rengel, Z. Na+/H+ antiporter activity of the SOS1 gene: Lifetime imaging analysis and electrophysiological studies on Arabidopsis seedlings. Physiol. Plant. 2009, 137, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Núnez-Ramírez, R.; Sánchez-Barrena, M.; Villalta, I.; Vega, J.; Pardo, J.M.; Quintero, F.J.; Martínez-Salazar, J.; Albert, A. Structural insights on the plant salt-overly-sensitive 1 (SOS1) Na+/H+ antiporter. J. Mol. Biol. 2012, 424, 283–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraile-Escanciano, A.; Kamisugi, Y.; Cuming, A.; Rodríguez-Navarro, A.; Benito, B. The SOS1 transporter of Physcomitrella patens mediates sodium efflux in planta. New Phytol. 2010, 188, 750–761. [Google Scholar] [CrossRef]
- Mottaleb, S.A.; Rodríguez-Navarro, A.; Haro, R. Knockouts of Physcomitrella patens CHX1 and CHX2 transporters reveal high complexity of potassium homeostasis. Plant Cell Physiol. 2013, 54, 1455–1468. [Google Scholar] [CrossRef] [Green Version]
- Jia, B.; Sun, M.Z.; Duan Mu, H.Z.; Ding, X.D.; Liu, B.D.; Zhu, Y.; Sun, X.L. GsCHX19.3, a member of cation/H+ exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Zhao, J.; Cheng, N.H.; Motes, C.M.; Blancaflor, E.B.; Moore, M.; Gonzales, N.; Padmanaban, S.; Sze, H.; Ward, J.M.; Hirschi, K.D. AtCHX13 is a plasma membrane K+ transporter. Plant Physiol. 2008, 148, 796–807. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Li, P.; Motes, C.M.; Park, S.; Hirschi, K.D. CHX14 is a plasma membrane K+-efflux transporter that regulates K+ redistribution in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 2223–2238. [Google Scholar] [CrossRef]
- Chanroj, S.; Padmanaban, S.; Czerny, D.D.; Jauh, G.Y.; Sze, H. K+ transporter AtCHX17 with its hydrophilic C tail localizes to membranes of the secretory/endocytic system: Role in reproduction and seed set. Mol. Plant 2013, 6, 1226–1246. [Google Scholar] [CrossRef] [Green Version]
- Padmanaban, S.; Czerny, D.D.; Levin, K.A.; Leydon, A.R.; Su, R.T.; Maugel, T.K.; Zou, Y.; Chanroj, S.; Cheung, A.Y.; Johnson, M.A. Transporters involved in pH and K+ homeostasis affect pollen wall formation, male fertility, and embryo development. J. Exp. Bot. 2017, 68, 3165–3178. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.; Evans, A.R.; Newbury, H.J.; Pritchard, J. Functional analysis of CHX21: A putative sodium transporter in Arabidopsis. J. Exp. Bot. 2006, 57, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Czerny, D.D.; Padmanaban, S.; Anishkin, A.; Venema, K.; Riaz, Z.; Sze, H. Protein architecture and core residues in unwound α-helices provide insights to the transport function of plant AtCHX17. Biochim. Biophys. Acta 2016, 1858, 1983–1998. [Google Scholar] [CrossRef] [Green Version]
- Chanroj, S.; Lu, Y.; Padmanaban, S.; Nanatani, K.; Uozumi, N.; Rao, R.; Sze, H. Plant-specific cation/H+ exchanger 17 and its homologs are endomembrane K+ transporters with roles in protein sorting. J. Biol. Chem. 2011, 286, 33931–33941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, D.; Zhou, J.; Yang, S.; Qi, W.; Yang, K.; Su, Y. Function and regulation of ammonium transporters in plants. Int. J. Mol. Sci. 2020, 21, 3557. [Google Scholar] [CrossRef]
- Søgaard, R.; Alsterfjord, M.; MacAulay, N.; Zeuthen, T. Ammonium ion transport by the AMT/Rh homolog TaAMT1;1 is stimulated by acidic pH. Pflüg. Arch.-Eur. J. Physiol. 2009, 458, 733–743. [Google Scholar] [CrossRef]
- Neuhäuser, B.; Ludewig, U. Uncoupling of ionic currents from substrate transport in the plant ammonium transporter AtAMT1;2. J. Biol. Chem. 2014, 289, 11650–11655. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Ramirez, C.; Mora, S.I.; Trejo, J.M.; Pantoja, O. PvAMT1;1, a highly selective ammonium transporter that functions as H+/NH4+ symporter. J. Biol. Chem. 2011, 286, 31113–31122. [Google Scholar] [CrossRef] [Green Version]
- Neuhäuser, B.; Dynowski, M.; Mayer, M.G.; Ludewig, U. Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol. 2007, 143, 1651–1659. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Liao, K.; Xu, X.; Gao, Y.; Wang, Z.; Zhu, X.; Jia, B.; Xuan, Y. Wheat ammonium transporter (AMT) gene family: Diversity and possible role in host–pathogen interaction with stem rust. Front. Plant Sci. 2017, 8, 1637. [Google Scholar] [CrossRef] [Green Version]
- Mayer, M.; Dynowski, M.; Ludewig, U. Ammonium ion transport by the AMT/Rh homologue LeAMT1;1. Biochem. J. 2006, 396, 431–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Hsu, P.; Tsay, Y. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Krapp, A.; David, L.; Chardin, C.; Girin, T.; Marmagne, A.; Leprince, A.; Chaillou, S.; Ferrario-Méry, S.; Meyer, C.; Daniel-Vedele, F. Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Naz, M.; Fan, X.; Xuan, W.; Miller, A.J.; Xu, G. Plant nitrate transporters: From gene function to application. J. Exp. Bot. 2017, 68, 2463–2475. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, Y.; Chen, K.; Tsay, Y. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Theodoulou, F.; Muldin, I.; Ingemarsson, B.; Miller, A.J. Cloning and functional characterization of a Brassica napus transporter that is able to transport nitrate and histidine. J. Biol. Chem. 1998, 273, 12017–12023. [Google Scholar] [CrossRef] [Green Version]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [Green Version]
- Chiu, C.; Lin, C.; Hsia, A.; Su, R.; Lin, H.; Tsay, Y. Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 2004, 45, 1139–1148. [Google Scholar] [CrossRef]
- Lin, S.H.; Kuo, H.F.; Canivenc, G.; Lin, C.S.; Lepetit, M.; Hsu, P.K.; Tillard, P.; Lin, H.L.; Wang, Y.Y.; Tsai, C.B.; et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 2008, 20, 2514–2528. [Google Scholar] [CrossRef] [Green Version]
- Almagro, A.; Lin, S.H.; Tsay, Y.F. Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 2008, 20, 3289–3299. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.M.; Koh, S.; Stacey, G.; Yu, S.M.; Lin, T.; Tsay, Y. Cloning and functional characterization of a constitutively expressed nitrate transporter gene, OsNRT1, from rice. Plant Physiol. 2000, 122, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Tang, Z.; Tan, Y.; Zhang, Y.; Luo, B.; Yang, M.; Lian, X.; Shen, Q.; Miller, A.J.; Xu, G. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. USA 2016, 113, 7118–7123. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Liu, D.; Crawford, N. The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. USA 1998, 95, 15134–15139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.Z.; Tian, W.H.; Liu, X.X.; Lin, X.Y.; Jin, C.W.; Zheng, S.J. Alleviation of proton toxicity by nitrate uptake specifically depends on nitrate transporter 1.1 in Arabidopsis. New Phytol. 2016, 211, 149–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Yu, M.; Du, X.; Wang, Z.; Wu, W.; Quintero, F.J.; Jin, X.; Li, H.; Wang, Y. NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell 2017, 29, 2016–2026. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Byrt, C.S.; Qiu, J.; Baumann, U.; Hrmova, M.; Evrard, A.; Johnson, A.; Birnbaum, K.; Mayo, G.; Jha, D.; et al. Identification of a stelar-localized transport protein that facilitates root-to-shoot transfer of chloride in Arabidopsis. Plant Physiol. 2015, 170, 1014–1029. [Google Scholar] [CrossRef] [PubMed]
- Longo, A.; Miles, N.W.; Dickstein, R. Genome mining of plant NPFs reveals varying conservation of signature motifs associated with the mechanism of transport. Front. Plant Sci. 2018, 9, 1668. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Bankston, J.R.; Payandeh, J.; Hinds, T.R.; Zagotta, W.N.; Zheng, N. Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 2014, 507, 73–77. [Google Scholar] [CrossRef] [Green Version]
- Ho, C.; Frommer, W. Fluorescent sensors for activity and regulation of the nitrate transceptor CHL1/NRT1.1 and oligopeptide transporters. eLife 2014, 3, e01917. [Google Scholar] [CrossRef]
- Jorgensen, M.E.; Olsen, C.E.; Geiger, D.; Mirza, O.; Halkier, B.A.; NourEldin, H.H. A functional EXXEK motif is essential for proton coupling and active glucosinolate transport by NPF2.11. Plant Cell Physiol. 2015, 56, 2340–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, J.L.; Li, C.; Brinth, A.; Wang, Z.; Vogeley, L.; Solcan, N.; Ledderboge-Vucinic, G.; Swanson, J.; Caffrey, M.; Voth, G.; et al. Proton movement and coupling in the POT family of peptide transporters. Proc. Natl. Acad. Sci. USA 2017, 114, 13182–13187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, J.L.; Newstead, S. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 2014, 507, 68–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Huang, C.; Tsay, Y. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 1999, 11, 865–874. [Google Scholar] [CrossRef]
- Mudge, S.R.; Rae, A.; Diatloff, E.; Smith, F. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Nussaume, L.; Kanno, S.; Javot, H.; Marín, E.; Pochon, N.; Ayadi, A.; Nakanishi, T.; Thibaud, M. Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2011, 2, 83. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Xu, Y.J.; Jiang, H.H.; Jiang, C.S.; Du, Y.B.; Gong, C.; Wang, W.; Zhu, S.W.; Han, G.M.; Cheng, B.J. Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int. J. Mol. Sci. 2016, 17, 930. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lv, S.; Jiang, P.; Li, Y. Roles, regulation, and agricultural application of plant phosphate transporters. Front. Plant Sci. 2017, 8, 817. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Deng, M.J.; Xu, J.M.; Zhu, X.L.; Mao, C.Z. Molecular mechanisms of phosphate transport and signaling in higher plants. Semin. Cell Dev. Biol. 2018, 74, 114–122. [Google Scholar] [CrossRef]
- Roch, G.V.; Maharajan, T.; Ceasar, S.A.; Ignacimuthu, S. The Role of PHT1 family transporters in the acquisition and redistribution of phosphorus in plants. Crit. Rev. Plant Sci. 2019, 38, 171–198. [Google Scholar] [CrossRef]
- Shin, H.; Shin, H.; Dewbre, G.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Bayle, V.; Arrighi, J.; Creff, A.; Nespoulous, C.; Vialaret, J.; Rossignol, M.; González, E.; Paz-Ares, J.; Nussaume, L. Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 2011, 23, 1523–1535. [Google Scholar] [CrossRef] [Green Version]
- Remy, E.; Cabrito, T.R.; Batista, R.A.; Teixeira, M.C.; Sá-Correia, I.; Duque, P. The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 2012, 195, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Gu, M.; Xia, Y.; Dai, X.; Dai, C.R.; Zhang, J.; Wang, S.; Qu, H.; Yamaji, N.; Ma, J.F.; et al. OsPHT1;3 mediates uptake, translocation, and remobilization of phosphate under extremely low phosphate regimes. Plant Physiol. 2018, 179, 656–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Yuan, J.; Chang, X.; Yang, M.; Zhang, L.; Lu, K.; Lian, X. The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLoS ONE 2015, 10, e0126186. [Google Scholar]
- Jia, H.; Ren, H.; Gu, M.; Zhao, J.; Sun, S.; Zhang, X.; Chen, J.; Wu, P.; Xu, G. The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 2011, 156, 1164–1175. [Google Scholar] [CrossRef] [Green Version]
- Preuss, C.; Huang, C.; Tyerman, S. Proton-coupled high-affinity phosphate transport revealed from heterologous characterization in Xenopus of barley-root plasma membrane transporter, HvPHT1;1. Plant Cell Environ. 2011, 34, 681–689. [Google Scholar] [CrossRef]
- Preuss, C.; Huang, C.; Gilliham, M.; Tyerman, S. Channel-like characteristics of the low-affinity barley phosphate transporter pht1;6 when expressed in Xenopus oocytes. Plant Physiol. 2010, 152, 1431–1441. [Google Scholar] [CrossRef] [Green Version]
- Rausch, C.; Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta 2002, 216, 23–37. [Google Scholar] [CrossRef]
- Młodzińska, E.; Zboińska, M. Phosphate uptake and allocation—A closer look at Arabidopsis thaliana L. and Oryza sativa L. Front. Plant Sci. 2016, 7, 1198. [Google Scholar] [CrossRef] [Green Version]
- Ullrich, C.I.; Novacky, A.J. Extra- and intracellular pH and membrane potential changes induced by K+, Cl−, H2PO4−, and NO3− uptake and fusicoccin in root hairs of Limnobium stoloniferum. Plant Physiol. 1990, 94, 1561–1567. [Google Scholar] [CrossRef] [Green Version]
- Mimura, T.; Yin, Z.H.; Wirth, E.; Dietz, K.J. Phosphate transport and apoplastic phosphate homeostasis in barley leaves. Plant Cell Physiol. 1992, 33, 563–568. [Google Scholar]
- Sakano, K.; Yazaki, Y.; Mimura, T. Cytoplasmic acidification induced by inorganic phosphate uptake in suspension cultured Catharanthus roseus cells–Measurement with fluorescent pH indicator and P-31 nuclear-magnetic-resonance. Plant Physiol. 1992, 99, 672–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedersen, B.P.; Kumar, H.; Waight, A.B.; Risenmay, A.J.; Roe-Zurz, Z.; Chau, B.H.; Schlessinger, A.; Bonomi, M.; Harries, W.; Sali, A.; et al. Crystal structure of a eukaryotic phosphate transporter. Nature 2013, 496, 533–536. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Li, C.; Gupta, M.; Verma, N.; Johri, A.; Stroud, R.; Voth, G. Key computational findings reveal proton transfer as driving the functional cycle in the phosphate transporter PiPT. Proc. Natl. Acad. Sci. USA 2021, 118, e2101932118. [Google Scholar] [CrossRef]
- Liao, Y.; Li, J.; Pan, R.; Chiou, T. Structure–function analysis reveals amino acid residues of Arabidopsis phosphate transporter AtPHT1;1 crucial for its activity. Front. Plant Sci. 2019, 10, 1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Gu, M.; Cao, Y.; Huang, X.; Zhang, X.; Ai, P.; Zhao, J.; Fan, X.; Xu, G. A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 2012, 159, 1571–1581. [Google Scholar] [CrossRef] [Green Version]
- Ai, P.; Sun, S.; Zhao, J.; Fan, X.; Xin, W.; Guo, Q.; Yu, L.; Shen, Q.; Wu, P.; Miller, A.J.; et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009, 57, 798–809. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Piñeros, M.; Wang, Z.; Wang, W.; Li, C.; Wu, Z.; Kochian, L.; Wu, P. Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 2014, 37, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhang, L.; Gao, Q.; Wang, J.; Li, X.; Wang, H.; Liu, Y.; Lin, H.; Liu, J.; Wang, X.; et al. A plasma membrane transporter coordinates phosphate reallocation and grain filling in cereals. Nat. Genet. 2021, 53, 906–915. [Google Scholar] [CrossRef]
- Mäser, P.; Thomine, S.; Schroeder, J.; Ward, J.; Hirschi, K.; Sze, H.; Talke, I.; Amtmann, A.; Maathuis, F.; Sanders, D.; et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667. [Google Scholar] [CrossRef] [Green Version]
- Véry, A.; Sentenac, H. Molecular mechanisms and regulation of K+ transport in higher plants. Annu. Rev. Plant Biol. 2003, 54, 575–603. [Google Scholar] [CrossRef]
- Véry, A.; Nieves-Cordones, M.; Daly, M.; Khan, I.; Fizames, C.; Sentenac, H. Molecular biology of K+ transport across the plant cell membrane: What do we learn from comparison between plant species? J. Plant Physiol. 2014, 17, 748–769. [Google Scholar] [CrossRef]
- Santa-María, G.; Oliferuk, S.; Moriconi, J.I. KT-HAK-KUP transporters in major terrestrial photosynthetic organisms: A twenty years tale. J. Plant Physiol. 2018, 226, 77–90. [Google Scholar] [CrossRef]
- Gupta, M.; Qiu, X.; Wang, L.; Xie, W.; Zhang, C.; Xiong, L.; Zhang, Q. KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol. Genet. Genom. 2008, 280, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, G.; Alli, A.; Yu, L. Plant HAK/KUP/KT K+ transporters: Function and regulation. Semin. Cell Dev. Biol. 2018, 74, 133–141. [Google Scholar] [CrossRef]
- Qi, Z.; Hampton, C.R.; Shin, R.; Barkla, B.J.; White, P.J.; Schachtman, D.P. The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J. Exp. Bot. 2008, 59, 595–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajappa, S.; Krishnamurthy, P.; Kumar, P.P. Regulation of AtKUP2 expression by bHLH and WRKY transcription factors helps to confer increased salt tolerance to Arabidopsis thaliana plants. Front. Plant Sci. 2020, 11, 1311. [Google Scholar] [CrossRef] [PubMed]
- Rigas, S.; Ditengou, F.A.; Ljung, K.; Daras, G.; Tietz, O.; Palme, K.; Hatzopoulos, P. Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytol. 2013, 197, 1130–1141. [Google Scholar] [CrossRef]
- Osakabe, Y.; Arinaga, N.; Umezawa, T.; Katsura, S.; Nagamachi, K.; Tanaka, H.; Ohiraki, H.; Yamada, K.; Seo, S.-U.; Abo, M. Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 2013, 25, 609–624. [Google Scholar] [CrossRef] [Green Version]
- Han, M.; Wu, W.; Wu, W.-H.; Wang, Y. Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol. Plant 2016, 9, 437–446. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Hu, Q.; Luo, L.; Yang, T.; Zhang, S.; Hu, Y.; Yu, L.; Xu, G. Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 2015, 38, 2747–2765. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zheng, C.; Kuang, B.; Wei, L.; Yan, L.; Wang, T. Receptor-like kinase RUPO interacts with potassium transporters to regulate pollen tube growth and integrity in rice. PLoS Genet. 2016, 12, e1006085. [Google Scholar] [CrossRef] [Green Version]
- Horie, T.; Sugawara, M.; Okada, T.; Taira, K.; Kaothien-Nakayama, P.; Katsuhara, M.; Shinmyo, A.; Nakayama, H. Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioeng. 2011, 111, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhang, S.; Hu, Y.; Wu, F.; Hu, Q.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Shen, L.; Shen, Z.; Jing, W.; Ge, H.; Zhao, J.; Zhang, W. The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice: OsHAK21 involvement in responses to salt stress. Plant Cell Environ. 2015, 38, 2766–2779. [Google Scholar] [CrossRef]
- Maathuis, F.J.; Sanders, D. Mechanism of high-affinity potassium uptake in roots of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1994, 91, 9272–9276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, D.J.; Leigh, R.A.; Miller, A.J. Potassium homeostasis in vacuolate plant cells. Proc. Natl. Acad. Sci. USA 1996, 93, 10510–10514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieves-Cordones, M.; Alemán, F.; Martínez, V.; Rubio, F. K+ uptake in plant roots: The systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 2014, 171, 688–695. [Google Scholar] [CrossRef]
- Rubio, F.; Arévaloa, L.; Caballero, F.; Botella, M.A.; Rubio, J.S.; García-Sánchez, F.; Martínez, V. Systems involved in K+ uptake from diluted solutions in pepper plants as revealed by the use of specific inhibitors. J. Plant Physiol. 2010, 167, 1494–1499. [Google Scholar] [CrossRef]
- Alemán, F.; Caballero, F.; Ródenas, R.; Rivero, R.M.; Martínez, V.; Rubio, F. The F130S point mutation in the Arabidopsis high-affinity K+ transporter AtHAK5 increases K+ over Na+ and Cs+ selectivity and confers Na+ and Cs+ tolerance to yeast under heterologous expression. Front. Plant Sci. 2014, 5, 430. [Google Scholar] [CrossRef]
- Rubio, F.; Nieves-Cordones, M.; Alemán, F.; Martínez, V. Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol. Plant 2008, 134, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Haro, R.; Sainz, L.; Rubio, F.; Rodríguez-Navarro, A. Cloning of two genes encoding potassium transporters in Neurospora crassa and expression of the corresponding cDNAs in Saccharomyces cerevisiae. Mol. Microbiol. 1999, 31, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Rivetta, A.; Allen, K.E.; Slayman, C.W.; Slayman, C.L. Coordination of K transporters in Neurospora: TRK1 is scarce and constitutive, while HAK1 is abundant and highly regulated. Euk. Cell 2013, 12, 684–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherzer, S.; Böhm, J.; Krol, E.; Shabala, L.; Kreuzer, I.; Larisch, C.; Bemm, F.; Al-Rasheid, K.A.; Shabala, S.; Rennenberg, H. Calcium sensor kinase activates potassium uptake systems in gland cells of Venus flytraps. Proc. Natl. Acad. Sci. USA 2015, 112, 7309–7314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tascón, I.; Sousa, J.S.; Corey, R.; Mills, D.; Griwatz, D.; Aumüller, N.; Mikušević, V.; Stansfeld, P.; Vonck, J.; Hänelt, I. Structural basis of proton-coupled potassium transport in the KUP family. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Yang, T.; Feng, H.; Zhang, S.; Xiao, H.; Hu, Q.; Chen, G.; Xuan, W.; Moran, N.; Murphy, A.S.; Yu, L.; et al. The potassium transporter OsHAK5 alters rice architecture via ATP-dependent transmembrane auxin fluxes. Plant Commun. 2020, 1, 100052. [Google Scholar] [CrossRef]
- Ródenas, R.; Ragel, P.; Nieves-Cordones, M.; Martínez-Martínez, A.; Amo, J.; Lara, A.; Martínez, V.; Quintero, F.J.; Pardo, J.M.; Rubio, F. Insights into the mechanisms of transport and regulation of the Arabidopsis high-affinity K+ transporter HAK5. Plant Physiol. 2021, 185, 1860–1874. [Google Scholar] [CrossRef]
- Senn, M.E.; Rubio, F.; Bãnuelos, M.A.; Rodríguez-Navarro, A. Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. J. Biol. Chem. 2001, 276, 44563–44569. [Google Scholar] [CrossRef] [Green Version]
- Garciadeblas, B.; Benito, B.; Rodríguez-Navarro, A. Molecular cloning and functional expression in bacteria of the potassium transporters CnHAK1 and CnHAK2 of the seagrass Cymodocea nodosa. Plant Mol. Biol. 2002, 50, 623–633. [Google Scholar] [CrossRef]
- Wang, M.Y.; Glass, A.; Shaff, J.E.; Kochian, L.V. Ammonium Uptake by Rice Roots (III. Electrophysiology). Plant Physiol. 1994, 103, 1259–11267. [Google Scholar] [CrossRef] [Green Version]
- Sakano, K. Proton/Phosphate Stoichiometry in uptake of inorganic phosphate by cultured cells of Catharanthus roseus (L.) G. Don. Plant Physiol. 1990, 93, 479–483. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Blatt, M.R.; Chen, Z.-H. Ion Transport at the Plant Plasma Membrane; John Wiley & Sons, Ltd.: Chichester, UK, 2018. [Google Scholar]
- Neuhäuser, B.; Dynowski, M.; Ludewig, U. Channel-like NH3 flux by ammonium transporter AtAMT2. FEBS Lett. 2009, 583, 2833–2838. [Google Scholar] [CrossRef] [Green Version]
- Ludewig, U.; Von Wirén, N.; Frommer, W.B. Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J. Biol. Chem. 2002, 277, 13548–13555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.Y.; Tian, W.H.; Zhou, M.; Zhu, Q.Y.; Du, W.X.; Zhu, Y.X.; Liu, X.X.; Lin, X.Y.; Zheng, S.J.; Jin, C.W. STOP1 activates NRT1.1-mediated nitrate uptake to create a favorable rhizospheric pH for plant adaptation to acidity. Plant Cell 2021, 226, koab226. [Google Scholar] [CrossRef] [PubMed]
- Miranda, R.; Mesquita, R.O.; Costa, J.H.; Alvarez-Pizarro, J.; Prisco, J.T.; Gomes-Filho, E. Integrative control between proton pumps and sos1 antiporters in roots is crucial for maintaining low Na+ accumulation and salt tolerance in ammonium-supplied Sorghum bicolor. Plant Cell Physiol. 2017, 8, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef] [Green Version]
- Tegeder, M.; Hammes, U.Z. The way out and in: Phloem loading and unloading of amino acids. Curr. Opin. Plant Biol. 2018, 43, 16–21. [Google Scholar] [CrossRef]
- Wang, L.; Chen, K.; Zhou, M. Structure and function of an Arabidopsis thaliana sulfate transporter. Nat. Commun. 2021, 12, 1–8. [Google Scholar]
- Santiago, J.P.; Ward, J.M.; Sharkey, T.D. Phaseolus vulgaris SUT1.1 is a high affinity sucrose-proton co-transporter. Plant Direct 2020, 4, e00260. [Google Scholar] [CrossRef]
| Protein Name | Regulation by pH | Key Residues of H+ Transfer Pathway |
|---|---|---|
| H+-ATPase family | ||
| AHA2 | Bell-shaped dependence on cytosolic pH, with maximal transport activity approaching at pH 6.6 [47] | D684, N106 and R655 [29,41,42,43] |
| AHA1&AHA3, NpPMA2 &NpPMA4, and rice H+-ATPases | Bell-shaped dependence on cytosolic pH, with maximal transport activity approaching at pH 6.0–6.6 [33,40,45,46,47,48] | |
| AHA7 | Active only when extracellular pH is ≥ 6.0 [44] | |
| NHX family | ||
| AtNHX7/SOS1 | Unclear [74,75] | |
| PpSOS1 | Stimulated by extracellular acidification [76] | |
| CHX family | ||
| AtCHX13 | Stimulated by extracellular acidification [79] | |
| AtCHX17 | Unclear [84] | |
| AMT family | ||
| PvAMT1;1 | Stimulated by extracellular acidification [89] | H211 [89] |
| AtAMT1;2 | Q67, W145 [88] | |
| NRT family | ||
| AtNRT1.1 | Stimulated by extracellular acidification [98,99,115] | (41)EXXER(45), H356 [110,111,114] |
| BnNRT1.2, AtNRT1.4, AtNRT1.5, AtNRT1.6, OsNRT1, OsNRT2.3b | Stimulated by extracellular acidification [97,100,101,102,103,104] | |
| OsNRT2.3b | Inhibited by cytosolic acidification [104] | |
| PT family | ||
| AtPHT1;1 | Stimulated by extracellular acidification [137] | D35, D38, R134 and D144 [137] |
| OsPHT1;1, OsPHT1;6, OsPHT1;8, OsPHT1;9, OsPHT1;10 | Bell-shaped dependence on cytosolic pH, with maximal transport activity approaching at pH 5.5–6.5 [127,138,139,140] | |
| KT/HAK/KUP family | ||
| AtHAK5 | E312 [169] | |
| DmHAK5, CnHAK1&CnHAK2, HvHAK1 & HvHAK2 | Stimulated by extracellular acidification [166,170,171] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.-Y.; Hao, D.-L.; Yang, G.-Z. Regulation of Cytosolic pH: The Contributions of Plant Plasma Membrane H+-ATPases and Multiple Transporters. Int. J. Mol. Sci. 2021, 22, 12998. https://doi.org/10.3390/ijms222312998
Zhou J-Y, Hao D-L, Yang G-Z. Regulation of Cytosolic pH: The Contributions of Plant Plasma Membrane H+-ATPases and Multiple Transporters. International Journal of Molecular Sciences. 2021; 22(23):12998. https://doi.org/10.3390/ijms222312998
Chicago/Turabian StyleZhou, Jin-Yan, Dong-Li Hao, and Guang-Zhe Yang. 2021. "Regulation of Cytosolic pH: The Contributions of Plant Plasma Membrane H+-ATPases and Multiple Transporters" International Journal of Molecular Sciences 22, no. 23: 12998. https://doi.org/10.3390/ijms222312998
APA StyleZhou, J.-Y., Hao, D.-L., & Yang, G.-Z. (2021). Regulation of Cytosolic pH: The Contributions of Plant Plasma Membrane H+-ATPases and Multiple Transporters. International Journal of Molecular Sciences, 22(23), 12998. https://doi.org/10.3390/ijms222312998





